Abstract
In response to environmental stress, the translation machinery of cells is reprogrammed. The majority of actively translated mRNAs are released from polysomes and driven to specific cytoplasmic foci called stress granules (SGs) where dynamic changes in protein-RNA interaction determine the subsequent fate of mRNAs. Here we show that the DEAH box RNA helicase RHAU is a novel SG-associated protein. Although RHAU protein was originally identified as an AU-rich element-associated protein involved in urokinase-type plasminogen activator mRNA decay, it was not clear whether RHAU could directly interact with RNA. We have demonstrated that RHAU physically interacts with RNA in vitro and in vivo through a newly identified N-terminal RNA-binding domain, which was found to be both essential and sufficient for RHAU localization in SGs. We have also shown that the ATPase activity of RHAU plays a role in the RNA interaction and in the regulation of protein retention in SGs. Thus, our results show that RHAU is the fourth RNA helicase detected in SGs, after rck/p54, DDX3, and eIF4A, and that its association with SGs is dynamic and mediated by an RHAU-specific RNA-binding domain.
Posttranscriptional regulation of gene expression is important and highly regulated in response to developmental, environmental, and metabolic signals (1). During stress conditions such as heat shock, oxidative stress, ischemia, or viral infection, mRNA translation is reprogrammed and allows the selective synthesis of stress response and repair proteins (2). Under these conditions, the translation of housekeeping genes is arrested, and untranslated mRNAs accumulate in cytoplasmic foci known as stress granules (SGs) (3). SG3 formation is triggered by translation initiation arrest involving eIF2α phosphorylation or by inhibition of eIF4A helicase function. Phosphorylation of eIF2α can be mediated by several stress-induced protein kinases (for a review, see Ref. 4), suggesting that the formation of SGs is highly regulated and that eIF2α plays a pivotal role in sensing stress. Upon translational arrest, polysome-free 48 S preinitiation complexes containing initiation factors, small ribosomal subunits, and poly(A)-binding protein 1 (PABP-1) aggregate into SGs (3, 5). Several known SG-associated mRNA-binding proteins have been identified and shown to induce or inhibit SG aggregation when overexpressed. It is presumed that the overexpression of mRNA-binding proteins, which are able to oligomerize, disturb the equilibrium of mRNA distribution between polysomes and polysome-free ribonucleoprotein (RNP) complexes and thus induce SG formation by their aggregation (6). Nevertheless some RNA-binding proteins do not induce SG formation upon overexpression, e.g. a zipcode-binding protein 1 (ZBP1), heterogeneous nuclear RNP A1, or PABP-1 (7-9). Under normal conditions, most RNA-binding proteins are involved in various aspects of mRNA metabolism, such as translation (TIA-1, TIA-1-related protein, PCBP2, Pumilio 2, and cytoplasmic polyadenylation element-binding protein), degradation (G3BP, TTP, Brf1, rck/p54, KH domain RNA binding protein [KSRP], and PMR1), stability (HuR), and specific intracellular localization (ZBP1, Staufen, Smaug, Caprin-1, and FMRP) (for a review, see Ref. 10). The differing flux of these SG-associated proteins and poly(A)+ mRNAs revealed by fluorescent recovery after photobleaching (FRAP) analysis suggests that SGs are dynamic foci (6, 9, 11-14). They are also considered to be sites at which RNPs undergo structural and compositional remodeling and may be temporally stored, returned to polysomes for translation, or packaged for degradation (6).
Immediately after transcription, RNA forms with RNA-binding proteins RNPs that are dynamic and take part in RNA metabolism. RNP remodeling, which is essential for the cellular localization, processing, function, and fate of RNA (15), is mainly regulated by a large family of proteins called RNA helicases. These enzymes use energy released by ATP hydrolysis to unwind secondary structures of RNA or displace proteins from RNA (16). The majority of RNA helicases are assigned to superfamily 2 (SF2), which is divided into three subfamilies named after the sequence in the helicase motif II: DEAD, DEAH, and DEXH (17). Several DEX(H/D) proteins have been shown to unwind double-stranded RNA in an ATP-dependent manner in vitro (16, 18), but most are involved in the ATP-dependent remodeling of RNPs (16). Although RNA helicases contain a highly conserved helicase core region, they are involved in all of the RNA processes ranging from transcription, pre-mRNA splicing, ribosome biogenesis, RNA export, and translation initiation to RNA decay (for reviews, see Refs. 19-21). The specific functions of individual enzymes are attributed to the less conserved N/C termini, which are responsible for substrate specificity, subcellular localization, and cofactor requirements (22-25).
RHAU is a DEAH box helicase (DHX36) originally identified as an RNA helicase associated with AU-rich element (ARE) of urokinase-type plasminogen activator (uPA) mRNA together with NF90 and HuR (26). RHAU is a nucleocytoplasmic shuttling protein found predominantly in the nucleus and to a lesser extent in the cytoplasm. As with other helicases (27-29), ATPase activity is necessary for RHAU function in the decay of uPA mRNA and for its nuclear localization (26, 30). Despite the role of RHAU as a factor destabilizing uPA mRNA, global analysis of gene expression in RHAU knockdown cells revealed that changes in steady-state levels of mRNAs were only partially influenced by mRNA decay regulation (30). Indeed the nuclear localization of RHAU and its guanine quadruplex (G4) DNA/RNA-resolving activity (31) may reflect that RHAU regulates gene expression at various steps other than mRNA decay.
Given that SGs are sites at which dynamic changes in protein-RNA interaction determine the fate of mRNAs during and after stress, it is surprising that the role of SG-associated RNA helicases with an essential function in remodeling protein-RNA interactions is largely unknown. Here we show that RHAU is a novel component of SGs and that its recruitment to SGs is mediated by an RNA interaction. Although identified as an ARE-associated protein involved in ARE-mediated mRNA decay of uPA, it was not immediately clear whether RHAU directly binds RNA. Here we show that RHAU physically interacts with RNA in vitro and in vivo via a unique N-terminal RNA-binding domain composed of a G-rich region and an RHAU-specific motif (RSM) that is highly conserved between RHAU orthologs. The same RNA-binding domain is necessary and sufficient for RHAU recruitment to SGs. Finally we show that the ATPase activity of RHAU is involved in the dynamic regulation of RHAU shuttling into and out of SGs.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—The plasmids pTER-shRHAU and pTER-shLuc were described previously (30). Plasmids EGFP-RHAU, EGFP-Nter, EGFP-HCR, and EGFP-Cter were based on pEGFP-C1 (Clontech) and EGFP-E335A (also termed DAIH in this study), which was derived from EGFP-RHAU by point mutation in motif II as described previously (30). Plasmids RHAU-EGFP, (50-1008)-EGFP, (105-1008)-EGFP, (1-130)-EGFP, and (1-105)-EGFP were based on pEGFP-N1 (Clontech) by inserting corresponding fragments into the BglII (XhoI for RHAU full length) and AgeI sites of the vector. Plasmids EGFP-(50-1008) and EGFP-(105-1008) were prepared by inserting the corresponding fragments between BglII and BamHI sites of pEGFP-C1. RHAU-FLAG, (50-1008)-FLAG, (105-1008)-FLAG, (1-105)-FLAG, and (1-130)-FLAG were prepared by inserting corresponding RHAU fragments into the BglII (NheI for RHAU full length) and AgeI sites of pIRES.EGFP-N1-FLAG. The vector was prepared by insertion of IRES.EGFP between AgeI and NotI of pEGFP-N1 (Clontech). The IRES.EGFP insert was designed by PCR using the pIRES.ECMV-EGFP vector, which was kindly provided by D. Schmitz Rohmer and B. A. Hemmings, as a template. The PCR product of the IRES.EGFP insert contained FLAG sequence with the AgeI site on the 5′ end and the NotI site on the 3′ end. Plasmids β-gal-(1-52)-EGFP, β-gal-(1-130)-EGFP, and β-gal-(52-200)-EGFP were prepared by inserting corresponding RHAU fragments into EcoRI and SalI sites of pβ-gal-EGFP.4 The GST-RHAU vector was designed as described previously (26). GST-Nter was prepared by inserting the fragment (1-200 aa) between BamHI and EcoRI sites of pGEX-2T (Amersham Biosciences). The oligonucleotides used in this work and detailed descriptions of the plasmid construction are available upon request.
