Abstract
MurM and MurN are tRNA-dependent ligases that catalyze the addition of the first (l-Ala/l-Ser) and second (l-Ala) amino acid onto lipid II substrates in the biosynthesis of the peptidoglycan layer of Streptococcus pneumoniae. We have previously characterized the first ligase, MurM (Lloyd, A. J., Gilbey, A. M., Blewett, A. M., De Pascale, G., El Zoeiby, A., Levesque, R. C., Catherwood, A. C., Tomasz, A., Bugg, T. D., Roper, D. I., and Dowson, C. G. (2008) J. Biol. Chem. 283, 6402–6417). In order to characterize the second ligase MurN, we have developed a chemoenzymatic route to prepare the lipid II-Ala and lipid II-Ser substrates. Recombinant MurN enzymes from penicillin-resistant (159) and -sensitive (Pn16) S. pneumoniae were expressed and purified as MBP fusion proteins and reconstituted using a radiochemical assay. MurN ligases from strains 159 and Pn16 both showed a 20-fold higher catalytic efficiency for lipid II-l-Ala over lipid II-l-Ser, with no activity against unmodified lipid II, and similar kinetic parameters were measured for MurN from penicillin-resistant and penicillin-sensitive strains. These results concur with the peptidoglycan analysis of S. pneumoniae, in which the major cross-link observed is l-Ala-l-Ala. The combined action of ligases MurM and MurN is therefore required in order to rationalize the high level of dipeptide cross-links in penicillin-resistant S. pneumoniae, with ligase MurM showing the major difference between penicillin-resistant and penicillin-sensitive strains.
The peptidoglycan layer of Streptococcus pneumoniae and other Gram-positive pathogens is cross-linked between Lys at position 3 of its pentapeptide side chain -l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala (2–4). In Gram-negative organisms, there are direct links between meso-diaminopimelic acid at position 3 and the fourth position d-Ala of a second pentapeptide chain (5). Some Gram-positive bacteria contain direct cross-links between l-lysine and d-alanine, but many Gram-positive bacteria contain a further peptide cross-link comprising one or more amino acids (3, 6). The composition of such branched peptidoglycan peptide cross-links varies between bacterial species, as shown in Table 1 (2, 7).
TABLE 1.
| Bacterial species | Peptide cross-bridge composition (from ε-l-Lys to d-Ala) |
|---|---|
| E. coli | None (direct cross-link: meso-diaminopimelic acid → d-Ala) |
| Staphylococcus aureus | Gly-Gly-Gly-Gly-Gly |
| S. pneumoniae | l-Ala-l-Ala or l-Ser-l-Ala |
| W. viridescens | l-Ala-l-Ser |
| Enterococcus faecalis | l-Ala-l-Ala |
| Enterococcus faecium | d-Asxa |
| Streptomyces coelicolor | Gly |
d-Aspartate or d-asparagine.
The addition of the branched peptide cross-link usually occurs at the stage of lipid intermediate II (although it occurs on UDP-MurNAc-pentapeptide in Weissella viridescens (8)). Residues are added sequentially to the ε-amino terminus of l-lysine, in the opposite direction to that of protein synthesis (9–13). The addition of the amino acid residues of the cross-link is catalyzed by membrane-associated ligases, which utilize aminoacyl-tRNAs as substrates (7, 13).
The genetic determinants of branched wall structure in S. pneumoniae are the murM and murN genes (14, 15). MurM catalyzes the addition of l-Ala or l-Ser, whereas the addition of the second l-Ala is catalyzed by MurN (16). S. pneumoniae cell walls contain a mixture of directly linked (unbranched) and indirectly linked (branched) peptidoglycan, but the murMN genes are not essential, since direct cross-links can be formed (7, 15, 17). However, these enzymes do have a role in the phenotype of penicillin resistance, since inactivation of murMN leads to a loss of penicillin resistance (16, 17). Clinical strains of penicillin-resistant S. pneumoniae require for the high level resistance phenotype 1) the presence of specific murMN sequences, responsible for dipeptide cross-link formation and 2) specific modified penicillin-binding protein sequences (16–20). However, certain laboratory S. pneumoniae strains containing resistant murMN alleles do not show penicillin resistance, since they lack high affinity penicillin-binding proteins (35).
The characterization of S. pneumoniae MurM ligases from a highly penicillin-resistant strain (159) and penicillin-susceptible strain (Pn16) has been recently carried out by Lloyd et al. (1), using enzymatically synthesized lipid II substrate (1, 21–23). The markedly different branching phenotype displayed by S. pneumoniae Pn16 and 159 is rationalized in vitro by the much higher specific activity of MurM159 over MurMPn16 with pneumococcal alanyl-tRNAAla and the higher activity with alanyl-tRNAAla than with seryl-tRNASer (1).
In order to better understand the molecular basis of penicillin resistance caused by MurM and MurN, we wished to kinetically characterize the second ligase MurN in two clinical isolates of S. pneumoniae, one highly penicillin-resistant (159) and the other penicillin-sensitive (Pn16). In order to reconstitute the MurN-catalyzed reaction, we have developed a chemoenzymatic method to prepare the aminoacyl-lipid II substrate for MurN, and we report the specificity of recombinant MurN for lipid II-Ala versus lipid II-Ser substrates.
EXPERIMENTAL PROCEDURES
UDP-MurNAc-pentapeptide Biosynthesis and Purification—Details of preparation and purification of the UDP-MurNAc-pentapeptide are reported in Ref. 1.
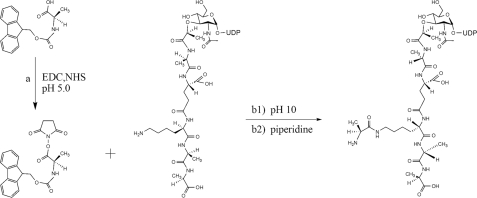
Synthesis of UDP-MurNAc-hexapeptide (l-Ala)—To 2 ml of 80% (v/v) acetonitrile in water were added 17.2 mg (90 μmol) of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride, 6.9 mg (60 μmol) of N-hydroxysuccinimide, 1.9 mg (10 μmol) of N-(ethylsulfite)-morpholine, and 7.5 mg (24 μmol) of l-alanine-Fmoc. The pH was adjusted to 5.0 if needed. After 20 min of stirring at room temperature, 100 μl of 20 mm UDP-MurNAc-pentapeptide (2.3 mg) in 500 mm NaHCO3 (pH 10.0) were added. The suspension was stirred at room temperature for 3 h, followed by the addition of 100 μl of ethanolamine and further incubated for 20 min before the addition of 100 μl of piperidine. After 30 min of incubation, 18 ml of H2O were added, and the solution was filtered with a nitrocellulose syringe filter (0.20-μm pore size). The UDP-MurNAc-hexapeptide (l-Ala) synthesis was achieved in 66% yield. The filtrate product was freeze-dried and stored at –20 °C.
