Abstract
MAPKs are key components of cell signaling pathways with a unique activation mechanism: i.e. dual phosphorylation of neighboring threonine and tyrosine residues. The ERK enzymes form a subfamily of MAPKs involved in proliferation, differentiation, development, learning, and memory. The exact role of each Erk molecule in these processes is not clear. An efficient strategy for addressing this question is to activate individually each molecule, for example, by expressing intrinsically active variants of them. However, such molecules were not produced so far. Here, we report on the isolation, via a specifically designed genetic screen, of six variants (each carries a point mutation) of the yeast MAPK Mpk1/Erk that are active, independent of upstream phosphorylation. One of the activating mutations, R68S, occurred in a residue conserved in the mammalian Erk1 (Arg-84) and Erk2 (Arg-65) and in the Drosophila ERK Rolled (Arg-80). Replacing this conserved Arg with Ser rendered these MAPKs intrinsically active to very high levels when tested in vitro as recombinant proteins. Combination of the Arg to Ser mutation with the sevenmaker mutation (producing Erk2R65S+D319N and RolledR80S+D334N) resulted in even higher activity (45 and 70%, respectively, in reference to fully active dually phosphorylated Erk2 or Rolled). Erk2R65S and Erk2R65S+D319N were found to be spontaneously active also when expressed in human HEK293 cells. We further revealed the mechanism of action of the mutants and show that it involves acquisition of autophosphorylation activity. Thus, a first generation of Erk molecules that are spontaneously active in vitro and in vivo has been obtained.
Mitogen-activated protein kinases (MAPKs)2 are key components in cell signaling pathways. They are highly conserved throughout evolution in all eukaryotes in both sequence and mechanism of activation. MAPKs are classified to subfamilies based on degree of homology, biological responses, and phosphorylation motif (1, 2). Mammalian subfamilies include the extracellular signal-regulated kinases (ERKs), the c-Jun amino-terminal kinases (JNKs), and the p38s (2, 3). Other known mammalian MAPKs are Erk5 (BMK1) and Erk7 (4). Most MAPKs are cytoplasmic proteins that following activation are capable of translocating to the nucleus where they phosphorylate and regulate nuclear proteins (5–8). In addition, they phosphorylate cytoplasmic and membrane proteins (2, 9). Some MAPKs are essential for embryonic development (10, 11), as well as for proper differentiation and functionality of the brain (12), muscle (13), and the immune system (14). Abnormal, high activity of MAPKs is associated with inflammatory diseases (15), degenerative diseases (mainly in the brain (16–18)), and cancer (19–21).
Members of all subfamilies are concomitantly activated (to different levels) in response to any of a variety of stimuli, including growth factors, cytokines, radiations, high osmolarity, and shear stress (1, 22). In cells not exposed to stimulation, the catalytic activity of MAPKs is kept off very efficiently. Exposure of cells to a given stimulus induces the relevant MAPK pathways, each composed of three kinases (MAPKKK, MAPKK, and a MAPK) that phosphorylate and activate one another in a hierarchical way (2, 4, 9, 23). MAPKs are unique with respect to phosphorylation-mediated activation because their activation requires dual phosphorylation of both a threonine residue and a neighboring tyrosine residue (a TXY motif) in their activation loop (24–26). Since many MAPK families are concomitantly activated, it is difficult to dissect the biochemical and biological role of each family and of each component within a family. An efficient strategy for following the exact function of each component is to activate it in the cell when all other components are not activated. This could be done by expressing intrinsically active variants of the molecules in question. Production of intrinsically active MAPKKKs or MAPKKs is relatively straightforward and was successfully executed by replacing the Thr/Ser phosphoacceptors with Glu or Asp (27, 28). This strategy is not useful for MAPKs because no amino acid can accurately mimic Tyr phosphorylation. Also, attempts to mimic phosphorylation by replacing the relevant Thr to Glu were not successful (25, 29). A large number of studies applying other strategies were also just partially successful (30). Several years ago, we took a genetic approach for isolation of intrinsically active mutants of the yeast MAPK Hog1. The essence of the approach was screening for Hog1 molecules that are biologically active in the absence of their MAPKK (for details on the rationale and techniques of the screens, see Ref. 31). Not only was the screen most successful and provided intrinsically active Hog1 variants (32), the activating mutations occurred in residues conserved in mammalian MAPKs and directly led to the production of intrinsically active variants of all members of the human p38 family (33–35).
The goal of the current study was to test whether it is possible to produce intrinsically active variants of the ERK MAP kinases. Hitherto, intrinsically active mutants were not available for this important family of kinases. Only mutations that slightly increase or just prolong the activity of Erk were so far isolated (30, 36–38). The most important mutation among those is the so-called, sevenmaker mutation ((38); D319N in Erk2), which was shown to reduce the affinity of Erk2 to phosphatases, thereby prolonging the activity of Erk following the exposure of the cell to epidermal growth factor (36, 39). It seems to have no effect on the catalytic activity of Erk per se (36).
The mammalian ERK family is composed of two genes, ERK1 and ERK2, and several splicing variants (3, 40–43). Although highly similar to each other (they share 83% amino acid identity), Erk1, Erk2, and their splicing variants have distinct biological functions (10, 42, 44). Erk proteins were found to be activated in cells transformed by oncogenic B-Raf, Ras, or Her2 (45, 46), as well as in the majority of acute leukemia cases studied (47). They were also shown to be hyperactive in cancer of the prostate (48, 49), breast (50), and lung (51) and to be in association with early Tau deposition in neuron and glial cells in Alzheimer disease (52, 53). However, it is not known whether Erks are mutated in these cases. Despite the strong correlation between the activity of Erks and the various pathologies, the exact role of Erks in these diseases is not clear. This role could be specifically studied with intrinsically active variants of Erks.
In this study, we describe our attempts to produce active variants of Erks. First, we inserted into ERK2 mutations identical to those that rendered Hog1 and p38 active. Mutations were inserted alone or in combinations, but intrinsic activity obtained was low. We, therefore, designed and implemented a genetic screen in yeast, aimed at isolation of intrinsically active (MEK-independent) Erks. The screen is analogous to the previous screen that led to the isolation of Hog1 mutants (32) but was applied on the yeast MPK1/ERK pathway (1, 22). The screen provided six different point mutations in Mpk1, each of them sufficient to render Mpk1 catalytically and biologically active, independent of upstream regulation.
Insertion of equivalent mutations to Erk1 and Erk2 revealed that one mutation (R84S in Erk1; R65S in Erk2) rendered these enzymes intrinsically active as measured in vitro. Unexpectedly, combination of the R65S mutation with the sevenmaker mutation (D319N) rendered Erk2 even more active in vitro. Intrinsic activity of Erk2R65S+D319N was about 45% of MEK1-activated Erk2WT. We further revealed the mechanism of action of the mutants and show that they acquire an efficient autophosphorylation activity on both the Thr and the Tyr residues of the phosphorylation lip. We also show that a similar mutation renders the Drosophila MAPK Rolled intrinsically active. Finally, we report that spontaneous phosphorylations and activities of Erk1R84S, Erk2R65S, and Erk2R65+D319N were also measured following transient expression of these mutants in HEK293T cells. Thus, a first series of mutants of the ERK family, including ERKs of yeast, flies, and mammals, that are spontaneously active in vitro and in vivo has been developed.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media—The Saccharomyces cerevisiae strains used in this study were: mkk1Δ/mkk2Δ (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YPL140c::kanMX4; mkk1::LEU2; this study) and mpk1Δ (BY4741; Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YHR030c::kanMX4; obtained from Euroscarf). Yeast cultures were maintained on YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose + 15 mm caffeine, where indicated) or on the synthetic medium YNB-URA (0.17% yeast nitrogen base without amino acids and NH4(SO4)2, 0.5% ammonium sulfate, 2% glucose, and 40 mg/liter adenine, histidine, tryptophan, lysine, leucine, and methionine).
Random Mutagenesis Procedures—The library of MPK1 mutants was produced in the bacterial strain LE30 according to (54). A pAES426-HA-MPK1 plasmid, carrying the MPK1 and URA3 genes, was introduced into LE30 cells. About 50,000 colonies were obtained and allowed to grow on LB + ampicillin plates for 24 h. All colonies were collected in a pool into 1 liter of LB medium (containing ampicillin) and further grown for 12 h. Then, the culture was diluted 1:5 and further grown for 15 h prior to plasmid preparation. (For more information, see Ref. 31.)
Site-directed mutagenesis was performed for some mutations via polymerase chain reaction and for others via the QuikChange kit (Stratagene) according to the recommendations of the manufacturer. The sequences of the primers used for mutagenesis are listed in Table 1. All mutated cDNAs were verified via DNA sequencing of the entire ERK1 or ERK2 cDNAs.