Antibodies—Mouse monoclonal anti-RHAU antibody (12F33) was generated against a peptide corresponding to the C terminus of RHAU, 991-1007 aa, as described previously (31). Commercially obtained antibodies were: mouse anti-green fluorescent protein (B-2, sc-9996), goat anti-TIA-1 (sc-1751), goat anti-eIF3b (N-20, sc-16377), mouse anti-HuR (3A2, sc-5261), and rabbit anti-eIF2α (FL-315, sc-11386) (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-actin (pan Ab-5, Thermo Fisher Scientific, Fremont, CA); rabbit monoclonal anti-eIF2α-P (Ser-51, 119A11, Cell Signaling Technology, Danvers, MA); and mouse anti-FLAG M2 (Sigma-Aldrich). The mouse antibodies were all monoclonal.
Cell Culture, Transfection, and Stress Treatments—HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37 °C in the presence of 5% CO2. T-REx-HeLa cells stably transfected with pTER-shRHAU were maintained as described previously (30). To induce short hairpin RNA expression and consequent depletion of endogenous RHAU, cells were treated with 1 μg/ml doxycycline (Sigma-Aldrich) for 7 days as described in supplemental Fig. 1. For immunofluorescence analysis, transient transfection of DNA plasmids using FuGENE 6 (Roche Applied Science) was performed according to the manual provided using 1 μg of plasmid DNA and 3 μl of FuGENE 6/35-mm dish. For immunoprecipitation analysis, cells were transfected by Lipofectamine 2000 (Invitrogen) in Opti-MEM I medium (Invitrogen). Briefly HeLa cells were seeded at 0.8 × 106 cells/35-mm dish and 24 h later were transfected with 4 μg of plasmid DNA and 10 μl of Lipofectamine 2000. The cells were used 24 h later for the following experiments. RNA interference silencing was induced by transient transfection of small interfering RNAs with INTERFERin (Polyplus Transfection, New York, NY) following the manual instructions. Small interfering RNA was added to give a final concentration of 2.5 nm in 2 ml of medium and 8 μl of INTERFERin for transfection of 40% confluent cells in each 35-mm dish. To test SG formation, 0.5 mm sodium arsenite (Sigma-Aldrich) or 1 μm hippuristanol (kindly provided by J. Tanaka (32)) was added in conditioned medium for 45 or 30 min, respectively. To induce SGs with the ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), cells were washed with 1× PBS- (PBS without Ca2+ and Mg2+) and cultured in glucose- and pyruvate-free Dulbecco's modified Eagle's medium containing 1 μm CCCP. Heat shock was performed at 44 °C in a 5% CO2 incubator for 45 min.
Immunocytochemistry and Image Processing—At 24 h after transfection by FuGENE 6, HeLa cells were replated in 12-well dishes with coverslips coated with 0.2% gelatin. The next day, cells were treated with the indicated stimuli, fixed with 3.8% paraformaldehyde in 1× PBS- for 10 min, permeabilized with 0.2% Triton X-100 in PHEM buffer (25 mm HEPES, 10 mm EGTA, 60 mm PIPES, 2 mm MgCl2, pH 6.9) for 30 min and blocked with 5% horse serum in PHEM buffer for 1 h. All steps were performed at room temperature. Samples were incubated with primary antibodies (goat anti-TIA-1 (1:200), mouse anti-HuR (1:200), and goat anti-eIF3b (1:200)) diluted in the blocking buffer overnight at 4 °C. Coverslips with fixed cells were washed three times with 0.2% Triton X-100 in PHEM buffer and incubated in the dark with the secondary antibody and 500 ng/ml DAPI (Santa Cruz Biotechnology) to identify the nuclei for 40 min at room temperature. Cy2-, Cy3-, or Cy5-conjugated donkey secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at dilutions of 1:200, 1:2,000, or 1:200 with 2.5% horse serum in PHEM buffer, respectively. The cells were mounted in FluoroMount reagent (SouthernBiotech, Birmingham, AL). Fluorescent images were captured with a confocal microscope (LSM 510 META, Carl Zeiss, Jena, Germany) as described previously (30) except that a Plan-NeoFluar 100×/1.3 oil differential interference contrast objective was used. The data obtained were processed using standard image software (Bitplane Imaris 5.7.1, Adobe Photoshop, and Adobe Illustrator). To quantify association of RHAU with SGs, at least 100 transfected cells were analyzed under the wide spectrum microscope (Axioplan 2, Carl Zeiss) and scored as positive when the green fluorescent protein signal was enriched and co-localized with TIA-1 in SGs. Three (n = 3) independent transfections were analyzed to calculate mean percentages and ±S.E.
Protein Extraction and Western Blotting—To prepare total cell lysates, cells were lysed with Nonidet P-40 buffer (50 mm Tris-HCl, pH 7.5, 120 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 5 mm Na3VO4, 5 mm NaF, 0.5 μg/ml aprotinin, 1 μg/ml leupeptin) on ice for 10 min and centrifuged at 20,000 × g for 15 min at 4 °C to remove cell debris. Typically 30 μg of the total cell lysate was loaded for Western blotting. The protein bands were visualized with the direct infrared fluorescence method or the chemiluminescence method as described previously (30).
Cross-linking Immunoprecipitation (CLIP)—RHAU and RNA interaction was determined by the previously reported CLIP method with slight modifications (33). HeLa cells (0.8 × 106/35-mm dish) were rinsed twice with ice-cold PBS and then UV irradiated (400 mJ/cm2) to induce cross-linking between protein and RNA. Cells were then lysed with 200 μl of RIPA buffer (1% Nonidet P-40, 1% deoxycholate, 0.1% SDS, 50 mm NaCl, 10 mm sodium phosphate, pH 7.2, 2 mm EDTA, 50 mm NaF, 0.2 mm sodium vanadate, 100 units/ml aprotinin)/well in a 6-well dish and shaken for 15 min at 4 °C. Pooled lysates from 6 wells were treated with 30 μl of RQ1 RNase-free DNase (1 unit/μl; Promega, Madison, WI) and with 31 units of RNase A (31 units/μl; USB Corp.) as described in the CLIP protocol (33). Treated samples were centrifuged at 20,000 × g for 20 min at 4 °C. The supernatants (600 μg) were incubated with 4.5 μg of a mouse anti-RHAU monoclonal antibody (12F33) or 10 μl (bed volume) of anti-FLAG M2 affinity gel (A2220, Sigma-Aldrich) with rotation for 2 h at 4 °C. Beads were washed twice with RIPA buffer, twice with high salt washing buffer (5× PBS, 0.1% SDS, 0.5% deoxycholate, 0.5% Nonidet P-40) and twice with 1× PNK buffer (50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 0.5% Nonidet P-40). The associated nucleic acids were radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase (Roche Applied Science) as described in the CLIP protocol (33), and RHAU-RNA complexes were resolved in a NuPAGE 4-12% bis-Tris gel (Invitrogen). Immunoprecipitated RHAU was detected by Coomassie Blue staining and in-gel Western blotting according to the manual of Odyssey In-Gel Western detection (LI-COR Biosciences). Half of the samples were transferred to a polyvinylidene difluoride membrane to facilitate better protein detection by Western blotting and to remove free RNA. The proteins were detected by the Odyssey infrared imager as described above. Radiolabeled RNA was detected by a phosphorimaging system, Typhoon 9400 (GE Healthcare), and analyzed using the ImageQuant TL program. To test whether RHAU associates with RNA, bound nucleic acids were isolated and radiolabeled according to the CLIP protocol (33). Nucleic acids were mixed with increasing amounts of RNase A (0.015, 0.15, 1.5, and 15 units) in H2O and 1 unit of RQ1 DNase in 1× RQ1 DNase reaction buffer. Reactions were incubated for 30 min at 37 °C and resolved by denaturing 8% PAGE in 1× Tris borate-EDTA buffer.