Synthesis of UDP-MurNAc-hexapeptide (l-Ser)—The synthesis was conducted as for UDP-MurNAc-hexapeptide (l-Ala), except that the incubations contained 7.9 mg (24 μmol) of l-serine-Fmoc instead of l-alanine. The UDP-MurNAc-hexapeptide (l-Ser) synthesis was achieved in 61% yield.
Purification of UDP-MurNAc-hexapeptides—To isolate UDP-MurNAc-hexapeptide products, the crude filtrates were resuspended in 100 ml of 10 mm ammonium acetate, pH 7.5, and loaded onto a Source 30Q column (26 × 120 mm; Amersham Biosciences) equilibrated in 10 mm ammonium acetate, pH 7.5, and the column was developed with an ammonium acetate gradient from 0 to 300 mm ammonium acetate over 7 column volumes at 15 ml min–1. UDP-MurNAc-peptide elution was followed at 254 nm. The UDP-MurNAc-peptide peak was collected, freeze-dried four times to remove trace amounts of the buffer, dissolved in water, and stored at –20 °C. Purification was achieved in 95% yield.
Synthesis of Lipid II-l-Ala and Lipid II-l-Ser—In order to form lipid II hexapeptide substrates, 30 μmol of UDP-GlcNAc (Sigma), 2.5 μmol of UDP-MurNAc-hexapeptide, 2.5 μmol of undecaprenyl phosphate (Larodan Fine Chemicals AB), and 4.5 mg of Micrococcus flavus membranes protein in 0.1 m Tris, pH 8,5mm MgCl2, 1% (w/v) Triton X-100 in a final volume of 1.5 ml was incubated at 37 °C for 3 h. The lipids were extracted and purified by DEAE-cellulose anion exchange chromatography, as described in Ref. 21. Synthesis of lipid precursor was confirmed by TLC on silica and by negative ion electrospray-mass spectrometry as in Ref. 21.
Escherichia coli Strains and Plasmids—Details of E. coli strains and plasmids in this study are indicated in the supplemental materials.
S. pneumoniae Strains and Isolation of Pneumococcal DNA—Details of S. pneumoniae strains and isolation methods of genomic DNA are described in Refs. 19 and 24.
Protein Analytical Methods—SDS-PAGE, protein assay, and Western blotting for histidine tags were performed according to Refs. 25 and 26.
Cloning, Overexpression, and Purification of MurN from S. pneumoniae—The murN genes from S. pneumoniae Pn16 and 159 strains were cloned into the expression vector pBADM-41 to allow expression of MurN fused to a maltose-binding protein (MBP)2 with N-terminal hexahistidine tag and C-terminal tobacco etch virus (TEV) protease cleavage site (27). The same primers were designed to amplify both alleles (Table S1). The initial start codon of murN was absent in murN-Nco1 Fw primer, and the murN stop codon was present in the MBP-murN-Xho1 Rv primer. The murN genes were amplified from S. pneumoniae Pn16 and 159 genomic DNA using Pfx polymerase (Invitrogen). The primers and conditions are detailed in Table S1. Amplified products were purified, restricted with NcoI and XhoI, and ligated into similarly restricted pBADM-41 as described in Refs. 26 and 28. Clones carrying the recombinant murN159 and murNPn16 genes were verified by sequencing, and one correct clone was retained for expression of each protein (pBADM-41::murN159 and pBADM-41::murNPn16).
To overexpress the MurN proteins, 1-liter cultures of E. coli TB1, harboring either pBADM-41::murN159 or pBADM-41::murNPn16 in LB plus 50 μg/ml ampicillin were grown at 37 °C to A600 of 0.7, when murN expression was induced by 0.04% (w/v) l-arabinose. E. coli cells were harvested after 4 h, and the cell pellets were washed at 4 °C in 50 mm HEPES, 1 mm MgCl2, pH 7.5, and 2 mm β-mercaptoethanol. The cells were lysed by sonication on ice and clarified by centrifugation at 10,000 × g. The supernatant was then transferred in fresh tubes and centrifuged at 50,000 × g at 4 °C. The subcellular fractions were analyzed by SDS-PAGE, and MBP-MurN159 and MBP-MurNPn16 were present in the soluble fraction.
N-His6-MBP-MurN159 and N-His6-MBP-MurNPn16 were purified from 50,000 × g supernatant by nickel-Sepharose affinity chromatography. 5 ml of prepacked nickel-Sepharose columns (GE Healthcare) were equilibrated in 50 mm HEPES, pH 7.5, 250 mm NaCl, 1 mm MgCl2, 10% (v/v) glycerol, and 5 mm imidazole (HEPES buffer A) and were loaded with soluble cell lysates. N-His6-MBP-MurN fusion proteins were then eluted isocratically by 50 mm HEPES, pH 7.5, 250 mm NaCl, 1 mm MgCl2, 10% (v/v) glycerol, and 500 mm imidazole. The collected fractions were analyzed by SDS-PAGE and Western blotting. Peak fractions were dialyzed overnight at 4 °C into 50 mm Tris, pH 8.0, 25 mm NaCl, 1 mm MgCl2, and 5% (v/v) glycerol. MBP-MurN159 and MBP-MurNPn16 were then further purified by anion exchange chromatography using a Source 30Q column (Amersham Biosciences). The column was developed with a sodium chloride gradient between 25 mm and 1 m sodium chloride in 50 mm Tris, pH 8.0, 25 mm NaCl, 1 mm MgCl2, and 5% (v/v) glycerol using over 20 column volumes. Fractions containing MBP-MurN159 and MBP-MurNPn16 were analyzed by SDS-PAGE and Western blotting. After two purification steps, the amount of MBP-MurN for a liter of culture was 5 mg/liter of culture for MBP-MurNPn16 and 4 mg/liter of culture for MBP-MurN159.
TEV Protease Cleavage—To attempt cleavage of MurN from its MBP fusion protein, the TEV protease cleavage reaction was carried out overnight at 4 °C in 50 mm Tris, pH 8.0, 25 mm NaCl, 1mm MgCl2, and 5% (v/v) glycerol. A ratio of 1:25 TEV/MBP-MurN was used, usually 4 μg of TEV and 100 μg of MBP-MurN in 1 ml. The cleavage reactions were analyzed by SDS-PAGE.