TABLE 1.
Sequences of primers used in polymerase chain reaction and site-directed mutagenesis reactions
| Primer name | Primer sequence |
|---|---|
| ERK1-F-R84S | 5′-gaa cat cag acc tac tgc cag agc acg ctc cg-3′ |
| ERK1-R-R84S | 5′-cgg agc gtg ctc tgg cag tag gtc tga tgt tc-3′ |
| ERK1-F-S139A | 5′-gaa aag cca gca gct ggc caa tga cca tat ctg c-3′ |
| ERK1-R-S139A | 5′-gca gat atg gtc att ggc cag ctg ctg gct ttt c-3′ |
| ERK1-F-Y222C | 5′-gaa ctc caa ggg ctg tac caa gtc cat cg-3′ |
| ERK1-R-Y222C | 5′-cga tgg act tgg tac agc cct tgg agt tc-3′ |
| ERK1-F-Y280C | 5′-cat gaa ggc ccg aaa ctg cct aca gtc tc-3′ |
| ERK1-R-Y280C | 5′-gag act gta ggc agt ttc ggg cct tca tg-3′ |
| ERK2-F-R65S | 5′-cag acc tac tgt cag agt acc ctg aga gag-3′ |
| ERK2-R-R65S | 5′-ctc tct cag ggt act ctg aca gta ggt ctg-3′ |
| ERK2-F-S120A | 5′-gct ctt gaa gac aca gca cct cgc caa tga tca tat ctg c-3′ |
| ERK2-R-S120A | 5′-gca gat atg atc att ggc gag gtg ctg tgt ctt caa gag c-3′ |
| ERK2-F-Y203C | 5′-gaa ttc caa ggg ttg tac caa gtc cat tga tat ttg g-3′ |
| ERK2-R-Y203C | 5′-cca aat atc aat gga ctt ggt aca acc ctt gga att c-3′ |
| ERK2-F-Y261C | 5′-gct aga aac tgt ttg ctt tct ctc ccg cac-3′ |
| ERK2-R-Y261C | 5′-gtg cgg gag aga aag caa aca gtt tct agc-3′ |
| ERK2-F-D319N | 5′-gca gta tta tga ccc aag taa tga gcc cat tgc tga agc-3′ |
| ERK2-R-D319N | 5′-gct tca gca atg ggc tca tta ctt ggg tca taa tac tgc-3′ |
| Rolled-F-R80S | 5′-cca aac tta ttg tca aag cac tct cag aga aat aac c-3′ |
| Rolled-R-R80S | 5′-ggt tat ttc tct gag agt gct ttg aca ata agt ttg g-3′ |
| Rolled-F-D334N | 5′-gca ata tta tga tcc tgg aaa tga gcc tgt cgc tg-3′ |
| Rolled-R-D334N | 5′-cag cga cag gct cat ttc cag gat cat aat att gc-3′ |
Screening of the MPK1 Mutant Library—Transformation of the library of MPK1 mutants into mkk1Δmkk2Δ yeast cells was performed as described by Schiestl and Gietz (55). Transformed cells were plated on selective YNB-URA plates. Colonies that grew (about 10,000/100-mm plate) were replicaplated onto YPD plates containing different concentrations of caffeine (12 and 15 mm). Plasmid loss assay, for positive colonies, was performed by streaking patches of positive colonies on YPD plates, allowing the colonies to grow for 24 h, and isolating single colonies. These single colonies were replica-plated onto YNB-URA plates as well as onto caffeine-containing YPD plates.
Only cultures that gave rise to colonies that showed an absolute linkage between growth on YNB-URA plates and growth on caffeine plates were further used for extraction of plasmids. Isolated plasmids were introduced into naive mkk1Δ/mkk2Δ cells. Plasmids that were found positive in this test were sequenced.
Protein Expression and Purification—Wild-type and mutant forms of ERKs were expressed in Escherichia coli cells as described earlier (56). Each expression plasmid was transformed into BL21(DE3)-pLysS cells (Invitrogen). Cell cultures were grown in volumes of 0.5 liters at 30 °C until they reached an A600 of 0.3–0.4. Protein expression was induced using 0.2 mm isopropyl-1-thio-β-d-galactopyranoside. The cells were pelleted by centrifugation after 5 h of induction. Cells were washed in a buffer containing 50 mm Tris-HCl, pH 8, 10 mm imidazole, and 0.3 m NaCl, centrifuged again, flash-frozen in liquid nitrogen, and stored at –80 °C. The frozen pellet was gently thawed on ice and suspended in a buffer containing 50 mm Tris-HCl, pH 8, 10 mm imidazole, 0.3 mm NaCl, and a protease inhibitor mixture (1 μg/ml leupeptin, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 100 μg/ml benzamidine, and 0.1 mm phenylmethylsulfonyl fluoride). After mechanical disruption of the cells using a Microfluidizer (model M-110 EHIS, Microrofluidics Corp., Newton, MA) at ∼70% strength, the lysate was centrifuged at 40,000 × g for 30 min at 4 °C. The supernatant containing the soluble protein was loaded on a 1.6-ml nickel-nitrilotriacetic acid agarose bead (Adar Biotech) gravity flow open column. The Erk1 and Erk2 proteins were washed using a buffer containing 50 mm Tris-HCl, pH 8, 30 mm imidazole, 0.3 mm NaCl and eluted from the column using 50 mm Tris-HCl, pH 8, 0.3 m NaCl, and 250 mm imidazole. The protein solution was then dialyzed overnight against 50 mm Tris-HCl, pH 8, 0.1 m NaCl, 10% glycerol, and 0.5 mm dithiothreitol. After dialysis, protein concentration was determined using a Bradford assay, and the purified protein was then divided into aliquots, flash-frozen in liquid nitrogen, and stored in –80 °C.
Paper-spotted Kinase Assay—Reactions were carried out in 96-well plates with conical bottoms. 0.2 μg of purified recombinant hexahistidine tag-Erks mutant and wild-type proteins were used in a final volume of 50 μl/well. The kinase assays were initialized by the addition of 45 μl of reaction mixture to 5 μl of ERK enzyme. Final reaction conditions were 20 mm Hepes, pH 8, 0.1 mm benzamidine, 10 mm MgCl2, 25 mm 2-glycerolphosphate, 1 mm Na3VO4, 0.1 mm dithiothreitol, 0.5 μg/μl myelin basic protein (MBP), 0.1 mm ATP, and 0.1 μCi of [γ-32P]ATP. The kinase reactions proceeded for 15 min at 30 °C and were terminated by the addition of 50 μl of 0.5 m EDTA, pH 8 (250 mm final) and placement on ice. Following reaction termination, aliquots of 85 μl from each well were spotted onto 3 × 3-cm Whatman No. 3MM paper squares and briefly air-dried. Each square was rinsed three times with 10% trichloroacetic acid and 3% sodium pyrophosphate (10 ml/square) for 1.5 h (each time) with gentle agitation, and a fourth wash for overnight was given without shaking. The following day, the squares were rinsed twice with 100% ethanol (4 ml/square) for 20 min each time and air-dried. The radioactivity of each square was counted using a scintillation counter running a 32P Cherenkov program. Experimental points were usually triplicates. In addition, Laemmli sample buffer was added to 12 μl from each reaction. All samples were boiled at 100 °C for 5 min and were separated on 10% SDS-PAGE. The gel was exposed to film.
To activate the recombinant Erk variants, 1 μg of a purified recombinant hexahistidine-tagged Erk protein and 10 ng of recombinant active MEK1 (Upstate Biotechnology, 14-429) were used. Final reaction conditions were 100 mm NaCl, 35 mm Tris-Cl, pH 8, 15 mm MgCl2, 5 mm 2-glycerolphosphate, 0.2 mm Na3VO4, 0.02 mm dithiothreitol, 1 mm EGTA, and 100 μm ATP. The activation reactions proceeded for 30 min at 30 °C and were terminated by placement on ice.
Western Blotting—100 ng of recombinant proteins or 30 μg of protein lysates from mammalian cells were separated by SDS-PAGE and subsequently transferred to a nitrocellulose membrane. After incubation of the membrane with the appropriate antibodies (Table 2), specific proteins were visualized using an enhanced chemiluminescence detection reagent.
TABLE 2.