Protein Purification—Escherichia coli BL21 (DE3) transformed with glutathione S-transferase (GST) or GST-Nter proteins were induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside for 12 h at 25 °C and purified by affinity chromatography with glutathione-Sepharose 4B (Amersham Biosciences) according to the manufacturer's instructions. GST-RHAU protein was expressed in Sf9 cells according to the supplier's instructions (BD Biosciences Pharmingen) and purified as above. The purity of recombinant proteins was analyzed by 10% SDS-PAGE and Coomassie Blue staining as reported in supplemental Fig. 2.
Double Filter RNA Binding Assay—5 μg of total RNA isolated from HeLa cells was alkali-treated with 0.1 m NaOH on ice for 10 min and then EtOH-precipitated. Redissolved RNA or poly(rU) (P9528, Sigma-Aldrich) was 5′-end-labeled using [γ-32P]ATP (Hartmann Analytic GmbH, Braunschweig, Germany) and T4 polynucleotide kinase (Roche Applied Science) at 37 °C for 30 min and passed through a G-50 column (GE Healthcare) to remove free nucleotides. Reaction mixtures (50 μl) containing varying amounts of recombinant proteins (0-150 nm as specified in text), radiolabeled RNA (poly(rU), 10,000 cpm; total RNA, 5,000 cpm) and 2 units of RNase inhibitor (RNAguard, Roche Applied Science) in the binding buffer (50 mm Tris-HCl, pH 8.0, 1 mm dithiothreitol, 5 mm NaCl) were incubated for 30 min at 37 °C. The double filter RNA binding assay was performed with a slot-blot apparatus using a 0.45-μm nitrocellulose (Protran, Whatman) and nylon membranes (positively charged; Roche Diagnostics) that was presoaked in different buffers as described previously (34). Loaded samples were washed three times with 200 μl of the binding buffer. Retained RNA was detected with a phosphorimaging system, Typhoon 9400, and analyzed using the ImageQuant TL program. The nitrocellulose membrane retains only RNA-protein complexes, and free RNAs are captured on the nylon membrane. The ratio of RNA that was bound to GST-RHAU or GST-Nter was calculate using the following formula: bound RNA (%) = 100((signalnitrocellulose)/(signalnitrocellulose + signalnylon)).
Bioinformatics—The program RNABindR (35) was used to predict RNA binding potential in amino acid sequences. Programs RISP (RNA-Interaction Site Prediction) and BindN+ were used to confirm the reliability of the RNABindR program: RISP (36) runs with 72.2% RNA binding prediction accuracy, and BindN+ (37) runs with 68% RNA binding prediction accuracy. For multiple sequence alignments of N termini, RHAU orthologs were identified by a BLASTP (version 2.2.18+) search of non-redundant protein entries in the NCBI data base using the entire sequence of RHAU as a query. Multiple sequence alignment was carried out with ProbCons (version 1.12) (38). Similarity of groups was generated using GeneDoc (version 2.7) with the BLOSUM62 scoring matrix.
Fluorescence Recovery after Photobleaching—HeLa cells (1.5 × 105/35-mm dish) were plated and transfected by FuGENE 6 on glass-bottomed dishes (Micro-Dish 35 mm, Fisher Scientific). FRAP experiments were carried out with a confocal microscope (LSM 510, Carl Zeiss) using a 63×/1.4 numerical aperture oil differential interference contrast objective. Bleaching was performed using the 488-nm lines from a 40-milliwatt argon laser operating at 75% laser power. Bleaching of circular regions of interest (ROI) was done with three scan iterations lasting a total of 1.5 s. Fluorescence recovery was monitored at low laser intensity (∼2% of the 40-milliwatt laser) at 0.5-s intervals. Two additional ROI were monitored in parallel to detect fluorescence fluctuations that were independent of bleaching (ROI of non-bleached SG) and to extract a background (ROI in the cytoplasm). Raw data of one protein were obtained from ≥10 different FRAP analyses from three independent transfections. Fluorescence recovery was normalized with the function ((It - Bt)/(Ct - Bt))/I0 where It is the mean intensity in ROI of bleached SGs at time point t, Bt is the mean intensity in ROI of the background at time point t, Ct is the mean intensity in ROI of control SGs at time point t, and I0 the average intensity in ROI of bleached SGs during prebleach. The average fluorescence intensities ±S.E. from at least ≥10 independent FRAP analyses were extrapolated to the graph as recovery curves.
RESULTS
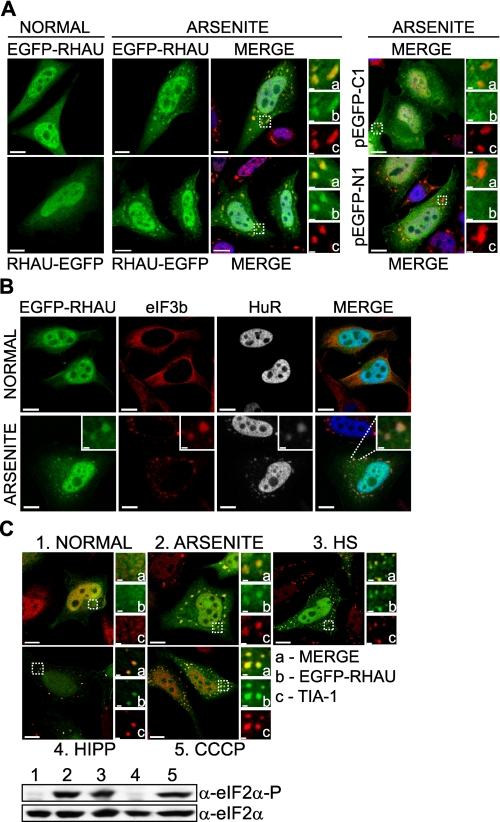
RHAU Protein Associates with SGs in Response to Arsenite-induced Stress—Various research groups have shown that different ARE-binding proteins, such as TTP, Brf1, HuR, TIA-1, or TIA-1-related protein, associate with SGs in cells exposed to environmental stress (8, 39, 40). To determine whether RHAU is also recruited to SGs, HeLa cells were transfected with EGFP-tagged RHAU and treated with arsenite, a typical inducer of SGs. As shown in Fig. 1A, the arsenite treatment induced, in addition to diffuse nuclear and cytoplasmic localizations, the accumulation of RHAU in distinct cytoplasmic foci that co-localized with TIA-1 protein, an SG marker. Because all available anti-RHAU antibodies, both commercial and our own preparation, were not suitable to visualize endogenous RHAU by immunofluorescence analysis, we examined the intracellular localization of RHAU by transiently transfecting cells. We used either EGFP-RHAU or RHAU-EGFP to eliminate the unwanted effects of EGFP on RHAU structure and subsequent subcellular localization. Indeed both EGFP-RHAU and RHAU-EGFP showed similar localization patterns in normal and stress conditions (Fig. 1A). EGFP itself did not accumulate in SGs (Fig. 1A). The localization of RHAU in arsenite-induced cytoplasmic foci was confirmed using other SG markers such as HuR and eIF3b (p116) (Fig. 1B), indicating that RHAU is a novel SG component.
FIGURE 1.
RHAU helicase accumulates in SGs in response to all tested stress stimuli. A, immunofluorescence analysis of RHAU localization in arsenite-induced SGs. HeLa cells, transiently transfected with EGFP-RHAU or RHAU-EGFP for 48 h, were treated with 0.5 mm sodium arsenite for 45 min or cultured in normal conditions. After fixation, cells were stained with DAPI (blue) to visualize nuclei and with anti-TIA-1 antibody (red) to detect SGs. Left, distribution of EGFP-RHAU or RHAU-EGFP (green) under normal and arsenite-treated conditions. Panels denoted as MERGE show the digital combination of EGFP, TIA-1, and DAPI images under arsenite-treated conditions; enlargements of boxed regions are shown in small panels indicating merge (a), EGFP (b), and TIA-1 (c). Right, merges of DAPI and TIA-1 with empty vectors pEGFP-C1 or pEGFP-N1; enlargements of boxed regions are shown in small panels as above. B, co-localization of RHAU with eIF3b and HuR in arsenite-induced SGs. HeLa cells were transfected with EGFP-RHAU and treated with or without arsenite as above. The extreme left panels show EGFP-RHAU (green), and the two middle panels are immunofluorescent images for eIF3b (red) and HuR (white; blue in merge), two SG markers. The extreme right panels are their merges. C, effects of different stress stimuli on eIF2α phosphorylation and RHAU localization in SGs. HeLa cells transiently transfected with EGFP-RHAU were cultured without (1) or with the following stress inducers: 0.5 mm arsenite for 45 min (2), heat shock (HS) at 42 °C for 45 min (3), 1 μm hippuristanol (HIPP) for 30 min (4), and 1 μm CCCP for 90 min (5). The images show merges of EGFP-RHAU (green) and TIA-1 (red). Enlargements of boxed regions are shown in small panels indicating merge (a), EGFP (b), and TIA-1 (c). The total eIF2α and phosphorylated eIF2α (eIF2α-P) were detected by Western blotting. Bar, 10 μm (1 μmin enlargements).