MurN Radiochemical Assay—To follow the transfer of label from [3H]alanyl-tRNAAla and [3H]seryl-tRNASer to the peptidogylcan precursor, the assay mix was typically in a final volume of 30 μl routinely containing 50 mm MOPS, pH 6.8, 30 mm KCl, 10 mm MgCl2, 1.5% (w/v) CHAPS, 1 mm dithiothreitol, 1 mm l-alanine, lipid substrate (as indicated under “Results,” between 5 and 200 μm), MurN (as indicated under “Results,” between 5 and 50 nm). Reactions were initiated by (unless otherwise indicated) the addition of 0.45 μm [3H]alanyl-tRNAAla (prepared as reported in Ref. 1) and were incubated at 37 °C for times specified below (although initial rate data were usually taken from the first 4 min of reaction). Reactions were terminated at the appropriate time by the addition of ice-cold 30 μl of 6 m pyridinium acetate, pH 4.5, and 60 μl of ice-cold n-butyl alcohol. The incubations were rapidly mixed and centrifuged for 5 min at 4 °C at 13,000 × g, after which the n-butyl alcohol phase was washed with 70 μl of water and then assayed for [3H]scintillation counting in 5 ml of Optiphase scintillation mixture (HiSafe′ 2 from PerkinElmer Life Sciences). To follow generation of 3H-serylated lipid precursors, exactly the same procedure was followed, except that the 1 mm alanine and 0.45 μm [3H]alanyl-tRNAAla were replaced with 1 mm serine and 0.45 μm [3H]seryl-tRNASer (prepared as reported in Ref. 1).
Kinetic Data Analysis—Nonlinear regression analyses of dependences of MurN initial velocity on substrate concentration were performed using GraphPad Prism 4 software.
RESULTS
Preparation of Lipid-linked Cell Wall Precursors—In order to study the enzymology of MurN, access to its natural substrates (lipid II-l-Ala or lipid II-l-Ser) was required. These substrates were synthesized using a chemoenzymatic route. UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala was synthesized enzymatically, as described by Lloyd et al. (1), and then chemically coupled with N-terminally protected l-alanine or l-serine to generate UDP-MurNAc-hexapeptide (l-Ala or l-Ser) (Fig. 1), which was then converted to lipid II-l-Ala or lipid II-l-Ser using M. flavus membranes (21).
FIGURE 1.
Synthesis of UDP-MurNAc-hexapeptide (l-Ala) from UDP-MurNAc-pentapeptide. Reagents and conditions were as follows: acetonitrile, EDC, NHS, l-alanine-Fmoc, pH 5.0, 20 min (a); UDP-MurNAc-pentapeptide, NaHCO3, pH 10.0, 3 h (b1); piperidine, 30 min (b2).
Synthesis of UDP-MurNAc-hexapeptides—The protocol to attach l-alanine or l-serine on the ε-NH2 of l-lysine of the UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala (Fig. 1) was based on a carbodiimide coupling reaction using 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) and promoted by N-hydroxysuccinimide (NHS) (29). EDC reacts with a carboxyl group on Fmoc-l-Ala or Fmoc-l-Ser, forming an amine-reactive O-acylisourea intermediate. The addition of NHS forms a water-soluble active ester, which is amine-reactive but more stable than the O-acylisourea EDC adduct, thus increasing the efficiency of EDC-mediated coupling reactions (29). The Fmoc group was then deprotected using piperidine.
To maximize the efficiency of UDP-MurNAc-hexapeptide formation, the pH, the incubation time, the concentrations of coupling reagent, and the protected amino acid were optimized. The activation of the protected amino acid with EDC/NHS was found to be optimal at pH 5.0, whereas the reaction of the resulting NHS ester with the amine of l-Lys of the UDP-MurNAc-pentapeptide was favored at higher pH, at which the ε-NH2 of l-Lys (pKa 10.3) is more deprotonated. For this reason, the reaction was performed in two stages, first the activation of the protected amino acid was carried at pH 5.0, followed by the addition of UDP-MurNAc-pentapeptide in sodium carbonate at pH 10.0. The coupling reaction time was varied from 30 min up to 24 h, where 3 h has been established to be the optimum time. The same time-dependent profile was obtained for the addition of Fmoc-l-Ser to the UDP-MurNAc-pentapeptide.
In order to achieve a good yield, a large excess of coupling reagents and protected amino acid was necessary. It was found that the UDP-MurNAc-pentapeptide must be free of ammonium acetate, due to the possible reaction of the ammonia with the activated amino acid. For this reason, some batches of UDP-MurNAc-pentapeptide were further purified by gel filtration. The best results were obtained using 45 eq of EDC, 30 eq of NHS, and 12 eq of protected amino acid with respect to the UDP-MurNAc-pentapeptide, with yields in the range 60–65%.
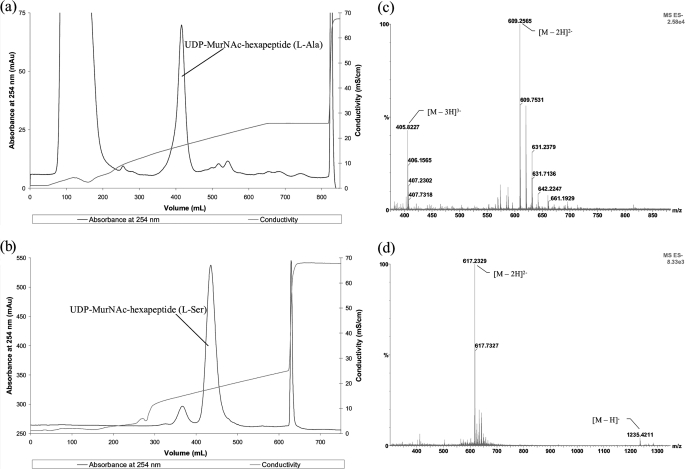
Purification and Characterization of UDP-MurNAc-hexapeptides—The UDP-MurNAc-hexapeptides were purified by Source 30Q anion exchange chromatography, using a 0–300 mm ammonium acetate gradient. The UDP-MurNAc-hexapeptide (l-Ala) was eluted at 195 mm NH4OAc (Fig. 2a). The UDP-MurNAc-hexapeptide (l-Ser) was eluted at 205 mm NH4OAc, with a small amount of unreacted UDP-MurNAc-pentapeptide (Fig. 2b). The peaks containing UDP-MurNAc-hexapeptides (l-Ala/l-Ser) were freeze-dried four times to remove trace amounts of the buffer and then dissolved in water.
FIGURE 2.