Antibodies that were used in this study
| Antibody name | Origin | Dilution | Manufacturer |
|---|---|---|---|
| Anti-phospho-ERK | Mouse | 1:2,000 | Santa Cruz Biotechnology (SC7383) |
| Anti-ERK | Rabbit | 1:1,000 | Santa Cruz Biotechnology (SC154) |
| Anti-phospho-Thr | Mouse | 1:3,000 | Cell Signaling (9386S) |
| Anti-phospho-Tyr | Mouse | 1:100 | Millipore (4G10) |
| Anti-HA (for Western blot) | Mouse | 1:1000 | Santa Cruz Biotechnology (12CA5) |
| Anti-HA (for immunoprecipitation) | Goat | Bethyl Laboratories (A190207A) | |
| Anti-His (for Western blot and immunoprecipitation) | Mouse | Sigma (H1029) |
Cell Culture and Treatment—HEK293 and NIH3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin (Biological Industries, Beit Ha'emek, Israel) and were incubated at 37 °C in 5% CO2. Transfections of HEK293 cells were performed using the calcium phosphate method. Transfections of NIH3T3 cells were performed using the ExGen 500 reagent (Fermentas), according to the manufacturer's instructions. All of the transfected recombinant ERK1 cDNAs carried a hexahistidine tag, and all of the recombinant ERK2 cDNAs carried an HA tag. They were all cloned into pCEFL vectors (Invitrogen). Unless otherwise stated, 48 h after transfection, the cells were harvested in two ways. (i) For Western blotting, the cells were washed with phosphate-buffered saline, and 60–250 μl of Laemmli's buffer were added. The cells were scraped using rubber policeman. (ii) For native lysis, all of the steps were performed on ice. The cells were washed twice with cold phosphate-buffered saline followed by the addition of 0.5 ml of lysis buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride 10 μg/ml leupeptin, 10 μg/ml trypsin inhibitor, 10 μg/ml pepstatin A, 313 μg/ml benzamidine, 1 mm Na3VO4, 1 mm p-nitrophenyl phosphate, and 10 mm μ-glycerol phosphate) and 30 min of incubation under shaking. The cells were scraped, frozen in liquid nitrogen, and thawed on ice. After a 10-min centrifugation at 20,000 × g, supernatant was collected.
Immunoprecipitations and Kinase Assay for Immunoprecipitated Erks—300 μg of lysate were incubated in lysis buffer (see above) with 20 μl of protein G-Sepharose beads (GE Healthcare) bound to 1 μg of anti-HA or 1 μg of anti-His antibodies for 12 h at 4 °C in a rotating wheel. The samples were then washed twice with 1 ml of lysis buffer and twice with 1 ml of kinase buffer (25 mm HEPES, pH 7.5, 20 mm MgCl2, 1 mm dithiothreitol, 20 mm β-glycerol phosphate, 5 mm p-nitrophenyl phosphate, and 0.1 mm Na3VO4). The supernatants were removed, and 30 μl of kinase buffer containing 20 μm ATP and 10 μCi of [γ-32P]ATP were added. Kinase reactions were performed in a 30 °C shaker for 30 min. The reactions were terminated by placing the tubes on ice and the addition of 10 μl of 4 × Laemmli's buffer. The samples were boiled for 3 min, and 30 μl of each sample were separated on 10% SDS-PAGE followed by transfer to nitrocellulose membrane. The membrane was exposed to film. To measure the levels of Erks molecules that were immunoprecipitated in each reaction, the same membrane was incubated in a blocking solution (Tris-buffered saline, 1% Tween 20, 5% low fat milk), and a Western blot was performed using anti-Erk and anti-HA/anti-His antibodies.
RESULTS
Mutations That Render Hog1 and p38 Intrinsically Active Have Just a Minor Effect on the Activity of ERK—Mutations that rendered Hog1 intrinsically active occurred in residues Tyr-68, Asp-170, Ala-314, Phe-318, Trp-320, Phe-322, Trp-332, and Asn-391 (32). Five of these sites are conserved in members of the p38 family, and we, therefore, mutated them in all p38 isoforms (33–35). Mutations in two sites, Asp-176 and Phe-327 (human p38α numbering), rendered p38α intrinsically active at high levels in vitro and in vivo (33, 35). A mutation in Phe-324 of p38δ rendered this kinase intrinsically active ((33, 34); Phe-318 of Hog1 is conserved in p38δ but not in p38α, p38β, and p38γ). Three of the mutations that rendered Hog1 intrinsically active occurred in sites that are also conserved in ERKs. These sites are Asp-173, Ala-323, and Phe-327. As MAP kinases are highly similar to each other and as mutating these sites in Hog1 and p38s rendered these kinases active, we speculated that mutating them in ERKs would render those MAPKs intrinsically active too. We also combined these mutations with mutations reported by others to render ERKs active to some degree (37, 38, 57). Altogether, combinations of the following mutations were constructed in the human ERK2: D173A, A323T, F327S, F327L, D319N, L73P, and S151D.
All mutated Erk2 proteins were expressed in E. coli, purified to homogeneity, and assayed for their catalytic activity using MBP as a substrate. The intrinsic activity manifested by the mutants was, in most cases, above the level shown by Erk2WT but very low when compared with the activity manifested by MEK1-activated Erk2WT (Table 3). Thus, mutations that render active MAPKs of the p38 family are not useful to the ERK family.
TABLE 3.
Catalytic activity of recombinant purified Erk2 proteins carrying the indicated mutations
Proteins were not activated with MEK. Activities are in reference to fully active, dually phosphorylated Erk2. Standard deviations are shown for cases in which experiments were repeated at least three times. In other cases, the average of two experiments, performed in duplicates, is shown.
| Erk2 protein | % Activity |
|---|---|
| Erk2WT | 0.1 |
| Erk2L73P | 1.15 ± 0.18 |
| Erk2L74P | 0.31 |
| Erk2D173A | 0.2 ± 0.13 |
| Erk2A323T | 0.39 |
| Erk2S151D | 1.35 ± 0.16 |
| Erk2D319N | 0.06 |
| Erk2F327S | 0.45 |
| Erk2F327L | 0.22 |
| Erk2L73P + F327L | 0.3 ± 0.12 |
| Erk2L73P + F327S | 1.68 ± 0.23 |
| Erk2L73P + D173A | 2.81 ± 0.5 |
| Erk2L73P + A323T | 1.76 ± 0.49 |
| Erk2L73P + D319N | 2.94 ± 0.21 |
| Erk2L73P + S151D | 3.62 ± 0.88 |
| Erk2S151D + F327L | 1.65 ± 0.33 |
| Erk2S151D + F327S | 1.36 ± 0.31 |
| Erk2S131D + D173A | 1.26 ± 0.28 |
| Erk2S131D + D319N | 1.12 ± 0.13 |
| Erk2S151D + A323T | 3.97 ± 0.98 |
| Erk2D173A + A323T | 0.36 |
| Erk2D173A + F327S | 0.53 ± 0.2 |
| Erk2D173A + F327L | 0.6 |
| Erk2D173A + D319N | 0.35 |
| Erk2L73P + S151D + D173A | 6.33 ± 1.31 |
| Erk2L73P + S151D + A323T | 3.76 ± 0.6 |
| Erk2L73P + S151D + F327L | 2.31 ± 0.5 |
| Erk2L73P + S151D + F327S | 3.18 ± 0.13 |
| Erk2L73P + S151D + D319N | 1.58 ± 0.08 |
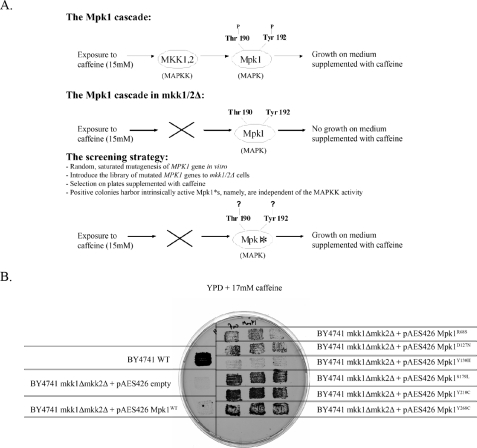
Isolation of Intrinsically Active Variants of the Yeast MAPK Mpk1 via a Specific Genetic Screen—As mutations identified previously by us and others to render some MAPKs active (32, 34, 37, 57) are not relevant to the ERK family, we searched for mutations that do render Erks active, through a genetic screen in yeast. We took advantage of the yeast MPK1 pathway, the MAPK of which, Mpk1, belongs to the ERK family of kinases (1, 22). Furthermore, the human Erk2 can functionally replace Mpk1 (68). The Mpk1 cascade is responsible for the integrity of the cell wall of the yeast (58–60). Therefore, exposure of yeast cells to drugs such as caffeine or to stimuli that damage the cell wall induces this pathway (61). Cells that are defective in components of the pathway, such as those lacking the MAPKKK Bck1, the redundant MAPKKs, Mkk1 and Mkk2, or the Mpk1 itself, cannot grow under these conditions. The phenotypes shown by mutants of the pathway allow designing a screen for intrinsically active (Mkk1/2-independent) Mpk1 molecules. The rationale of the screen is similar to that used for isolating intrinsically active (Pbs2-independent) Hog1 molecules (32) and is described schematically in Fig. 1A. Briefly, the idea is that only intrinsically active Mpk1 molecules, which escaped the requirement of upstream activation, could rescue mkk1Δmkk2Δ cells when exposed to drugs that damage the cell wall, such as caffeine (Fig. 1A). To identify such intrinsically active, Mkk1/2-independent Mpk1 molecules, we introduced a library of randomly mutated MPK1 molecules into cells lacking both MKK1 and MKK2. Transformants were allowed to form colonies on selective medium (YNB with no uracil, to select for plasmids). Then, colonies were replica-plated to plates supplemented with 15 mm caffeine. Out of ∼100,000 transformants, 100 were able to grow in the presence of caffeine. The linkage between growth on caffeine and the plasmid library was verified through a plasmid loss assay. Clones that passed these tests were considered true positives, and the plasmids they harbored were isolated and sequenced. Altogether, the screen yielded six independent clones containing Mpk1 molecules that executed their function in cells lacking Mkk1 and Mkk2 (Fig. 1B). Plasmids isolated from each of the clones contained a single point mutation in the coding sequence of Mpk1. The mutations were: R68S, D127N, Y130H, S179L, Y210C, and Y268C (Fig. 2). To verify that the capability of the Mpk1 mutants to rescue mkk1Δmkk2Δ cells is a result of an increased activity, we measured the phosphorylation status of the Mpk1 mutants expressed in either mpk1Δ or mkk1Δmkk2Δ cells, using anti-phospho-Erk antibodies. These antibodies cross-react with Mpk1. We observed that Mpk1R68S, Mpk1S179L, Mpk1Y210C, and Mpk1Y268C are spontaneously dually phosphorylated in mkk1Δmkk2Δ cells. Mpk1WT is essentially not phosphorylated in these cells. In mpk1Δ cells, these mutant proteins are spontaneously phosphorylated to higher levels then Mpk1WT (data not shown). Unexpectedly, Mpk1D127N and Mpk1Y130H were phosphorylated at levels similar to those of Mpk1WT (data not shown), and therefore, the mechanism through which they rescue mkk1Δmkk2Δ cells is currently enigmatic.