Although the formation of SGs was not abolished or reduced in T-Rex-HeLa cells after RNA interference-mediated down-regulation of endogenous RHAU (supplemental Fig. 1), we asked whether the recruitment of RHAU to SGs is restricted to a specific type of stress. To address this question, we treated EGFP-RHAU transfected HeLa cells with different SG-inducing stimuli, such as arsenite, CCCP (a mitochondrial inhibitor), heat shock, and hippuristanol (an inhibitor of eIF4A RNA binding activity known to induce SG aggregation independently of eIF2α phosphorylation) (32). As shown in Fig. 1C, EGFP-RHAU co-localized with TIA-1 in SGs in response to all stimuli tested. The phosphorylation of eIF2α was detected after treatment with arsenite, heat shock, and CCCP but, as expected, not with hippuristanol (Fig. 1C). These results indicate that the formation of SGs, rather than phosphorylation of eIF2α, is required for RHAU protein recruitment to SGs.
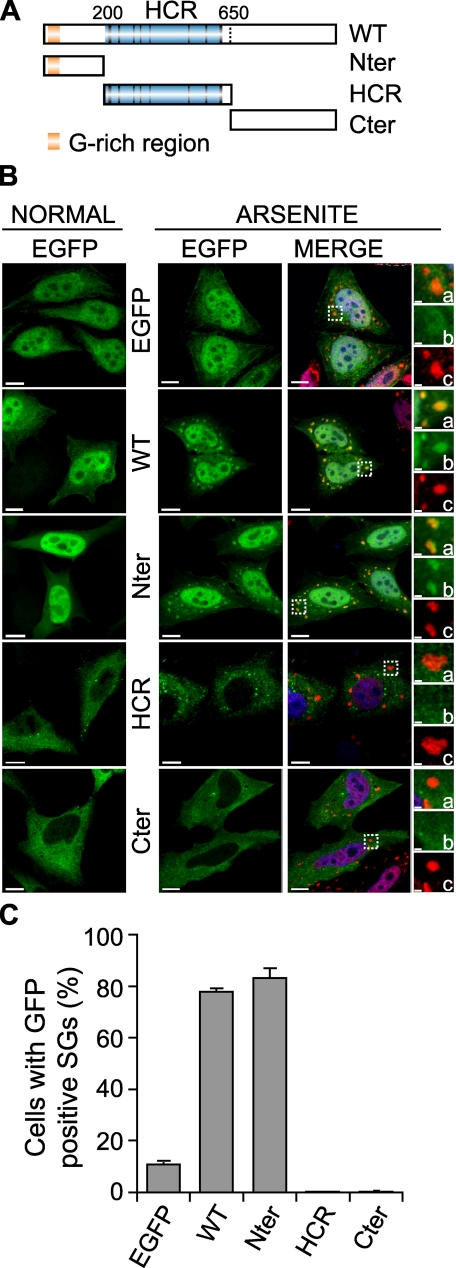
The N-terminal Domain Recruits RHAU to SGs—RHAU protein consists of three main domains: the unique N terminus (Nter), the evolutionarily conserved helicase core region (HCR), and the C terminus (Cter) partially conserved among DEAH subfamily members. To identify domains essential for localization of RHAU in SGs, we generated deletion mutants of RHAU: EGFP-Nter (1-200 aa), EGFP-HCR (201-650 aa), and EGFP-Cter (651-1008 aa) (Fig. 2A). As shown in Fig. 2B, the full-length RHAU (WT) as well as the N terminus alone accumulated in arsenite-induced SGs. Unlike EGFP-WT and EGFP-Nter, EGFP-HCR and EGFP-Cter were excluded from the nucleus and diffused in the cytoplasm as described previously (30). After arsenite treatment, they showed low level accumulation in small foci other than SGs (Fig. 2B). A similar proportion of cells positive for EGFP signals in arsenite-induced SGs was observed with EGFP-WT (78%)- and EGFP-Nter (83%)-transfected cells, but no positive EGFP signals in arsenite-induced SGs were detected after transfection with EGFP-HCR and EGFP-Cter (Fig. 2C). Taken together, these results indicate that the N terminus of RHAU is essential for stress-induced accumulation of RHAU in SGs.
FIGURE 2.
The N-terminal domain of RHAU localizes in SGs. A, scheme of wild-type RHAU and its fragments. Nter, 1-200 aa; HCR, 200-650 aa; Cter, 200-1008 aa. B, intracellular localization of different parts of RHAU. HeLa cells were transfected with an empty pEGFP-C1 vector or plasmids expressing EGFP-fused RHAU (WT) or fragments of RHAU, Nter, HCR, and Cter. After 48 h, cells were cultured in normal conditions or treated with 0.5 mm arsenite for 45 min, fixed, and stained for TIA-1 (red). Nuclei were visualized with DAPI (blue). Small panels show enlargements of boxed regions: merge (a), EGFP-fused fragments of RHAU (b) and TIA-1 (c). C, data obtained by quantitative immunofluorescence analysis showing the percentage of transfected cells in which EGFP signals were detected in SGs. Values ± S.E.M. (standard errors of means) were derived from three independent experiments. Bar, 10 μm (1 μmin enlargements). GFP, green fluorescent protein.
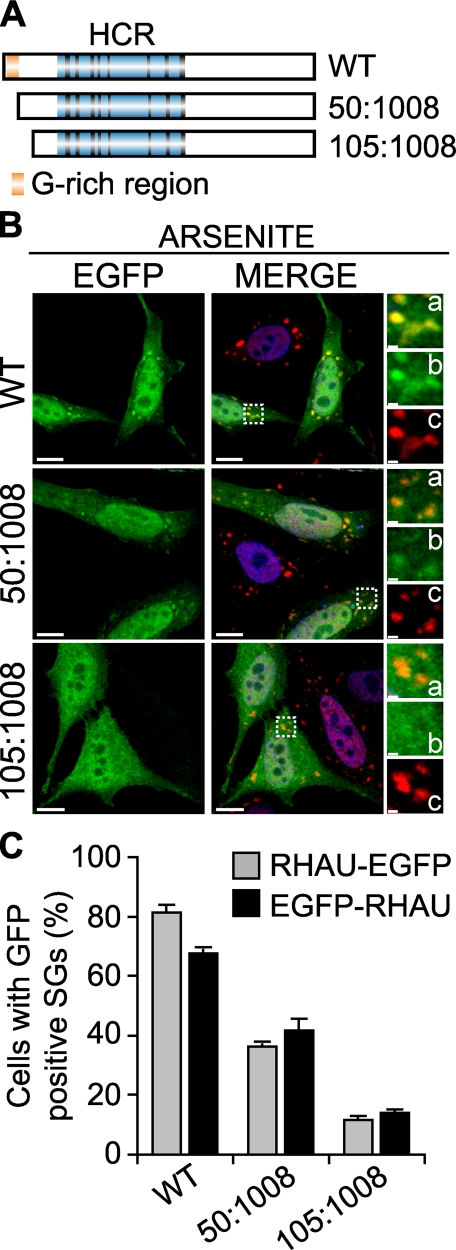
Because only the first 200 amino acids of RHAU were necessary for SG localization, we further studied functional domains within the N terminus. For this we generated two new N-terminally truncated forms of EGFP-fused RHAU by stepwise deletion as depicted in Fig. 3A where the first deletion mutant (50-1008 aa) lacked a G-rich region. These truncated mutants and WT were transfected into HeLa cells. The association of RHAU and its mutants with arsenite-induced SGs was assessed by immunofluorescence microscopy (Fig. 3B) and quantified (Fig. 3C). Deletion of the G-rich region significantly decreased RHAU association with SGs from 81 to 36% of transfected cells. The truncation of amino acids 1-105 further reduced RHAU accumulation in SGs to 12%. As shown in a graph (Fig. 3C), EGFP fusion at the N or C terminus with the WT or the mutants did not affect their localization efficiency in SGs. Taken together, these data suggest that the first 105 amino acids of the N-terminal domain are essential for the recruitment of RHAU protein to SGs and that an additional site, other than the G-rich region, plays a role in this recruitment process.