UDP-MurNAc-Hexapeptide purification and characterization by mass spectrometry. Shown is purification of UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys(ε-l-Ala)-d-Ala-d-Ala (a) and UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys(ε-l-Ser)-d-Ala-d-Ala (b) by Source 30Q anion exchange. Also shown is negative ion electrospray mass spectrum of UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys(ε-l-Ala)-d-Ala-d-Ala (c)(m/z ratio of 609.25 for the [M – 2H]2– ion and 405.82 for the [M – 3H]3– ion) and UDP-MurNAc-l-Ala-γ-d-Glu-l-Lys(ε-l-Ser)-d-Ala-d-Ala (d)(m/z ratio of 1235.42 for the [M – H]– ion and 617.23 for the [M – 2H]2– ion). Methods are described under “Experimental Procedures.”
The UDP-MurNAc-hexapeptides were analyzed by negative ion electrospray mass spectrometry. Electrospray MS/MS fragmentation was used to verify the position where l-Ala or l-Ser was attached to the peptide chain. The mass spectrum for UDP-MurNAc-hexapeptide (l-Ala) showed m/z 609.25 for the [M – 2H]2– ion (calc. 609.20) and 405.82 for the [M – 3H]3– ion (calc. 405.80), shown in Fig. 2c. The mass spectrum for UDP-MurNAc-hexapeptide (l-Ser) showed m/z 1235.42 for the [M – H]– ion (calc. 1235.38) and 617.23 for the [M – 2H]2– ion (calc. 617.19), shown in Fig. 2d. The fragmentation patterns confirm that l-Ala/l-Ser are attached to the ε-NH2 of l-lysine of the UDP-MurNAc-pentapeptide (supplemental Figs. S1 and S2). No contamination by UDP-MurNAc-pentapeptide was observed.
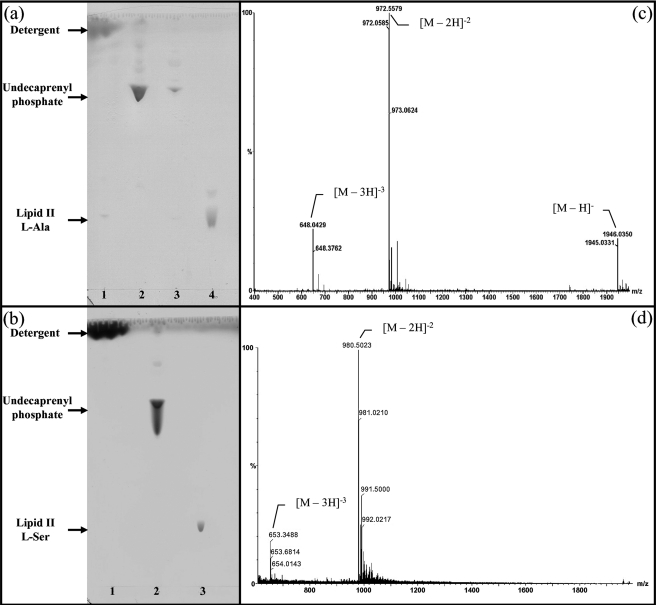
Synthesis of Lipid II-l-Ala and Lipid II-l-Ser—The UDP-MurNAc-hexapeptides (l-Ala/l-Ser) were converted into lipid II-l-Ala and lipid II-l-Ser using a membrane preparation of M. flavus, supplemented with undecaprenyl phosphate and the appropriate UDP-activated amino sugars, using the method of Breukink et al. (21). Lipid II-l-Ala and lipid II-l-Ser were purified on a DEAE-cellulose column, using the method of Breukink et al. (21), and were analyzed by thin layer chromatography (Fig. 3). The lipid II-l-Ala and lipid II-l-Ser products were analyzed by negative ion electrospray mass spectrometry. For lipid II-l-Ala, peaks were observed at m/z 1945.03 ([M – H]– ion, calc. 1945.09), 972.05 ([M – 2H]2– ion, calc. 972.04), and 647.70 ([M – 3H]3– ion, calc. 647.69), as shown in Fig. 3c. For lipid II-l-Ser, peaks were observed at m/z 980.50 ([M – 2H]2– ion, calc. 980.54) and 653.34 ([M – 3H]3– ion, calc. 653.36) as shown in Fig. 3d. The yield after purification was 45–50% based on UDP-MurNAc-hexapeptides, in batches of about 2 mg of lipid II-l-Ala and lipid II-l-Ser from each reaction.
FIGURE 3.
Branched lipid purification and characterization by mass spectrometry. Shown is purification of lipid II-l-Ala (a) and lipid II-l-Ser (b) by DEAE-cellulose chromatography as described under “Experimental Procedures.” The TLC plates show the collected fractions at different percentagesof buffer B (chloroform, methanol, 1 m ammonium bicarbonate (2:3:1)) in buffer A (chloroform/methanol/water (2:3:1)). a, lane 1, buffer A wash; lane 2, 15% buffer B wash; lane 3, 20% buffer B wash; lane 4, 100% buffer B wash. b, lane 1, buffer A wash; lane 2, 20% buffer B wash; lane 3, 100% buffer B wash. Silica gel TLC plates developed in chloroform, methanol, water, ammonia (88:48:10:1). Shown are negative ion electrospray mass spectra of lipid II-l-Ala (c) (m/z ratio of 1945.03 determined for the [M – H]– ion, 972.05 determined for the [M – 2H]2– ion, and 647.70 determined for the [M – 3H]3– ion) and lipid II-l-Ser (d)(m/z ratio 980.50 determined for the [M – 2H]2– ion and 653.34 determined for the [M – 3H]3– ion). Methods are described under “Experimental Procedures.”
Overexpression and Purification of MurN159 and MurNPn16—To overexpress MurN from S. pneumoniae 159 and Pn16, murN genes were cloned into several expression vectors: (i) pET-33b to allow expression of MurN fused to a C-terminal hexahistidine tag (30); (ii) pMW172 to allow expression of MurN without a tag (31); (iii) pET33-MurN to allow expression of the MurM fused to MurN and a C-terminal hexahistidine tag; (iv) pBADM-41 (32) to allow expression of MurN fused to N-His6-MBP with TEV protease cleavage site between the proteins. Small scale expression trials were performed using four E. coli expression strains: BL21Star (DE3), B834 (DE3), C41 (DE3), and TB1 harboring plasmid for rare tRNA (pRare or pRareII, Novagen) or for chaperones (pGKJE8; Takara) (33, 34). The expression of MurN was analyzed by SDS-PAGE and Western blotting, except the expression of pMW172-murN that was not analyzed by Western blotting due to the absence of any tag. The only construct giving expression was pBADM-41-MurN into E. coli expression host TB1.
MBP-MurN159 and MBP-MurNPn16 were purified from the soluble fraction using a nickel-Sepharose column, followed by Source 30Q anion exchange chromatography. The yield of purified protein was 5 mg of MBP-MurNPn16/liter of culture and 4 mg of MBP-MurN159/liter of culture, judged to be >95% purity by SDS-PAGE.