FIGURE 1.
A, general scheme and rationale of the genetic screen, aimed at isolating intrinsically active (MEK-independent) Mpk1 molecules. The asterisk indicates a Mkk1/Mkk2-independent, intrinsically active Mpk1. B, Mpk1 molecules harboring the point mutations isolated in the screen allow mkk1Δmkk2Δ cells to grow in the presence of 17 mm caffeine. Patches of the indicated strains were allowed to grow on a YNB-URA plate for 48 h and then replica-plated to the plate shown, containing YPD supplemented with 17 mm caffeine. Patches of three different colonies are shown for each mutant. Single patches are shown for controls. All patches grew similarly in the absence of caffeine (data not shown).
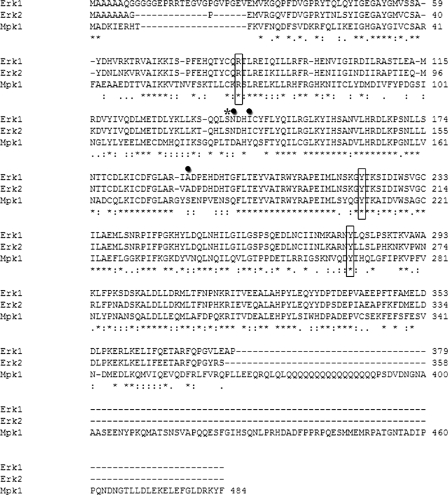
FIGURE 2.
Three of the six Mpk1-activating mutations identified in the genetic screen are conserved in human Erks. The sequence alignment of the yeast Mpk1 and the mammalian Erk1 and Erk2 MAPKs is shown. Conserved mutation sites are shown in rectangles. Non-conserved mutation sites are shown in circles. Thr-123 of Mpk1 (Ser-139 in Erk1 and Ser-120 of Erk2) is marked by an asterisk.
Design, Expression, and Purification of ERKs Active Mutants—Sequence alignment of the yeast Mpk1 and the mammalian Erks (we used the human Erk1 cDNA (National Center for Biotechnology Information (NCBI) accession number X60188) and rat Erk2 cDNA (NCBI accession number M64300)) reveals that these MAPKs share about 50% identity (Fig. 2). Therefore, we decided to introduce the mutations that rendered Mpk1 active into Erks. Of the six activating mutations found in Mpk1, only three occurred in residues conserved in Erk1 and Erk2 (Fig. 2). We mutated these residues in ERK1 and ERK2 to the same residues that rendered Mpk1 active, generating the mutants Erk1R84S, Erk1Y222C, Erk1Y280C, Erk2R65S, Erk2Y203C, and Erk2Y261C. We further produced two additional mutants, Erk1S139A and Erk2S120A, which were not directly derived from the screen in yeast but are conserved with Thr-123 in p38. It was reported that phosphorylation of Thr-123 in p38α, by GRK2, leads to inactivation of p38α (62). If the phosphorylation of this residue inactivates ERKs, as happens in p38α, it is conceivable that disruption of the serine may enable activity.
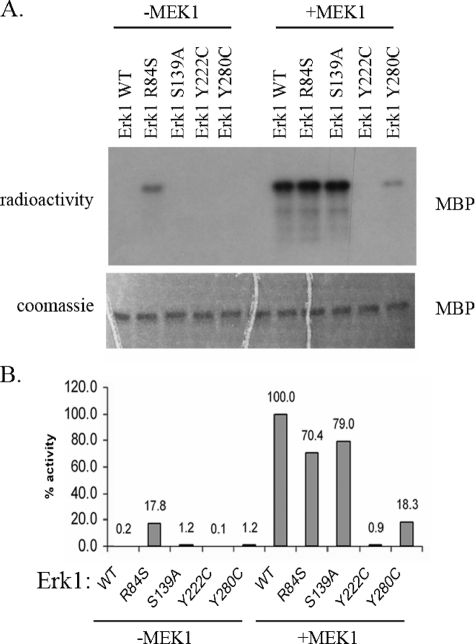
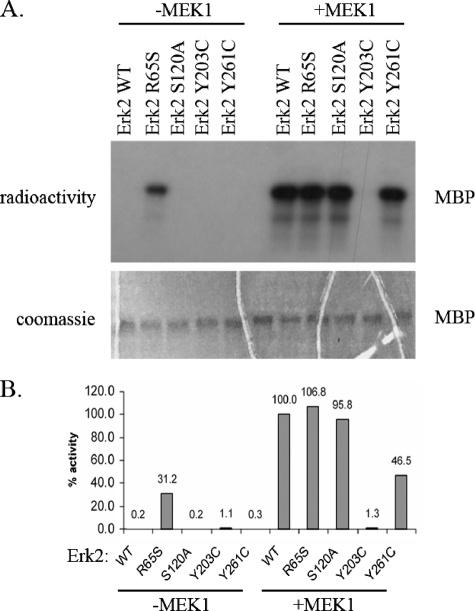
Activity Assay of Erks Proteins—The new mutated Erk1 and Erk2 molecules, as well as the wild-type Erk1 and Erk2 proteins, were expressed in E. coli as hexahistidine-tagged proteins and purified utilizing affinity chromatography. The purified proteins were assayed for their capability to phosphorylate MBP. As expected, Erk1WT and Erk2WT did not show any activity in this assay. Some of the mutants also showed no activity (Figs. 3A and 4A). In contrast, the Erk1R84S and Erk2R65S proteins exhibited significant activity (Figs. 3A and 4A). Because the activity was assayed with purified recombinant proteins expressed in E. coli, it is clear that the activity is intrinsic and does not depend on upstream activation. To compare the activities of the mutants with that of the MEK1-activated Erk1WT and Erk2WT and to examine whether the Erk mutants could be further activated by MEK1-mediated dual phosphorylation, we incubated all Erk proteins with an activated MEK1 and ATP. Subsequent to MEK1 treatment, the ErksWT and the mutants were subjected to a standard in vitro kinase assay with MBP and radioactive ATP (Figs. 3A and 4A). The results show that the mutants Erk1R84S, Erk1S139A, Erk2R65S, and Erk2S120A could be further activated by MEK1-mediated phosphorylation, to the same level of the MEK1-activated ErksWT. However, Erk1Y280C and Erk2Y261C were activated to lower levels than the activated ErksWT, and Erk1Y222C and Erk2Y203C could not be activated at all. We further established and validated a quantitative paper-spotted kinase assay that allowed the performance of quantitative kinase assays in an accurate way. Using this approach, we quantified the activities of ErksWT and mutants (Figs. 3B and 4B) and found that the activity of Erk1R84S and Erk2R65S mutants, not treated with any MEK, reached values of 18 and 30%, respectively, relative to those manifested by the MEK1-activated ErksWT.