FIGURE 3.
The first N-terminal 105 amino acids of RHAU are necessary for RHAU localization in SG. A, scheme of wild-type RHAU and its deletion mutants. B, intracellular localization of RHAU N-terminal deletion mutants. HeLa cells were transfected with plasmids expressing EGFP-fused RHAU (WT) or N-terminal deletion mutants of RHAU, 50-1008 and 105-1008. After 48 h, cells were treated with 0.5 mm arsenite for 45 min, fixed, and stained for TIA-1 (red). Nuclei were visualized with DAPI (blue). Small panels show enlargements of boxed regions: merge (a), EGFP-fused fragments of RHAU (b), and TIA-1 (c). C, quantitative immunofluorescence analysis showing the percentage of transfected cells in which EGFP signals were detected in SGs. Values ± S.E.M. (standard errors of means) were derived from three independent experiments. Bar, 10 μm (1 μmin enlargements a-c). GFP, green fluorescent protein.
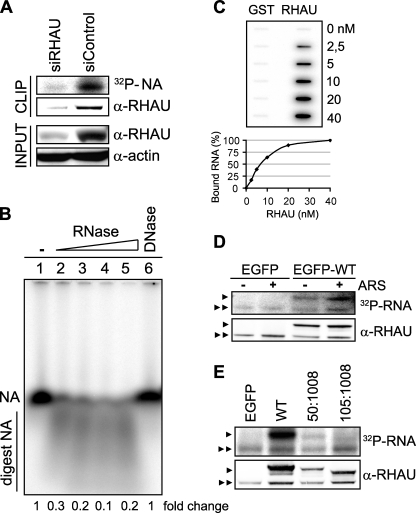
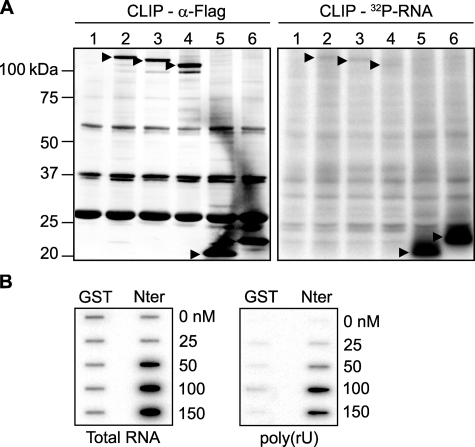
RHAU Binds to RNA via the N-terminal Domain—The recruitment of proteins to SGs was reported to be mediated by protein-protein or RNA-protein interactions (10). Because RHAU is an RNA helicase, we first examined whether RHAU binds to RNA and, if so, whether RNA binding activity is required for its accumulation in SGs. To test this, we used adapted CLIP (33), utilizing irreversible UV cross-linking between protein and RNA to purify protein-RNA complexes. The tightly associated RNA molecules migrating together with immunoprecipitated protein were radiolabeled with [γ-32P]ATP and detected with a phosphorimaging system. Endogenous RHAU was immunoprecipitated with monoclonal anti-RHAU antibody. Phosphorimaging analysis of radiolabeled nucleic acids showed a prominent signal at 120 kDa corresponding to RHAU (Fig. 4A). The signal was absent when RHAU was depleted by RNA interference before immunoprecipitation. This indicated that under normal conditions the strong signal at 120 kDa was derived from RHAU-associated nucleic acids. UV irradiation not only cross-links proteins and RNA but also proteins and DNA. As the CLIP method includes a stringent DNase treatment prior to immunoprecipitation, it is likely that the detected signals involve RNA. Nevertheless to test this further, we purified co-immunoprecipitated nucleic acids, radiolabeled them, and examined their sensitivity to an increasing concentration of RNase (lanes 2-5) and 1 unit of DNase (lane 6). RHAU-associated nucleic acids were digested by RNase but not by DNase (Fig. 4B) indicating that RHAU interacts with RNA in vivo.
FIGURE 4.
RHAU associates with RNA through its N terminus. A, CLIP method to identify interactions of RHAU with nucleic acids (NA). HeLa cells were transiently transfected with small interfering RNAs against RHAU (siRHAU) or luciferase (siControl). Cells were UV irradiated 72 h later and harvested. Immunoprecipitation was performed with a RHAU antibody. Bound nucleic acids were radiolabeled and detected as a band of ∼120 kDa corresponding to RHAU (lane siControl). However, this band was strongly reduced when endogenous RHAU was depleted (lane siRHAU). Western blot (INPUT) shows efficient RHAU depletion by small interfering RNAs against RHAU. Actin was used as a loading control. B, identification of associated nucleic acids as RNA. After immunoprecipitation, the nucleic acids bound to RHAU were isolated, radiolabeled, and treated with increasing concentrations (0.015, 0.15, 1.5, and 15 units) of RNase A and 1 unit of RQ1DNase. Lane1, isolated and non-treated nucleic acids(NA); lanes 2-5, isolated nucleic acids treated with increasing concentrations of RNase; lane 6, nucleic acids treated with DNase. -Fold changes in intensity ratio (NA/digest NA) are depicted relative to the intensity of non-treated nucleic acids in lane 1, set as 1. Note that nucleic acids are RNase-sensitive but DNase-insensitive. C, RHAU binds to RNA in vitro. 0, 2.5, 5, 10, 20, and 40 nm GST or GST-RHAU (RHAU) was incubated with 5,000 cpm 5′-end-labeled RNA for 30 min at 37 °C and filtered through nitrocellulose and nylon membranes. The percentage of bound RNA was plotted as a function of increasing concentration of GST-RHAU. Note that dose-dependent RNA binding was seen only with GST-RHAU. D, effect of stress on RHAU association with RNA in vivo. HeLa cells were transfected with EGFP alone or EGFP-WT (a full-length RHAU). Half were treated 24 h later with arsenite (0.5 mm for 45 min) followed by UV irradiation and radiolabeling of RHAU-associated nucleic acids. RHAU and associated RNA were analyzed by Western blotting and a phosphorimaging system to detect levels of protein expression and the amount of radiolabeled RNA, respectively. Note that the arsenite treatment (+) did not abolish or dramatically change the RNA binding activity of RHAU. ARS, arsenite. E, comparison of RNA binding activities of wild-type RHAU and its N-terminal deletion mutants. HeLa cells were transfected with EGFP, EGFP-WT, EGFP-(50-1008), or EGFP-(105-1008). Protein and associated RNA were analyzed as in A. Radioactivity of bound RNA was normalized to the expression levels of corresponding proteins. In sharp contrast to constant RNA binding of endogenous RHAU in each lane, N-terminal deletion mutants showed stepwise reduction of RNA interaction compared with WT. ▸, overexpressed EGFP-RHAU or its N-terminal deletion mutants; ▸▸, endogenous RHAU.