In order to generate native MurN, the MBP fusion proteins were successfully cleaved with TEV protease. However, attempts to separate the cleaved MurN from MBP using affinity chromatography, anion exchange, hydrophobic interaction, or ammonium sulfate precipitation were unsuccessful, suggesting a tight association between MurN and MBP. Possibly, MurN is stabilized by protein-protein interaction, which might explain the earlier difficulties in expressing MurN. In vivo MurN may interact with MurM; however, attempts to co-express MurM and MurN gave no improved expression (data not shown). Due to the difficulties in separating MurN from MBP, the full-length MBP-MurN159 and MBP-MurNPn16 fusion protein were used to characterize MurN activities in vitro, since subsequent assays verified that the MBP-MurN fusion proteins were fully active. The specific activities of the purified MurN fusion proteins with lipid II-l-Ala were 8.7 and 11.0 nmol min–1 mg–1, respectively, for MBP-MurNPn16 and MBP-MurN159.
Kinetic Characterization of MurN159 and MurNPn16—The MurN assay follows the transfer of radiolabel from [3H]alanyl-tRNAAla to the peptidoglycan precursor. The assay consisted of two steps; first the labeled amino acid was charged on the respective tRNA, and then the purified [3H]aminoacyl-tRNA was incubated with MurN and lipid II-Ala/Ser, and the [3H]-lipid II-dipeptide was extracted into n-butyl alcohol.
MurN Dependence upon tRNA for the Transfer of l-Alanine to Lipid II-l-Ala and Lipid II-l-Ser—To determine if MurN depended on tRNA for the acylation of lipid II-l-Ala and lipid-II-l-Ser, the precharged [3H]alanyl-tRNAAla from S. pneumoniae and M. flavus were tested with MurN159 and MurNPn16, using the vastly enhanced solubility of lipid II species in n-butyl alcohol relative to [3H]alanyl-tRNAAla to separate the [3H]acyl-tRNA substrate from the 3H-acylated lipid II product.
MurN159 catalyzed the transfer of [3H]alanyl groups from S. pneumoniae [3H]alanyl-tRNAAla to lipid II-l-Ala and lipid-II-l-Ser, as evidenced by the incorporation of 81 and 74% of the 3H added to the assay, into n-butyl alcohol-soluble material in the complete incubation (Table 2). In control experiments without MurN159 or lipid II-l-Ala, only 3% of the 3H added to the incubation was accumulated into n-butyl alcohol-extractable products, demonstrating the essential requirement of alanyl-tRNAAla for MurN activity. Similar results were obtained with lipid II-l-Ser (Table 2). To demonstrate the requirement of MurN for the tRNA portion of the [3H]alanyl-tRNAAla substrate, complete reactions were treated with 0.1 mg ml–1 RNase A, which reduced incorporation of 3H into lipid products to 3%, comparable with control values obtained without lipid II-l-Ala/l-Ser or MurN159 (Table 2). Similar results were obtained with MurNPn16 (Table 2). No detectable differences in MurN activity emerged using M. flavus alanyl-tRNAs or S. pneumoniae alanyl-tRNAs. For example, the incorporation percentage of 3H added to the assay into n-butyl alcohol-soluble material in the MurN Pn16 reaction with lipid II-l-Ala was 82% using S. pneumoniae tRNAs and 80% using M. flavus tRNAs (Table 2). Therefore, M. flavus tRNA was used to characterize MurN activity in vitro due to the convenience of tRNA from M. flavus.
TABLE 2.
Demonstration of MurN activity with aminoacyl tRNA from S. pneumoniae Pn16 and 159 and comparison between tRNA isolated from S. pneumoniae and M. flavus
The incubation time for these assays was 40 min in 30 ml of 25 mm lipid II-l-Ala or lipid II-l-Ser, 25 nm MurN 159 or Pn16, 0.45 mm M. flavus [3H]alanyl-tRNAAla (6183 cpm/assay; 458 cpm/pmol), or 0.45 mm S. pneumoniae [3H]alanyl-tRNAAla (6210 cpm/assay; 460 cpm/pmol). Butanol-1-extracted counts/min are averages of duplicate incubations that varied by ≤10%. Background was 9 cpm.
|
Lipid substrate |
Incubation |
MurN origin |
Butanol-soluble 3H |
Label in butanol phase |
||
|---|---|---|---|---|---|---|
| S. pneumoniae tRNA | M. flavus tRNA | S. pneumoniae tRNA | M. flavus tRNA | |||
| cpm | % | % | ||||
| Lipid II-l-Ala | Complete | 159 | 5012 | 5121 | 81 | 82 |
| Pn 16 | 5131 | 4996 | 82 | 80 | ||
| Complete + RNase A | 159 | 168 | 175 | 3 | 3 | |
| Pn 16 | 159 | 173 | 3 | 3 | ||
| No lipid II-l-Ala | 159 | 171 | 201 | 3 | 3 | |
| Pn 16 | 164 | 175 | 3 | 3 | ||
| No MurN | None | 190 | 157 | 3 | 2 | |
| Lipid II-l-Ser | Complete | 159 | 4589 | 4612 | 74 | 74 |
| Pn 16 | 4605 | 4501 | 74 | 72 | ||
| Complete + RNase A | 159 | 174 | 168 | 3 | 3 | |
| Pn 16 | 179 | 197 | 3 | 3 | ||
| No lipid II-l-Ser | 159 | 181 | 204 | 3 | 3 | |
| Pn 16 | 173 | 202 | 3 | 3 | ||
| No MurN | None | 190 | 157 | 3 | 2 | |
Comparison of the Activity of Full-length and Cleaved MBP-MurN—It was not possible to separate MurN from MBP after TEV protease cleavage. The full-length MBP-MurN and the cleaved MBP-MurN were assayed to determine if the TEV protease cleavage affects the activity of MurN. We also assayed, as control, only MBP in order to verify that the observed aminoacyl ligase activity was only due to the action of MurN. A time course experiment was performed with 20 nm full-length MBP-MurNs, a 20 nm concentration of the cleaved version, and 40 nm MBP. From these experiments, no difference between the full-length MBP-MurN species and the cleaved fusion protein was observed, and MBP control did not show any aminoacyl ligase activity. Therefore, the full-length MBP-MurNPn16 and MBP-MurN159 have been used to fully characterize MurN activity.
MurN Substrate Specificity Studies—Felipe et al. (14, 16, 35) demonstrated that MurN adds an alanine residue to a previously acylated stem peptide. To correlate this in vivo finding with the enzymatic properties of MurN, we assayed MurN activity with lipid II and seryl-tRNASer.