FIGURE 3.
ERK1R84S mutant is catalytically active in vitro. The Erk1 mutants, purified from E. coli, were treated (+MEK1) or not treated (–MEK1) with active MEK1 and ATP. Then, they were subjected to a standard kinase assay with MBP as a substrate. A, a fixed volume from each reaction was subjected to SDS-PAGE. Coomassie Brilliant Blue staining verified that equal amounts of substrate were loaded (lower panel). Then, the gel was exposed to x-ray film (upper panel). Note that the Erk1R84S variant is active independently of MEK1 phosphorylation. B, using the paper-spotted kinase assay technique, we quantified and normalized the activities of the mutants to that of the MEK1-activated Erk1WT that was defined as 100%. The Erk1R84S exhibited 18% activity relative to active Erk1WT. Results shown are the average of two independent experiments, each performed in triplicates.
FIGURE 4.
ERK2R65S mutant is catalytically active in vitro. The Erk2 mutants, purified from E. coli, were treated (+MEK1) or not treated (–MEK1) with active MEK1 and ATP. Then, they were subjected to a standard kinase assay with MBP as a substrate. A, a fixed volume from each reaction was subjected to SDS-PAGE. Coomassie Brilliant Blue staining verified that equal amounts of substrate were loaded (lower panel). Then, the gel was exposed to x-ray film (upper panel). Note that the Erk2R65S variant is active independently of MEK1 phosphorylation. B, using the paper-spotted kinase assay technique, we quantified and normalized the activities of the mutants to that of the MEK1-activated Erk2WT that was defined as 100%. The Erk2R65S exhibited 30% activity relative to active Erk2WT. Results shown are the average of two independent experiments, each performed in triplicates.
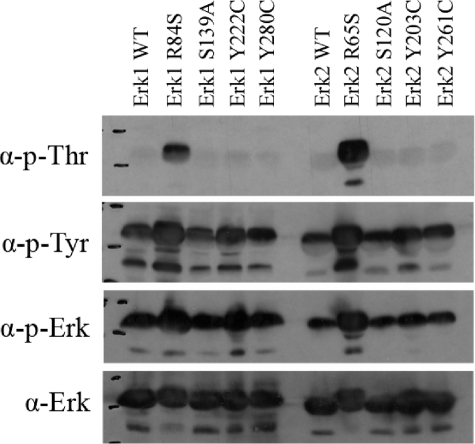
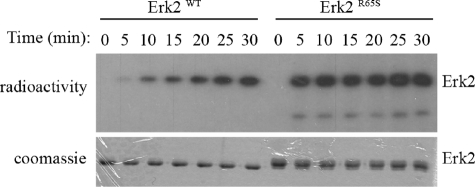
The Active Variants Have Acquired Efficient Autophosphorylation Activity—Just like all MAPKs, ERKs are catalytically active only when dually phosphorylated on their TEY motif (25). To test whether the active mutants bypassed this requirement or are rather somehow phosphorylated although purified from E. coli cells, we performed a Western blot analysis using anti-phospho-Erk antibody. All the mutants, as well as the ErksWT, reacted with these antibodies, suggesting that all are phosphorylated, but the active variants, Erk1R84S and Erk2R65S, seem to be phosphorylated to a somewhat higher level (Fig. 5). A similar result was obtained using anti-phospho-Tyr antibody. However, by using the anti-phospho-Thr antibody, we revealed that the active variants, Erk1R84S and Erk2R65S, were phosphorylated to a much higher levels than all other molecules tested (Fig. 5). We conclude that Erk1R84S and Erk2R65S are strongly dually phosphorylated, whereas all other proteins are phosphorylated mainly on the Tyr residue of the TEY motif. To confirm this notion unambiguously, we analyzed the recombinant Erk2R65S protein using mass spectrometry. We found that the protein (not treated with any MAPKK) was indeed phosphorylated on both the Tyr-185 and the Thr-183 residues. As E. coli cells do not possess any MEK activity, it is conceivable that the phosphorylation of Erks is a result of autophosphorylation activity. To test the capability of the Erk molecules to autophosphorylate, we incubated Erk2WT and Erk2R65S in a kinase assay buffer without a substrate. Both the wild-type and the mutant proteins showed autophosphorylation activity, but that of the active variant was more efficient and with a faster kinetic (Fig. 6). The autophosphorylation activity of Erk2WT, particularly on Tyr-185, is in agreement with previously described results (63). However, Erk2R65S autophosphorylates on Thr-183, too (Fig. 5). Thus, the mutants acquired an enhanced autophosphorylation activity that explains their independence of upstream activation.
FIGURE 5.
Active variants of Erks are spontaneously phosphorylated in vitro on both Thr and Tyr residues. The recombinant purified wild-type and active mutants of Erks were subjected to a Western blot analysis. Antibodies that recognize the dually phosphorylated form of ERK1/2 (p-ERK), the phosphorylated-Tyr residues (p-Tyr), or the phosphorylated-Thr residues (p-Thr) were used. Antibody against ERK1/2 (α-ERK) was applied as well. It is apparent that all proteins are equally phosphorylated on Tyr, but the active mutants, Erk1R84S and Erk2R65S, were also phosphorylated on Thr at a very high level.
FIGURE 6.
The Erk2R65S mutant exhibits efficient autophosphorylation activity in vitro. Purified recombinant Erk2WT and Erk2R65S were incubated in a kinase assay mixture without a substrate. Samples were removed from the assay at the indicated time points and subjected to SDS-PAGE. Coomassie Brilliant Blue staining (lower panel) verified the amount of enzyme in each lane. The radiograph (upper panel) reveals higher and faster phosphorylation of Erk2R65S in comparison with Erk2WT.
The Activating Mutation Also Rendered Rolled MAPK Active—Sequence alignment revealed that Arg-68 of Mpk1 is also conserved in the Drosophila melanogaster MAPK, Rolled. To test whether mutating this site may render active many members of the ERK family through evolution, we constructed the mutant RolledR80S. In addition, we constructed the mutant RolledD334N, which is known as the gain-of-function sevenmaker mutant (38). Finally, we also constructed a combined mutant, RolledR80S+D334N.
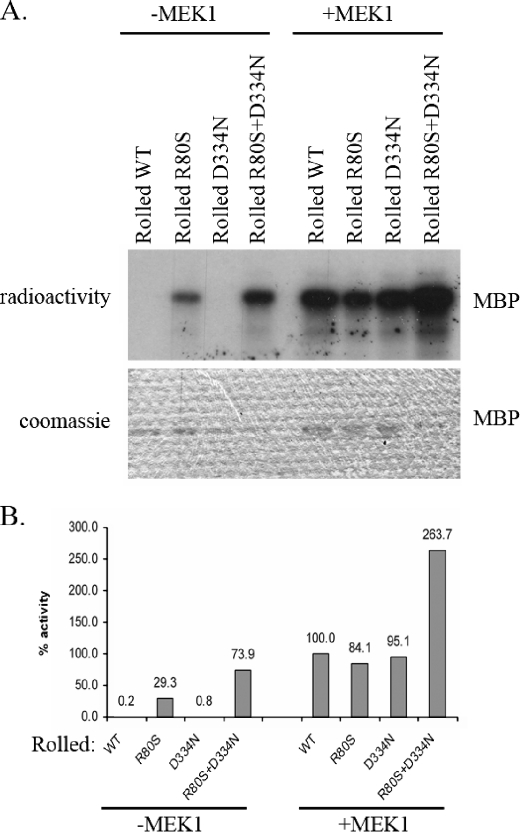
The purified Rolled proteins were assayed for their capability to phosphorylate MBP in an in vitro kinase assay. As expected, RolledWT and RolledD334N did not show any activity in this assay. However, RolledR80S exhibited significant activity (Fig. 7A). Surprisingly, the combined mutant RolledR80S+D334N exhibited even a higher activity than the RolledR80S, although RolledD334N had no activity at all and is considered to have no effect on catalysis per se but rather on the kinase affinity to phosphatases (36, 39).