To confirm our in vivo observation, the binding of RHAU to RNA was examined in vitro using purified, recombinant GST and GST-RHAU by double filter RNA binding assays (supplemental Fig. 2A and Fig. 4C). As we do not know yet the in vivo RNA targets of RHAU, we incubated radiolabeled, total RNA (5,000 cpm) from HeLa extract with increasing amounts of GST or GST-RHAU (0-40 nm) and filtered the mixture through nitrocellulose and nylon membranes. As shown in Fig. 4C, unlike GST itself, GST-RHAU was retained with associated RNA on nitrocellulose, and moreover it displayed dose-dependent RNA binding, suggesting that RHAU can directly associate with RNA. We express the relative binding affinity (KD) as the concentration of GST-RHAU at which 50% of RNA was bound. The apparent KD of GST-RHAU was 7 nm (Fig. 4C).
Next we examined the influence of arsenite stress on the RHAU-RNA interaction by treating the EGFP- or EGFP-WT-transfected HeLa cells with arsenite before UV irradiation and comparing the RNA binding activity of RHAU in normal and stress conditions. After immunoprecipitation by a monoclonal anti-RHAU antibody, two radiolabeled RNA bands per the lane were detected; the lower and upper bands correspond to endogenous RHAU and transfected EGFP-WT, respectively (Fig. 4D). The association of endogenous RHAU with RNA was used as a loading control. The intensity of the RNA signals normalized over RHAU protein levels was similar in control and arsenite-treated conditions, suggesting that the amount of RHAU-associated RNA was not influenced by stress. The RNA band migrating in the region corresponding to EGFP-WT was not present in extracts of EGFP-transfected cells, verifying that the RNA signals were derived from RHAU-associated RNA.
Given the apparent significance of the first 105 amino acids for RHAU localization in SGs, we examined whether the N terminus was also required for RHAU-RNA interaction. After cells were transiently transfected with EGFP-WT and its N-terminal deletion mutants, the extent of RNA binding to RHAU and mutants was examined as described above and normalized over RHAU protein levels. Although the CLIP method is not a suitable quantitative assay, in six independent assays EGFP-(50-1008) and EGFP-(105-1008) showed a reproducible decrease in RNA binding activity of ∼50 and 20%, respectively, compared with EGFP-WT (Fig. 4E). A stepwise decrease in the RNA binding activity of the two deletion mutants correlated with the decreased RHAU localization in arsenite-induced SGs (see Fig. 3C). The small amount of RNA remaining associated with EGFP-(105-1008) may reflect residual RNA binding activity derived from the helicase core domain of RHAU. Thus, the majority of RNA associated with RHAU is bound to its N terminus.
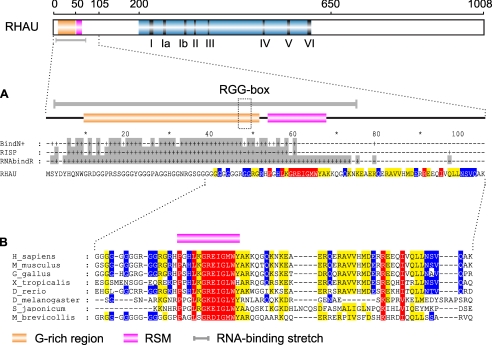
Bioinformatics Analysis of the N Terminus Revealed a Putative RNA-binding Domain—Knowing that RHAU interacts with RNA via the first 105 amino acids, we submitted the RHAU protein sequence to RNABindR, a computational program that predicts RNA binding potential of amino acid residues (35). This program is focused on sequence-based predictions and runs on a naïve Bayes classifier. According to this program, residues from 3 to 75 were highly positive for the RNA interaction (Fig. 5A). A similar prediction was obtained by two other programs based on different algorithms, BindN+ and RISP (36, 37) (Fig. 5A). Moreover sequence alignment within the first 105 amino acids of RHAU proteins from choanoflagellate to humans revealed a novel highly conserved domain (54-66 aa) termed RSM next to the already known G-rich region (10-50 aa) with one RGG box motif (47-49 aa) (Fig. 5B). Although RGG box motifs have been reported to be involved in RNA binding (41, 42), RSM has no similarity to any known functional sequence elements. Thus, RHAU likely harbors an atypical conserved RNA-binding domain in the N terminus.
FIGURE 5.
Bioinformatics analysis of the RHAU N-terminal domain. A, computational analysis for RNA-binding residues within the RHAU N terminus. The human RHAU amino acid sequence was subjected to three different programs, BindN+, RISP, and RNABindR, available on line. Residues predicted to be positive for RNA binding are indicated by “+” and depicted above the RHAU amino acid sequence. The gray shading corresponds to RNA binding consensus per one amino acid in all three programs. Note that the predicted RNA-binding domain is a long stretch from 3 to 75 aa (gray bar) that overlaps with the G-rich region containing an RGG box motif and with the highly conserved RSM. B, conservation of the RSM through evolution. Orthologs of RHAU used for the multiple sequence alignment were identified as described under “Experimental Procedures.” Similarity of groups was estimated using GeneDoc (version 2.7) with the BLOSUM62 scoring matrix. Red, 90-100% similarity; yellow, 70-90% similarity; and blue, 50-70% similarity. Species and NCBI and Ensembl (in case of X. tropicallis) accession numbers are as follows: human (Homo sapiens, NP_065916), mouse (Mus musculus, NP_082412), chicken (Gallus gallus, XP_422834), frog (Xenopus tropicalis, ENSXETP00000016953), zebrafish (Danio rerio, CAM56669), fruit fly (Drosophila melanogaster, NP_610056), trematoda (Schistosoma japonicum, AAW25528), and choanoflagellate (Monosiga brevicollis, EDQ87802).
The N-terminal RNA-binding Domain Is Essential and Sufficient for RNA Interaction and Localization of RHAU in SGs—To further test whether these 105 N-terminal residues are sufficient for RNA binding and subsequent RHAU recruitment to SGs, we designed FLAG-tagged truncated forms of the RHAU protein: WT-FLAG, (50-1008)-FLAG, (105-1008)-FLAG, (1:105)-FLAG and (1:130)-FLAG. For the RNA binding experiment, CLIP analyses were performed using a monoclonal mouse anti-FLAG M2 antibody coupled to agarose beads. Western blotting confirmed the successful expression and immunoprecipitation of all fragments with (1-105)-FLAG and (1-130)-FLAG at higher protein levels (Fig. 6A, left panel). To analyze the RNA binding activities of these proteins, the measured radiolabeled RNA levels were normalized over protein levels. As shown in Fig. 6A (right panel), compared with WT-FLAG (lane 2), the amounts of RNA associated with (50-1008)-FLAG (lane 3) and (105-1008)-FLAG (lane 4) were stepwise reduced to ∼70 and 30%, respectively. The high levels of RNA binding (before normalization) with (1-105)-FLAG and (1-130)-FLAG (lanes 5 and 6) may partly reflect their expression levels (Fig. 6A, right panel); however, they still showed ∼10-fold higher RNA binding activity than WT-FLAG after normalization. Thus, these results demonstrate that the first 105 amino acids are essential and sufficient for RHAU to bind RNA.
FIGURE 6.
The N-terminal RNA-binding domain is essential and sufficient for RNA interaction in vivo and in vitro. A, CLIP method to show RNA binding activity of the N-terminal 105 amino acid residues of RHAU. HeLa cells were transfected with an empty vector (lane 1), vectors expressing FLAG-tagged RHAU wild type (lane 2), or its fragments, (50-1008) (lane 3), (105-1008) (lane 4), (1-105) (lane 5), and (1-130) (lane 6). Cells were UV irradiated 24 h later, harvested, and immunoprecipitated by FLAG antibody. Bound RNA was labeled with [γ-32P]ATP and detected by a phosphorimaging system as described under “Experimental Procedures.” Left panel, Western blot of immunoprecipitated FLAG-tagged RHAU and its fragments. Right panel, bound RNA. The amount of associated RNA was normalized to the expression level of proteins.▸, FLAG-tagged RHAU and its fragments. B, the N terminus of RHAU binds to RNA in vitro. 0, 25, 50, 100, and 150 nm GST or GST-Nter (1-200 aa) was incubated with 10,000 cpm 5′-end-labeled total RNA (left panel) or poly(rU) (right panel) for 30 min at 37 °C and filtered through nitrocellulose and nylon membranes. Note that the dose-dependent RNA binding, both total RNA and poly(rU), was detected only with GST-Nter.