In order to determine if lipid II, the substrate of MurM, was also a MurN substrate, the activity of this enzyme was assayed between 0 and 250 μm lipid II. The assays were incubated for 6 min at 37 °C, using 50 nm MurN159 and 0.6 μm [3H]alanyl-tRNAAla from M. flavus. No MurN activity was detected using lipid II as a substrate. A time course experiment (from 2 to 120 min, at 200 μm lipid II and 50 nm MurN 159) was performed in case the MurN reaction with lipid II was particularly slow. However, even under these conditions, it was not possible to detect any MurN activity with lipid II as substrate. Similar results were obtained with MurNPn16. These data were consistent with the in vivo behavior of this enzyme.
MurN was tested with [3H]seryl-tRNASer from S. pneumoniae in order to determine if MurN could attach serine to lipid II-l-Ala and lipid II-l-Ser. A time course experiment (from 5 to 60 min) was performed, using 50 nm MurN 159 or Pn16, 100 μm lipid II-l-Ala, and 0.5 μm [3H]seryl-tRNASer from S. pneumoniae. The MurN 159 and Pn16 activity with [3H]seryl-tRNASer were equal to the minus MurN control, indicating the complete absence of the addition of l-serine to lipid II-l-Ala by MurN. Similar results were obtained with lipid II-l-Ser as substrate. Also, these data were consistent with the in vivo behavior of this enzyme.
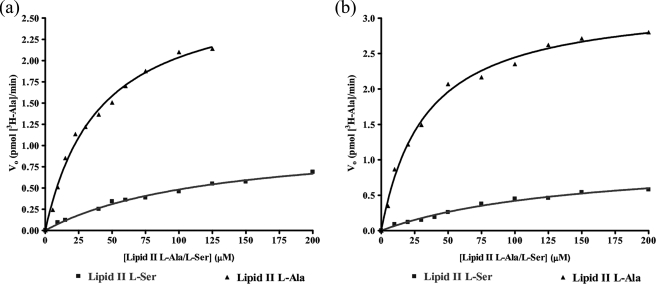
Dependence of MurN159 and MurNPn16 Activity on Branched Lipid Substrates—In order to determine whether there was any preference displayed by MurN159 and MurNPn16 toward lipid II-l-Ala or lipid II-l-Ser, both peptidoglycan precursors were tested as MurN substrates in the n-butyl alcohol extraction assay. The assays were performed for 4 min at 37 °C with 0.5 μm [3H]alanyl-tRNAAla from M. flavus and 25 nm MurN159 or 35 nm MurNPn16 in a final volume of 30 μl. The lipid II-l-Ala/ l-Ser concentrations were varied from 0 to 200 μm.
Dependences of MurN159 and MurNPn16 on lipid II l-Ala or lipid II l-Ser were hyperbolic and were fitted by nonlinear regression to the Michaelis-Menten equation (Fig. 4). MurN159 utilized lipid II l-Ala as substrate more efficiently than lipid II l-Ser. Comparison of kcat(app)/Km(app) for lipid II l-Ala and lipid II l-Ser (Table 3) suggested that the lipid II l-Ser substrate reduced its catalytic efficiency 20-fold. Likewise, MurNPn16 displayed a preference for lipid II l-Ala as substrate over lipid II l-Ser. The MurNPn16 catalytic efficiency (kcat(app)/Km(app)) was 11-fold higher for lipid II l-Ala than lipid II l-Ser (Table 3). No differences in activity were observed between MurN159 and MurNPn16; the two enzymes were comparable in terms of catalytic efficiency with each substrate (Table 3).
FIGURE 4.
Kinetics of dependence of MurN159 (a) and MurNPn16 (b) activity on lipid substrates. The initial velocity (Vo) is plotted versus [lipid II-l-Ala/l-Ser]. Data were fitted by nonlinear regression to the Michaelis-Menten equation, using GraphPad Prism 4 software.
TABLE 3.
Kinetic parameters for MurN159 and MurNPn16
The specific activities were calculated from initial rates (10 mm branched lipid at 37 °C, methods described under “Experimental Procedures”). The kinetic constants were determined according to the Michaelis-Menten equation. Fitting data was performed by nonlinear regression, using GraphPad Prism 4 software.
| Peptidoglycan precursor substrate | MurN species | Km(app) | Vmax(app) | kcat(app) | kcat(app)/Km(app) | Specific activity |
|---|---|---|---|---|---|---|
| mm | pmol min-1 | min-1 | m-1 s-1 | nmol min-1 mg-1 MurN | ||
| Lipid II l-Ala | 159 | 33.1 ± 2.8 | 3.2 ± 0.1 | 4.3 ± 0.2 | 2186 ± 185 | 11.0 ± 0.9 |
| Lipid II l-Ser | 159 | 146 ± 25 | 1.0 ± 0.1 | 1.0 ± 0.1 | 112 ± 19 | 0.7 ± 0.1 |
| Lipid II l-Ala | Pn16 | 40 ± 4 | 2.8 ± 0.1 | 3.81 ± 0.14 | 1579 ± 157 | 8.7 ± 0.8 |
| Lipid II l-Ser | Pn16 | 125 ± 15 | 1.08 ± 0.07 | 1.03 ± 0.06 | 138 ± 17 | 0.9 ± 0.1 |
DISCUSSION
MurN is an aminoacyl ligase that adds alanine as the second amino acid of a dipeptide branch to the stem peptide lysine of the pneumococcal peptidoglycan (14, 16). Studies in whole pneumococcal cells suggested that the addition of the dipeptide branch does not occur in the cytoplasmic steps of peptidoglycan biosynthesis (as it does in W. viridescens (8, 36, 37)) but in the lipid-linked stages, indicating that lipid II-l-Ala or lipid II-l-Ser might be the likely substrate for MurN (17).
The MurN gene product shares 26% sequence identity with Staphylococcus aureus FemA, which catalyzes the addition of the second and third glycine residues in the branched muropeptide of S. aureus (38, 39). Disruption of the femA gene abolishes methicillin resistance in this organism (40). The FemABX proteins have a requirement for aminoacyl-charged tRNAs as substrates for the nonribosomal peptide bond formation of the pentaglycine bridge (40). The functional and sequence similarity of MurN to these proteins suggested that it also uses aminoacyl-tRNAs as substrates.
We have recently reported the reconstitution and kinetic characterization of MurM from penicillin-resistant and penicillin-sensitive S. pneumoniae (1). The reconstitution and kinetic characterization of both MurM and MurN therefore provides a better understanding of the peptide bridge biosynthesis in S. pneumoniae.