FIGURE 7.
An equivalent mutation to that which rendered active Mpk1, Erk1, and Erk2 also rendered active the Drosophila Rolled. The indicated Rolled mutants, purified from E. coli, were treated (+MEK1) or not treated (–MEK1) with active MEK1 and ATP. Then, they were subjected to a standard kinase assay with MBP as a substrate. A, a fixed volume from each reaction was subjected to SDS-PAGE. Coomassie Brilliant Blue staining verified that equal amounts of substrate were loaded (lower panel). Then, the gel was exposed to x-ray film (upper panel). Note that the RolledR80S and the RolledR80S+D334N variants are active independently of MEK1 phosphorylation. B, using the paper-spotted kinase assay technique, we quantified and normalized the activities of the mutants to that of the MEK1-activated RolledWT that was defined as 100%. The RolledR80S exhibited 30% activity relative to that of MEK1-activated RolledWT. The RolledR80S/D334N exhibited 75% activity relative to that of the MEK1-activated RolledWT. Results shown are the average of two independent experiments, each performed in triplicates.
We further incubated each of the mutants as well as RolledWT with an activated MEK1 and non-radioactive ATP. Subsequent to MEK1 treatment, RolledWT and the mutants were subjected to a standard in vitro kinase assay with MBP as a substrate and radioactive ATP (Fig. 7A). The results show that all mutants could be further activated by MEK1-mediated phosphorylation. Quantification of the activities of RolledWT and the mutants, in a paper-spotted kinase assay, revealed that the activity of RolledR80S and RolledR80S+D334N mutants, not treated with MEK, reached values of 30 and 74%, respectively, relative to the MEK1-activated RolledWT (Fig. 7B). Activity of MEK1-treated RolledR80S+D334N was 264% of MEK1-treated RolledWT.
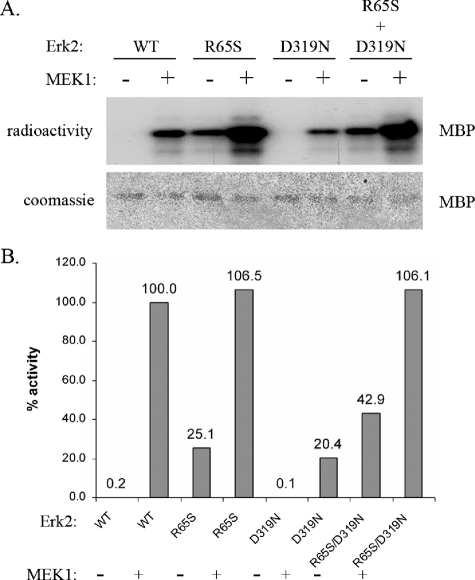
The Double Mutant Erk2R65S+D319N Is Significantly More Active Than the Single Mutant Erk2R65S—The results with the Drosophila Rolled protein raised the possibility that in mammalian Erks, the sevenmaker mutation would also synergize with the R65S-activating mutation. Thus, we generated the double mutant Erk2R65S+D319N. As observed in Rolled, the double mutant Erk2R65S+D319N is significantly more active than the single mutant Erk2R65S and manifests about 43% activity, relative to the MEK1-activated Erk2WT (Fig. 8). Just like Erk2R65S, this double mutant was also found to be phosphorylated on both the Tyr and the Thr residues of the TEY motif, as was validated using mass spectrometry analysis.
FIGURE 8.
The double mutant Erk2R65S+D319N has higher activity than the single mutant Erk2R65S. The indicated Erk2 mutants, purified from E. coli, were treated (+MEK1) or not treated (–MEK1) with active MEK1 and ATP. Then, they were subjected to a standard kinase assay with MBP as a substrate. A, a fixed volume from each reaction was subjected to SDS-PAGE. Coomassie Brilliant Blue staining verified that equal amounts of substrate were loaded (lower panel). Then, the gel was exposed to x-ray film (upper panel). Note that the Erk2R65S and the Erk2R65S+D319N variants are active independently of MEK1 phosphorylation. B, using the paper-spotted kinase assay technique, we quantified and normalized the activities of the mutants to that of the MEK1-activated Erk2WT that was defined as 100%. The Erk2R65S+D319N exhibited 45% activity relative to that of the MEK1-activated Erk2WT. This activity is higher than the 30% activity exhibited by the single mutant Erk2R65S. Results shown are the average of two independent experiments, each performed in triplicates.
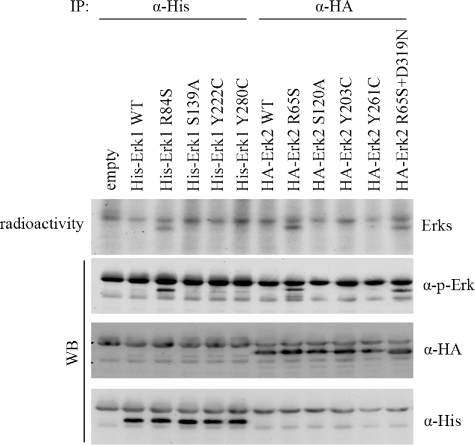
Erk1R84S, Erk2R65S, and Erk2R65S+D319N Are Also Spontaneously Phosphorylated and Active in Mammalian Cells—The observation that the new Erk mutants are intrinsically active raised the hope that they may be spontaneously active in vivo. It could also be that even mutants that were not active in vitro would be active in vivo. To test this notion, we transiently expressed the cDNAs of ERKsWT and the various mutants in human HEK293 cells. Cells were allowed to proliferate normally under optimal growth conditions and were not exposed to any signal. 24 h after transfection, cells were lysed, and the Erk proteins were monitored via Western blot analysis. Using anti-phospho-Erk antibody, we observed that only mutants that were active in vitro (i.e. Erk1R84S, Erk2R65S, and Erk2R65S+D319N) were spontaneously phosphorylated. All other mutants were not phosphorylated just as Erk1WT and Erk2WT were. Next, Erk proteins were immunoprecipitated, and their capability to autophosphorylate was examined. As shown in Fig. 9 (upper panel), only Erk1R84S, Erk2R65S, and Erk2R65S+D319N manifested an autophosphorylation activity. Thus, the Erk mutants that showed an intrinsic activity in vitro are also spontaneously phosphorylated and are autophosphorylating in vivo.
FIGURE 9.
Active variants of Erks are also spontaneously active in mammalian cells. The cDNAs encoding the specified mutants were introduced into HEK293 cells. Erk1 proteins were fused to a poly-His tag, and Erk2 proteins were fused to a HA tag. 24 h after transfection, cells were harvested. Erk molecules were immunoprecipitated from lysates, and immunoprecipitates were subjected to a kinase assay without a substrate. Assay mixtures were separated on SDS-PAGE and transferred to nitrocellulose membranes that were exposed to x-ray film. The upper panel shows the autoradiograms. The lower panels show Western blots (WB) of the same nitrocellulose membranes, using anti-phosphorylated-Erk (p-Erk), anti-HA, and anti-His antibodies.
DISCUSSION
This report describes the development of the first series of intrinsically active variants of the Erk MAPKs. The variants that were isolated in this work show significant high activity when compared with the unphosphorylated ErksWT and also when compared with the MEK1-activated form (between 18 and 74% activity of the maximal activity). The activating mutations of Mpk1/Erk were identified via an unbiased genetic screen, which follows the rationale of a previous screen performed in our laboratory that gave rise to active variants of Hog1 and p38s (32).
The activating mutations of the Mpk1/Erk system are totally different from those found to activate the Hog1/p38 system. For example, mutating an Asp residue of the phosphorylation lip to Ala (Asp-170 in Hog1, Asp-176 in p38α) rendered Hog1, p38α, p38β, and p38γ intrinsically active (32–35). Although this Asp, as well as the entire phosphorylation lip, is conserved between Hog1/p38 and Mpk1/Erk, mutating the relevant Asp (Asp-173) in Erk2 did not render it active (Table 1). Our genetic screen for active Mpk1 identified another mutation site in the phosphorylation lip, Ser-179 that is not conserved in mammalian ERKs. Thus, the phosphorylation lip could be a hot spot for activating mutations, but different mutations are required for Erks and for p38s. Similarly, many of the activating mutations of Hog1/p38 occurred in the L16 domain, but none of the Mpk1-activating mutations occurred there.