Because the N-terminal fragment of RHAU (1-105)-FLAG has been shown to bind RNA in vivo, we again performed a double filter RNA binding assay using the recombinant GST-Nter fragment (1-200 aa). Purified, recombinant GST and GST-Nter (supplemental Fig. 2B) were incubated with 5′-end-radiolabeled total RNA or with poly(rU) and then filtered through nitrocellulose and nylon membranes. As shown in Fig. 6B, the higher dose-dependent amount of RNA, both total RNA and poly(rU), was associated with GST-Nter rather than GST. The apparent affinities of GST-Nter for total RNA and poly(rU) were 45 and 80 nm, respectively. These data confirmed the in vivo data that the N terminus is essential and sufficient for RHAU association with RNA.
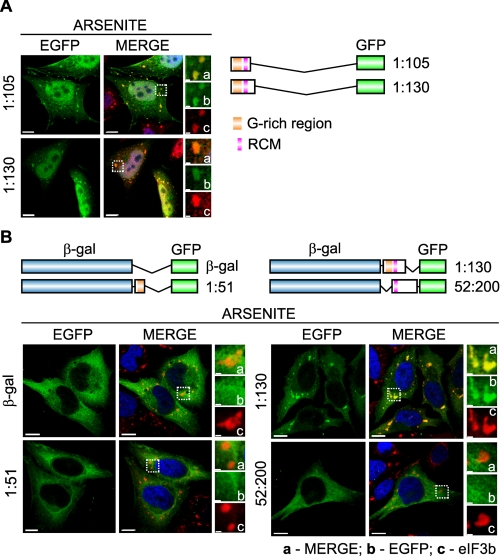
To determine whether the N terminus is also sufficient for RHAU recruitment to SG, we transfected HeLa cells with N-terminal mutants fused with the EGFP tag at their C termini as shown in Fig. 7A. Immunofluorescence analysis revealed that both tested mutants, (1-105)-EGFP and (1-130)-EGFP, co-localized with TIA-1 protein in SGs (Fig. 7A). Further we tested whether the N terminus can bring about SG localization of a protein that does not normally associate with SGs in response to stress. We fused different fragments of the RHAU N-terminal domain to β-gal-EGFP constructs (Fig. 7B) and transfected them into HeLa cells. In response to arsenite treatment, β-gal-EGFP by itself did not accumulate in SGs but was diffusely distributed in the cytoplasm as in normal conditions. However, when β-gal-EGFP was fused to the N-terminal 1-130 amino acids, including the complete putative RNA-binding domain with the G-rich region and the RSM, the fusion protein localized to SGs. In contrast, SG localization was not detected when β-gal-EGFP was fused with either the G-rich region (β-gal-EGFP(1-51 aa)) or the RSM (β-gal-EGFP(52-200 aa)) alone, indicating that only the complete RNA-binding domain is required for RHAU recruitment to SGs.
FIGURE 7.
The N-terminal RNA-binding domain is also essential and sufficient for SG localization of RHAU. A, immunofluorescent images of HeLa cells transfected with vectors expressing EGFP-tagged N-terminal fragments of RHAU, (1-105) and (1-130). After 48 h of transfection, cells were treated with arsenite (0.5 mm, 45 min) to induce SGs. The merge shows the co-localization of EGFP-tagged RHAU fragments (green) and TIA-1 (red). DAPI (blue) stains nuclei. B, immunofluorescent images of HeLa cells expressing EGFP-β-galactosidase double tagged RHAU N-terminal fragments. Cells were treated as above to induce SGs. The co-localization of EGFP (green) and eIF3b (red) is shown in the merge together with DAPI (blue). Note that only the (1-130) fragment containing an intact RNA-binding domain recruits EGFP-β-galactosidase to SGs. Enlargements of boxed regions are shown in small panels indicating merge (a), EGFP-fused RHAU N-terminal fragment (b), and eIF3b (c). Bar, 10 μm (1 μmin enlargements). GFP, green fluorescent protein.
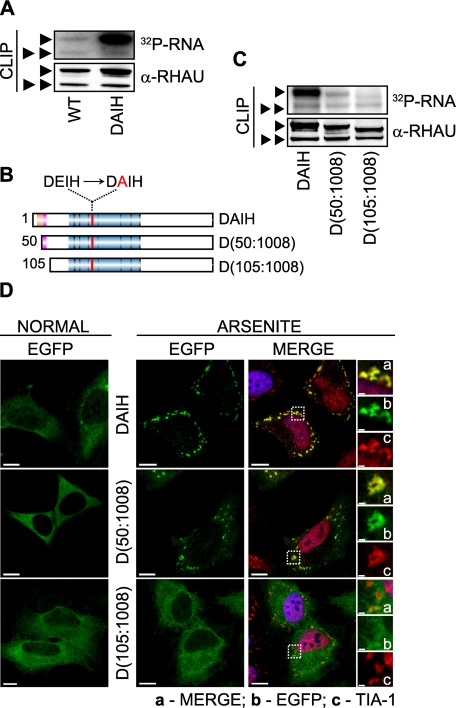
ATP Hydrolysis Plays a Role in RNA Binding and Kinetics of RHAU in SGs—Previously we showed that an ATPase-deficient RHAU mutant with a DEIH to DAIH point mutation is unable to accelerate uPA mRNA decay (26) and is excluded from the nucleus (30). This suggested that the ATPase activity of RHAU has profound effects on its biological activity and subcellular localization. Therefore, we examined the role of RHAU ATPase activity on its RNA binding activity and its localization in SGs. To elucidate the RNA binding potential of ATPase-deficient RHAU mutants, we performed CLIP analyses. After UV cross-linking and subsequent immunoprecipitation of EGFP-WT and EGFP-DAIH with an anti-RHAU antibody, the amount of associated RNA was normalized to corresponding protein levels. As shown in Fig. 8A, a higher amount of RNA was pulled down with the ATPase-deficient DAIH mutant than with EGFP-WT. Based on results of Tanaka and Schwer (34) where Prp22, a yeast DEAH box helicase, dissociated from the RNA upon ATP hydrolysis resulting in apparent RNA binding activity reduction, we concluded that ATPase activity of RHAU is involved in releasing RHAU from RNA rather than in binding to RNA.
FIGURE 8.
Role of RHAU ATPase activity in RNA binding and retention of protein in SGs. A, RNA binding activity of WT and the ATPase-deficient RHAU mutant DAIH. Levels of RHAU-associated RNA were measured by CLIP as described under “Experimental Procedures” 24 h after transfecting HeLa cells with EGFP-fused WT or DAIH. The level of RNA bound to the DAIH mutant was much higher than that bound to WT. Note that the level of RNA bound to endogenous RHAU is comparable in cells expressing exogenous WT or DAIH. B, schematic representation of N-terminal deletion, DAIH mutants: EGFP-DAIH, EGFP-DAIH(50-1008) (D(50:1008)), and EGFP-DAIH(105-1008) (D(105: 1008)). C, comparison of RNA binding between mutants shown in B. HeLa cells were transfected with vectors expressing EGFP-DAIH, EGFP-DAIH(50-1008) (D(50:1008)), or EGFP-DAIH(105-1008) (D(105:1008)), and 24 h later the levels of RNA associated with these mutants and endogenous RHAU were assessed by CLIP as in A. Note that N-terminal deletion mutants show stepwise reduction of RNA binding compared with DAIH full length (such as WT and its deletion mutants in Fig. 4E). D, intracellular localization of EGFP-tagged DAIH mutants listed in B in control and arsenite-treated cells. Transfected HeLa cells were treated without (NORMAL) or with 0.5 mm sodium arsenite for 45 min. Images denoted MERGE are the merger of EGFP (green), TIA-1 (red), and DAPI (blue). Enlargements of boxed regions show merge (a), EGFP-tagged DAIH mutants (b), and TIA-1 (c). Bar, 10 μm (1 μmin enlargements). ▸, EGFP-tagged DAIH and its deletion mutants; ▸▸, endogenous RHAU.