We have developed and optimized for the first time a chemoenzymatic method to prepare the substrates lipid II-l-Ala and lipid II-l-Ser, using a carbodiimide coupling onto the most reactive ε-NH2 group of l-lysine. Using this method, it was possible to prepare 10–12 mg of UDP-MurNAc-hexapeptide (l-Ala/l-Ser) per reaction with a yield between 40 and 65%. This synthesis is versatile, and other amino acid (glycine) or dipeptides (l-Ala-l-Ala and l-Ser-l-Ala) were also successfully attached on the ε-NH2 of l-lysine of the UDP-MurNAc-pentapeptide (data not shown). The UDP-MurNAc-hexapeptides (l-Ala/l-Ser) were successfully converted into lipid II-l-Ala and lipid II-l-Ser by M. flavus membranes, using the method of Breukink et al. (21). This method could, in principle, be applied to the synthesis of a variety of lipid II-peptide conjugates, which will be very useful to study the enzymology of the FemABX/MurMN ligase family, and for future work on penicillin-binding proteins from Gram-positive bacteria containing indirect peptidoglycan cross-links.
MurN from S. pneumoniae 159 and Pn16 strains could be expressed as MBP-MurN fusion proteins, but attempts to separate cleaved MurN from MBP were unsuccessful, suggesting a tight association between MurN and MBP. In vivo MurN probably interacts with MurM, as suggested by the work on S. aureus FemX, FemA, and FemB (13, 41), and this phenomenon may reflect the high affinity between MurN and MBP.
This is the first reconstitution of an aminoacyl-tRNA ligase with a modified lipid II-X substrate, the Fem ABX ligases having been reconstituted together (13). This allows an assessment of kinetic parameters and the substrate specificity of each ligase enzyme. MurN requires an aminoacyl-tRNA substrate for the transfer of l-alanine to lipid II-l-Ala and lipid II-l-Ser, as found by Lloyd et al. for MurM (1), but tRNA from M. flavus could be conveniently used in place of S. pneumoniae tRNA.
MurN159 and MurNPn16 do not use lipid II as substrate in vitro. This result is consistent with the in vivo observation that the peptidoglycan of S. pneumoniae MurM null mutant (Pen6ΔmurM) showed no branched structured stem peptides, thus MurN cannot utilize the MurM substrate (17).
Serine could not be transferred in vitro from [3H]seryl-tRNASer to lipid II l-Ala and lipid II l-Ser by MurN159 and MurNPn16. This result is in accordance with the cell wall analysis in S. pneumoniae, where only alanine has been found in position 2 of the branched peptide stem (1, 14).
It is not clear whether the amino acid selectivity of MurN was due to specific interaction with the tRNA or the amino acid moiety. Recent studies on S. pneumoniae MurM (1) and W. viridescens FemX (8) have shown that these enzymes primarily recognize only the acceptor stem and TΨC loop of tRNA and that other regions of the tRNA are not required for the peptidyltransferase activity (1, 8). In addition, as previously described, MurN uses tRNAs from M. flavus as well as the S. pneumoniae tRNA. These results could lead to the hypothesis that the tRNA is essential for activity but not for amino acid selectivity, but further studies are required.
No significant differences in kinetic parameters were observed between recombinant MurN from S. pneumoniae 159 and Pn16 strains (see Table 3). This result is entirely consistent with the relative absence of polymorphism in the murN gene, which is highly conserved and shows little sequence variation between resistant and susceptible S. pneumoniae strains (MurN 159 and Pn16 differ in only three amino acids: R212Q, E115Q, and S225T). A low level of divergence (between 1 and 2%) also emerged from the comparison of murN genes from clinical isolates and laboratory strains of S. pneumoniae. The absence of polymorphism in the murN gene could also explain the invariable addition of alanine to the second position of the peptide cross-link in S. pneumoniae.
Both MurN enzymes show a kinetic preference for lipid II l-Ala over lipid II l-Ser as substrate: a 20-fold and 11-fold difference in catalytic efficiency for MurN159 and MurNPn16, respectively (Table 3). In the resistant strain 159, this preference matches that of MurM159, which is 7-fold more active with alanyl-tRNAAla than with seryl-tRNASer (1). This result therefore rationalizes the peptidoglycan analysis of S. pneumoniae 159 strain, in which the majority of the peptidoglycan is branched and the predominant cross-link is l-Ala-l-Ala (1). In the sensitive strain Pn16, MurMPn16 is slightly more active with seryl-tRNASer (1); however, MurNPn16 has a somewhat higher catalytic efficiency with lipid II l-Ser as substrate than MurMPn16. Therefore, it is likely that MurNPn16 would be able to convert the proportion of lipid II l-Ser generated by MurMPn16, which rationalizes the presence of a proportion of l-Ser-l-Ala cross-links in sensitive strains.
The biochemical characterization of MurM and MurN therefore confirms and rationalizes the earlier genetic observations (14–18), giving a better understanding of the pneumococcal stem-peptide biosynthesis. Ligase MurM is selective for the addition of the first amino acid (alanine or serine) to the lipid II in position 3, and ligase MurN adds only alanine as the second amino acid to lipid II-l-Ala and lipid II-l-Ser. MurM enzymes from penicillin-resistant and -sensitive S. pneumoniae strains have different amino acid selectivity and specific activity; in contrast, the two MurN enzymes do not show any significant kinetic differences. This is in accordance with the presence of extensive sequence polymorphism in the murM gene, which is absent in the murN gene, and the relative abundance of branched peptidoglycan in those strains. In conclusion, MurM is clearly the major determinant in the occurrence and sequence of the dipeptide cross-link, and the role of MurN is to complete the biosynthesis of the dipeptide initiated by MurM.
Supplementary Material
Acknowledgments
We thank Dr. T. Clarke (Department of Biological Sciences, University of Warwick) for assistance with UDP-MurNAc-peptide synthesis and Dr. A. Blewett (Department of Biological Sciences, University of Warwick) for assistance with aminoacyl-tRNA synthetase cloning and purification.
This work was supported by Medical Research Council Grant G0400848, Wellcome Trust Grant 066443, a Biotechnology and Biological Sciences Research Council studentship, the Medical Research Fund (University of Warwick), and the European Community Framework 6 COBRA network. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
Footnotes
The abbreviations used are: MBP, maltose-binding protein; EDC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide; NHS, N-hydroxysuccinimide; MOPS, 3-(N-morpholino)propane sulfonic acid; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; TEV, tobacco etch virus; Fmoc, N-(9-fluorenyl)methoxycarbonyl.