Two mutation sites in Mpk1, Asp-127 and Tyr-130, are located at a helix surrounded by the CD and ED domain. Another mutation, Tyr-210, is at loop 195–205, which undergoes significant conformational changes when Erk2 is phosphorylated (64). Tyr-268 is located within the MAP kinase insertion. None of these domains were mutated in the Hog1/p38 screen. Finally, the mutation site that gave the most dramatic results in the yeast Mpk1, the human Erks, and the Drosophila Rolled, Arg-65 (of Erk2), occurred at the C-helix. In the Hog1 screen, another site of the C-helix was identified (Tyr-68) but was not relevant to p38α (32, 35). Thus, although highly similar in structure and although activated via the same mechanism of dual phosphorylation, it seems that different conformational changes are required to render p38s and Erks active. Notably, we recently inserted into Jnk1 and Jnk2 some of the mutations that rendered p38 and Hog1 active. In this case, too, only a minor increase in intrinsic activity was measured (data not shown).
The most important mutation identified in our screen for activating Mpk1 is R68S. So far, mutating the equivalent arginines residues to serine led to high intrinsic activity in four MAPKs of the ERK family: the yeast MAPK Mpk1, the D. melanogaster MAPK Rolled, and the mammalian MAPKs Erk1 and Erk2. This residue also exists in Erk5 and Erk8. Arg-65 of Erk2 was proposed as an important residue for catalysis, taking part in stabilizing the active site. This stabilization is achieved when Arg-65 contacts phospho-Thr-183, via a water molecule, in the dually phosphorylated form of Erk2 (64–66). It seems that a Ser residue in this location allows, at least in part, reorientation of the C-helix toward the phosphorylation lip, even in the absence of MEK-mediated phosphorylation. It could also be that Arg-65 is part of a structural feature that acts as a constraint on the autophosphorylation activity of Erk. Converting Arg-65 to Ser relieves this restriction, but on the other hand, also interferes with stabilizing the active site, via Arg-65 interaction with phospo-Thr. We think, however, that other residues in the C-helix compensate for Arg-65 with respect to stabilizing the active site. Strikingly, the conformation of Arg-65 in the published crystal structures of Erk2 is not clear, preventing reliable modeling of the effect of Ser at this position. Of the 18 deposited structures of Erk2 in the Protein Data Bank, 10 were crystallized using similar conditions and acquired identical space group and highly similar cell parameters, allowing structural comparison. Such comparison of the 10 structures revealed high conformational diversity of Arg-65 (supplemental Fig. S1). The maximal distance of the distal Arg side chain (Nη-Nη) reaches 7.8 Å. Clearly, modeling in this case for the possible conformation of Ser at this position would not be highly reliable. Importantly, Arg-65 was suggested to function similar to His-87 of catalytic unit of cAMP-dependent protein kinase in stabilizing an active site following phosphorylation of Thr-197 in that enzyme (equivalent to Thr-183 of Erk2) (67). Thus, mutations in this area may be relevant to many kinases, including non-MAPKs.
The striking synergistic effect of the R65S + D334N is totally unexpected because the D334N mutation is supposed to affect the properties of the enzyme only in vivo (because it reduces the affinity to phosphatases (36)). Accordingly, although RolledD334N has biological activity (38), it manifests no intrinsic catalytic activity. Its homolog, Erk2D319N, has no intrinsic catalytic either (37) (Fig. 8). Our findings suggest that the D319N mutation affects, in fact, the catalysis of Erks, too, not only its affinity to phosphatases. The mechanism of this effect is currently under study through a structural approach.
For a very long time and despite many efforts to obtain intrinsically active variants of Erks (30, 37), such proteins could not be achieved. Now that such molecules are available for yeast, flies, and mammals, novel approaches are open for investigating, in a specific manner, the biological and biochemical functions of each isoform and splicing variants of these kinases.
Supplementary Material
This study was supported by U.S-Israel Binational Science Foundation Grant 2005002 and by the Israel Science Foundation Grant 656/06. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: MAP, mitogen-activated protein; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPK kinase kinase; ERK, extracellular signal-regulated kinase; MEK, MAPK/ERK kinase; JNK, c-Jun NH2-terminal kinase; HA, hemagglutinin; WT, wild type; MBP, myelin basic protein.
References
- 1.Gustin, M. C., Albertyn, J., Alexander, M., and Davenport, K. (1998) Microbiol. Mol. Biol. Rev. 62 1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyriakis, J. M., and Avruch, J. (2001) Physiol. Rev. 81 807–869 [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., Gibson, T. B., Robinson, F., Silvestro, L., Pearson, G., Xu, B., Wright, A., Vanderbilt, C., and Cobb, M. H. (2001) Chem. Rev. 101 2449–2476 [DOI] [PubMed] [Google Scholar]
- 4.Pearson, G., Robinson, F., Beers Gibson, T., Xu, B. E., Karandikar, M., Berman, K., and Cobb, M. H. (2001) Endocr. Rev. 22 153–183 [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez, F. A., Seth, A., Raden, D. L., Bowman, D. S., Fay, F. S., and Davis, R. J. (1993) J. Cell Biol. 122 1089–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traverse, S., Gomez, N., Paterson, H., Marshall, C., and Cohen, P. (1992) Biochem. J. 288 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanghera, J. S., Peter, M., Nigg, E. A., and Pelech, S. L. (1992) Mol. Biol. Cell 3 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R. H., Sarnecki, C., and Blenis, J. (1992) Mol. Cell. Biol. 12 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis, T. S., Shapiro, P. S., and Ahn, N. G. (1998) Adv. Cancer Res. 74 49–139 [DOI] [PubMed] [Google Scholar]
- 10.Yao, Y., Li, W., Wu, J., Germann, U. A., Su, M. S., Kuida, K., and Boucher, D. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura, K., Sudo, T., Senftleben, U., Dadak, A. M., Johnson, R., and Karin, M. (2000) Cell 102 221–231 [DOI] [PubMed] [Google Scholar]
- 12.Kuan, C. Y., Yang, D. D., Samanta Roy, D. R., Davis, R. J., Rakic, P., and Flavell, R. A. (1999) Neuron 22 667–676 [DOI] [PubMed] [Google Scholar]
- 13.Gredinger, E., Gerber, A. N., Tamir, Y., Tapscott, S. J., and Bengal, E. (1998) J. Biol. Chem. 273 10436–10444 [DOI] [PubMed] [Google Scholar]
- 14.Hou, X. S., Goldstein, E. S., and Perrimon, N. (1997) Genes Dev. 11 1728–1737 [DOI] [PubMed] [Google Scholar]
- 15.van Montfrans, C., Peppelenbosch, M., te Velde, A. A., and van Deventer, S. (2002) Biochem. Pharmacol. 64 789–795 [DOI] [PubMed] [Google Scholar]
- 16.Garcia, M., Vanhoutte, P., Pages, C., Besson, M. J., Brouillet, E., and Caboche, J. (2002) J. Neurosci. 22 2174–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuperstein, F., and Yavin, E. (2002) Eur. J. Neurosci. 16 44–54 [DOI] [PubMed] [Google Scholar]
- 18.Pei, J. J., Braak, E., Braak, H., Grundke-Iqbal, I., Iqbal, K., Winblad, B., and Cowburn, R. F. (2001) J. Alzheimer's Dis. 3 41–48 [DOI] [PubMed] [Google Scholar]
- 19.Cripe, L. D., Gelfanov, V. M., Smith, E. A., Spigel, D. R., Phillips, C. A., Gabig, T. G., Jung, S. H., Fyffe, J., Hartman, A. D., Kneebone, P., Mercola, D., Burgess, G. S., and Boswell, H. S. (2002) Leukemia (Basingstoke) 16 799–812 [DOI] [PubMed] [Google Scholar]