References
- 1.Lloyd, A. J., Gilbey, A. M., Blewett, A. M., De Pascale, G., El Zoeiby, A., Levesque, R. C., Catherwood, A. C., Tomasz, A., Bugg, T. D., Roper, D. I., and Dowson, C. G. (2008) J. Biol. Chem. 283 6402–6417 [DOI] [PubMed] [Google Scholar]
- 2.Bugg, T. D. (1999) in Comprehensive Natural Products Chemistry (Pinto, M., ed) pp. 241–294, Elsevier Science Publishers B.V., Oxford
- 3.Ghuysen, J. M. (1968) Bacteriol. Rev. 32 425–464 [PMC free article] [PubMed] [Google Scholar]
- 4.Schleifer, K. H., and Kandler, O. (1972) Bacteriol. Rev. 36 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glauner, B., Holtje, J. V., and Schwarz, U. (1988) J. Biol. Chem. 263 10088–10095 [PubMed] [Google Scholar]
- 6.Ghuysen, J. M., Bricas, E., Lache, M., and Leyh-Bouille, M. (1968) Biochemistry 7 1450–1460 [DOI] [PubMed] [Google Scholar]
- 7.Bouhss, A., Josseaume, N., Allanic, D., Crouvoisier, M., Gutmann, L., Mainardi, J. L., Mengin-Lecreulx, D., van Heijenoort, J., and Arthur, M. (2001) J. Bacteriol. 183 5122–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegde, S. S., and Blanchard, J. S. (2003) J. Biol. Chem. 278 22861–22867 [DOI] [PubMed] [Google Scholar]
- 9.Hegde, S. S., and Shrader, T. E. (2001) J. Biol. Chem. 276 6998–7003 [DOI] [PubMed] [Google Scholar]
- 10.Matsuhashi, M., Dietrich, C. P., and Strominger, J. L. (1965) Proc. Natl. Acad. Sci. U. S. A. 54 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petit, J. F., Strominger, J. L., and Soll, D. (1968) J. Biol. Chem. 243 757–767 [PubMed] [Google Scholar]
- 12.Plapp, R., and Strominger, J. L. (1970) J. Biol. Chem. 245 3667–3674 [PubMed] [Google Scholar]
- 13.Schneider, T., Senn, M. M., Berger-Bachi, B., Tossi, A., Sahl, H. G., and Wiedemann, I. (2004) Mol. Microbiol. 53 675–685 [DOI] [PubMed] [Google Scholar]
- 14.Filipe, S. R., Pinho, M. G., and Tomasz, A. (2000) J. Biol. Chem. 275 27768–27774 [DOI] [PubMed] [Google Scholar]
- 15.Filipe, S. R., Severina, E., and Tomasz, A. (2000) J. Bacteriol. 182 6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filipe, S. R., Severina, E., and Tomasz, A. (2001) Microb. Drug Resist. 7 303–316 [DOI] [PubMed] [Google Scholar]
- 17.Filipe, S. R., Severina, E., and Tomasz, A. (2001) J. Biol. Chem. 276 39618–39628 [DOI] [PubMed] [Google Scholar]
- 18.Filipe, S. R., and Tomasz, A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4891–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barcus, V. A., Ghanekar, K., Yeo, M., Coffey, T. J., and Dowson, C. G. (1995) FEMS Microbiol. Lett. 126 299–303 [DOI] [PubMed] [Google Scholar]
- 20.Smith, A. M., and Klugman, K. P. (2001) Antimicrob. Agents Chemother. 45 2393–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breukink, E., van Heusden, H. E., Vollmerhaus, P. J., Swiezewska, E., Brunner, L., Walker, S., Heck, A. J., and de Kruijff, B. (2003) J. Biol. Chem. 278 19898–19903 [DOI] [PubMed] [Google Scholar]
- 22.El Zoeiby, A., Sanschagrin, F., Havugimana, P. C., Garnier, A., and Levesque, R. C. (2001) FEMS Microbiol. Lett. 201 229–235 [DOI] [PubMed] [Google Scholar]
- 23.Reddy, S. G., Waddell, S. T., Kuo, D. W., Wong, K. K., and Pompliano, D. L. (1999) J. Am. Chem. Soc. 121 1175–1178 [Google Scholar]
- 24.Whatmore, A. M., Barcus, V. A., and Dowson, C. G. (1999) J. Bacteriol. 181 3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd, A. J., Brandish, P. E., Gilbey, A. M., and Bugg, T. D. (2004) J. Bacteriol. 186 1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., New York
- 27.Kapust, R. B., Tozser, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D., and Waugh, D. S. (2001) Protein Eng. 14 993–1000 [DOI] [PubMed] [Google Scholar]
- 28.Promega (1996) Protocols and Applications Guide, 3rd Ed., Promega, Madison, WI
- 29.Staros, J. V., Wright, R. W., and Swingle, D. M. (1986) Anal. Biochem. 156 220–222 [DOI] [PubMed] [Google Scholar]
- 30.Studier, F. W., and Moffatt, B. A. (1986) J. Mol. Biol. 189 113–130 [DOI] [PubMed] [Google Scholar]
- 31.Way, M., Pope, B., Gooch, J., Hawkins, M., and Weeds, A. G. (1990) EMBO J. 9 4103–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, S. K., and Keasling, J. D. (2005) Appl. Environ. Microbiol. 71 6856–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishihara, K., Kanemori, M., Kitagawa, M., Yanagi, H., and Yura, T. (1998) Appl. Environ. Microbiol. 64 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishihara, K., Kanemori, M., Yanagi, H., and Yura, T. (2000) Appl. Environ. Microbiol. 66 884–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filipe, S. R., Severina, E., and Tomasz, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1550–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biarrotte-Sorin, S., Maillard, A. P., Delettre, J., Sougakoff, W., Arthur, M., and Mayer, C. (2004) Structure 12 257–267 [DOI] [PubMed] [Google Scholar]
- 37.Maillard, A. P., Biarrotte-Sorin, S., Villet, R., Mesnage, S., Bouhss, A., Sougakoff, W., Mayer, C., and Arthur, M. (2005) J. Bacteriol. 187 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohrer, S., Ehlert, K., Tschierske, M., Labischinski, H., and Berger-Bachi, B. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9351–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tschierske, M., Mori, C., Rohrer, S., Ehlert, K., Shaw, K. J., and Berger-Bachi, B. (1999) FEMS Microbiol. Lett. 171 97–102 [DOI] [PubMed] [Google Scholar]
- 40.Berger-Bachi, B., Barberis-Maino, L., Strassle, A., and Kayser, F. H. (1989) Mol. Gen. Genet. 219 263–269 [DOI] [PubMed] [Google Scholar]
- 41.Rohrer, S., and Berger-Bachi, B. (2003) Antimicrob. Agents Chemother. 47 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.