- 20.Esparis-Ogando, A., Diaz-Rodriguez, E., Montero, J. C., Yuste, L., Crespo, P., and Pandiella, A. (2002) Mol. Cell. Biol. 22 270–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, S. Y., Liang, Y. C., Ho, Y. S., Tsai, S. H., Pan, S., and Lee, W. S. (2002) Mol. Carcinog. 35 21–28 [DOI] [PubMed] [Google Scholar]
- 22.Widmann, C., Gibson, S., Jarpe, M. B., and Johnson, G. L. (1999) Physiol. Rev. 79 143–180 [DOI] [PubMed] [Google Scholar]
- 23.Marshall, C. J. (1995) Cell 80 179–185 [DOI] [PubMed] [Google Scholar]
- 24.Cobb, M. H., and Goldsmith, E. J. (2000) Trends Biochem. Sci. 25 7–9 [DOI] [PubMed] [Google Scholar]
- 25.Robbins, D. J., Zhen, E., Owaki, H., Vanderbilt, C. A., Ebert, D., Geppert, T. D., and Cobb, M. H. (1993) J. Biol. Chem. 268 5097–5106 [PubMed] [Google Scholar]
- 26.Schuller, C., Brewster, J. L., Alexander, M. R., Gustin, M. C., and Ruis, H. (1994) EMBO J. 13 4382–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, S., Jiang, Y., Li, Z., Nishida, E., Mathias, P., Lin, S., Ulevitch, R. J., Nemerow, G. R., and Han, J. (1997) Immunity 6 739–749 [DOI] [PubMed] [Google Scholar]
- 28.Cowley, S., Paterson, H., Kemp, P., and Marshall, C. J. (1994) Cell 77 841–852 [DOI] [PubMed] [Google Scholar]
- 29.Bell, M., and Engelberg, D. (2003) J. Biol. Chem. 278 14603–14606 [DOI] [PubMed] [Google Scholar]
- 30.Askari, N., Diskin, R., Avitzour, M., Yaakov, G., Livnah, O., and Engelberg, D. (2006) Mol. Cell. Endocrinol. 252 231–240 [DOI] [PubMed] [Google Scholar]
- 31.Engelberg, D., and Livnah, O. (2006) Methods (Amst.) 40 255–261 [DOI] [PubMed] [Google Scholar]
- 32.Bell, M., Capone, R., Pashtan, I., Levitzki, A., and Engelberg, D. (2001) J. Biol. Chem. 276 25351–25358 [DOI] [PubMed] [Google Scholar]
- 33.Askari, N., Diskin, R., Avitzour, M., Capone, R., Livnah, O., and Engelberg, D. (2007) J. Biol. Chem. 282 91–99 [DOI] [PubMed] [Google Scholar]
- 34.Avitzour, M., Diskin, R., Raboy, B., Askari, N., Engelberg, D., and Livnah, O. (2007) FEBS J. 274 963–975 [DOI] [PubMed] [Google Scholar]
- 35.Diskin, R., Askari, N., Capone, R., Engelberg, D., and Livnah, O. (2004) J. Biol. Chem. 279 47040–47049 [DOI] [PubMed] [Google Scholar]
- 36.Bott, C. M., Thorneycroft, S. G., and Marshall, C. J. (1994) FEBS Lett. 352 201–205 [DOI] [PubMed] [Google Scholar]
- 37.Emrick, M. A., Hoofnagle, A. N., Miller, A. S., Ten Eyck, L. F., and Ahn, N. G. (2001) J. Biol. Chem. 276 46469–46479 [DOI] [PubMed] [Google Scholar]
- 38.Brunner, D., Oellers, N., Szabad, J., Biggs, W. H., III, Zipursky, S. L., and Hafen, E. (1994) Cell 76 875–888 [DOI] [PubMed] [Google Scholar]
- 39.Chu, Y., Solski, P. A., Khosravi-Far, R., Der, C. J., and Kelly, K. (1996) J. Biol. Chem. 271 6497–6501 [DOI] [PubMed] [Google Scholar]
- 40.Boulton, T. G., Nye, S. H., Robbins, D. J., Ip, N. Y., Radziejewska, E., Morgenbesser, S. D., DePinho, R. A., Panayotatos, N., Cobb, M. H., and Yancopoulos, G. D. (1991) Cell 65 663–675 [DOI] [PubMed] [Google Scholar]
- 41.Boulton, T. G., Yancopoulos, G. D., Gregory, J. S., Slaughter, C., Moomaw, C., Hsu, J., and Cobb, M. H. (1990) Science 249 64–67 [DOI] [PubMed] [Google Scholar]
- 42.Aebersold, D. M., Shaul, Y. D., Yung, Y., Yarom, N., Yao, Z., Hanoch, T., and Seger, R. (2004) Mol. Cell. Biol. 24 10000–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yung, Y., Yao, Z., Aebersold, D. M., Hanoch, T., and Seger, R. (2001) J. Biol. Chem. 276 35280–35289 [DOI] [PubMed] [Google Scholar]
- 44.Pages, G., Guerin, S., Grall, D., Bonino, F., Smith, A., Anjuere, F., Auberger, P., and Pouyssegur, J. (1999) Science 286 1374–1377 [DOI] [PubMed] [Google Scholar]
- 45.Chong, H., Vikis, H. G., and Guan, K. L. (2003) Cell. Signal. 15 463–469 [DOI] [PubMed] [Google Scholar]
- 46.Mercer, K. E., and Pritchard, C. A. (2003) Biochim. Biophys. Acta 1653 25–40 [DOI] [PubMed] [Google Scholar]
- 47.Platanias, L. C. (2003) Blood 101 4667–4679 [DOI] [PubMed] [Google Scholar]
- 48.Gioeli, D., Mandell, J. W., Petroni, G. R., Frierson, H. F., Jr., and Weber, M. J. (1999) Cancer Res. 59 279–284 [PubMed] [Google Scholar]
- 49.Price, D. T., Della Rocca, G., Guo, C., Ballo, M. S., Schwinn, D. A., and Luttrell, L. M. (1999) J. Urol. 162 1537–1542 [PubMed] [Google Scholar]
- 50.Donovan, J. C., Milic, A., and Slingerland, J. M. (2001) J. Biol. Chem. 276 40888–40895 [DOI] [PubMed] [Google Scholar]
- 51.Vicent, S., Lopez-Picazo, J. M., Toledo, G., Lozano, M. D., Torre, W., Garcia-Corchon, C., Quero, C., Soria, J. C., Martin-Algarra, S., Manzano, R. G., and Montuenga, L. M. (2004) Br. J. Cancer 90 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrer, I., Blanco, R., Carmona, M., Ribera, R., Goutan, E., Puig, B., Rey, M. J., Cardozo, A., Vinals, F., and Ribalta, T. (2001) Brain Pathol. 11 144–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato, N., Kamino, K., Tateishi, K., Satoh, T., Nishiwaki, Y., Yoshiiwa, A., Miki, T., and Ogihara, T. (1997) Biochem. Biophys. Res. Commun. 232 637–642 [DOI] [PubMed] [Google Scholar]
- 54.Silhavy, T. J., Berman, M. L., and Enquist, L. W. (eds) (1984) Experiments With Gene Fusions, pp. 75–78, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 55.Schiestl, R. H., and Gietz, R. D. (1989) Curr. Genet. 16 339–346 [DOI] [PubMed] [Google Scholar]
- 56.Wang, Z., Harkins, P. C., Ulevitch, R. J., Han, J., Cobb, M. H., and Goldsmith, E. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2327–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall, J. P., Cherkasova, V., Elion, E., Gustin, M. C., and Winter, E. (1996) Mol. Cell. Biol. 16 6715–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costigan, C., Gehrung, S., and Snyder, M. (1992) Mol. Cell. Biol. 12 1162–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levin, D. E., Fields, F. O., Kunisawa, R., Bishop, J. M., and Thorner, J. (1990) Cell 62 213–224 [DOI] [PubMed] [Google Scholar]
- 60.Verna, J., Lodder, A., Lee, K., Vagts, A., and Ballester, R. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13804–13809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin, H., Castellanos, M. C., Cenamor, R., Sanchez, M., Molina, M., and Nombela, C. (1996) Curr. Genet. 29 516–522 [DOI] [PubMed] [Google Scholar]
- 62.Peregrin, S., Jurado-Pueyo, M., Campos, P. M., Sanz-Moreno, V., Ruiz-Gomez, A., Crespo, P., Mayor, F., Jr., and Murga, C. (2006) Curr. Biol. 16 2042–2047 [DOI] [PubMed] [Google Scholar]
- 63.Seger, R., Ahn, N. G., Boulton, T. G., Yancopoulos, G. D., Panayotatos, N., Radziejewska, E., Ericsson, L., Bratlien, R. L., Cobb, M. H., and Krebs, E. G. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 6142–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canagarajah, B. J., Khokhlatchev, A., Cobb, M. H., and Goldsmith, E. J. (1997) Cell 90 859–869 [DOI] [PubMed] [Google Scholar]
- 65.Cobb, M. H., and Goldsmith, E. J. (1995) J. Biol. Chem. 270 14843–14846 [DOI] [PubMed] [Google Scholar]
- 66.Zhang, F., Strand, A., Robbins, D., Cobb, M. H., and Goldsmith, E. J. (1994) Nature 367 704–711 [DOI] [PubMed] [Google Scholar]
- 67.Goldsmith, E. J., and Cobb, M. H. (1994) Curr. Opin. Struct. Biol. 4 833–840 [DOI] [PubMed] [Google Scholar]
- 68.Levin-Salomon, V., Maayan, I., Avrahami-Moyal, L., Marbach, I., Livnah, O., and Engelberg, D. (2008) Biochem. J., in press [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.