Considering the possibility that the N terminus and ATP hydrolysis contribute to the RNA interaction of RHAU, we investigated whether the N-terminal RNA-binding domain is also required for RNA binding of the DAIH mutant and its accumulation in SGs. For this, we introduced a point mutation (DEIH to DAIH) into motif II of the helicase core region to generate DAIH(50-1008) and DAIH(105-1008) fused with EGFP at their N termini (Fig. 8B). First the truncated fragments were used in the CLIP analysis. While being aware of CLIP limitations in quantitative analysis, stepwise deletion of the N-terminal domain of the DAIH mutant to DAIH(50-1008) and DAIH(105-1008) nevertheless decreased the amount of associated RNA to ∼50 and 15%, respectively, compared with the DAIH full-length mutant (Fig. 8C). The stepwise decrease in RNA interaction seen with the two DAIH deletion mutants corresponds to that seen with RHAU deletion mutants (see Fig. 4E). Thus, enhanced RNA binding of the DAIH mutant still required the N terminus.
Having shown that RHAU recruitment to SGs is dependent on its RNA interaction (see Figs. 3C and 4E), we tested whether the ATPase-deficient DAIH mutant accumulates in SGs more than wild type. Indeed the DAIH mutant localized in SGs (Fig. 8D) to a greater extent than the wild type, increasing the number of transfected cells with EGFP-positive SGs from 81 to 100%. Unlike DAIH full length and DAIH(50-1008), DAIH(105-100) was not detected in SGs. Therefore, RHAU recruitment to SGs is independent of its ATPase activity.
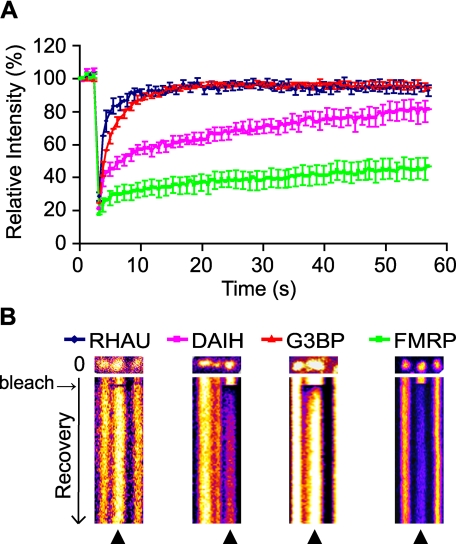
Having established that both RHAU wild type and the DAIH mutant accumulate in SGs, we used the FRAP method to examine whether the dynamics of shuttling into and out of SGs differs between the two proteins. Cells were transfected with EGFP-WT, EGFP-DAIH, EGFP-G3BP, or EGFP-FMRP, and the FRAP analyses were performed 48 h after transfection. It has been shown that TIA-1 and G3BP recover rapidly and completely in SGs within 30 s after bleaching (6, 13). In contrast, PABP-1, another marker for mRNA shuttling, showed only 60% fluorescence recovery after 30 s (3, 6). In our study, we used EGFP-G3BP as a control for the rapid fluorescence recovery (Fig. 9, A and B). Although FMRP has never been tested by FRAP analysis, we included this protein as a control as it is known to localize in SGs (43). As shown in Fig. 9A, the fluorescence signal of G3BP recovered rapidly and completely within 10 s as reported previously (6), but the FMRP signal did not recover fully within 60 s, suggesting that FMRP is a scaffold SG component. In this analysis, the wild type and DAIH signals showed different recovery patterns. The former displayed rapid recovery (≥90%) of fluorescence after 8 s similar to G3BP with the same recovery after 11 s. However, the recovery of the EGFP-DAIH mutant fluorescent signal was much slower with only 60% of fluorescence recovery after 30 s of bleaching similar to PABP-1. The recovery rate reflects the strength of binding whereby slower recoveries correspond to tighter binding. Thus, in comparison with the wild type, the ATPase-deficient mutant, which cannot release RNA because of its inability to hydrolyze ATP, is retained longer in SGs, resulting in slower recovery kinetics. Combining these results, we conclude that the ATPase activity of RHAU is required for its active shuttling into and out of SGs and may also be important for remodeling and/or recruitment of RNPs in SGs.
FIGURE 9.
ATP hydrolysis takes part in dynamic RHAU shuttling into and out of SGs. A, fluorescent recovery patterns of RHAU, ATP-deficient RHAU mutant (DAIH), G3BP, and FMRP. HeLa cells transfected with EGFP-tagged RHAU, DAIH, G3BP, or FMRP were treated with arsenite and analyzed by the FRAP method. Bleaching of selected SGs was performed with three scan iterations for a total of 1.5 s. Fluorescence recovery was monitored at 0.5-s intervals for 50 s, and results were analyzed as described under “Experimental Procedures.” Data were collected from at least 10 independent experiments, and each curve represents average fluorescence intensity ±S.E.M. (standard errors of means) over time. B, representative result of FRAP analysis of EGFP-tagged RHAU, DAIH, G3BP, or FMRP. SGs were photobleached, and subsequent fluorescence recoveries are shown in a fire mode in the time scale of recovery. ▴ indicates SGs bleaching.
DISCUSSION
In the present study, we have shown that the DEAH box helicase RHAU is a novel SG-associated protein that is recruited to SGs via RNA. Its N terminus is necessary and sufficient for RHAU localization in SGs and simultaneously for its binding to RNA. Bioinformatics analysis of the RHAU peptide sequence corroborated the experimental data, revealing that the N terminus of RHAU harbors a unique RNA-binding domain consisting of two abutting motifs: the G-rich region and the RSM. We have also shown that ATPase activity is involved in RHAU association with RNA and markedly influences the kinetics of RHAU recovery in SGs after photobleaching.
The list of proteins identified as components of SGs, co-localizing with known SG-associated proteins such as TIA-1, is still expanding. However, a method for isolating SGs to a significantly pure level has not yet been established, biochemical analysis of SGs is very difficult, and thus there is no consensus on their detailed structure and molecular composition. Furthermore it has not been systematically examined whether SGs induced by different stresses are qualitatively different. The presence of qualitatively different SGs is suggested by the fact that the recruitment of TTP to SGs is induced by carbonyl cyanide m-fluorophenylhydrazone but not by arsenite, whereas TIA-1 is recruited to SGs by both stimuli (39). Our results clearly demonstrate that RHAU is one of the SG-associated proteins and that its recruitment to SGs does not depend on the type of stress. Thus, RHAU appears to be a general SG component like TIA-1 or eIF3b. Although RHAU is recruited to SGs, it does not participate actively in SG formation as RNA interference-mediated down-regulation of RHAU had no effect on arsenite-induced SG formation (supplemental data). Similarly a further SG-associated protein, ZBP1, did not induce or inhibit SG formation when overexpressed or knocked down, respectively (7). Although ZBP1 does not modulate SG formation, its significance for SGs is attributed to its function in mRNA turnover regulation. Interestingly the stability of ZBP1 target mRNAs is not regulated by their recruitment to SGs but by their retention in SGs to prevent their rapid translocation and subsequent decay in processing bodies or by the exosome (7). Knowing that RHAU is a cis-acting factor involved in ARE-mediated decay of uPA mRNA and that its association with SG is regulated by RNA interaction, we speculate that RHAU acts in SGs in a manner similar to ZBP1.
Given that several RNA-binding proteins such as FMRP, TIA-1, G3BP, ZBP1, or Caprin-1 are mostly targeted to SGs via mRNA (7, 41, 43, 44), we performed CLIP, nitrocellulose filter binding assays, and bioinformatics analyses to elucidate whether the N terminus of RHAU, which is necessary and sufficient for RHAU accumulation in SGs, physically interacts with RNA. The analyses revealed that RHAU does associate with RNA both in vitro and in vivo and that only the N-terminal 105 amino acids harbor significant RNA binding activity. In addition, this region of RHAU contains the computationally predicted RNA-binding domain (3-75 aa) composed of a unique, previously unidentified RNA-binding motif. The domain is located in an unstructured and flexible sequence composed of two adjacent motifs, the G-rich region (10-50 aa) and the RSM (54-66 aa). The latter is highly conserved among RHAU orthologs, but it was not found in other proteins, suggesting that this motif confers a unique function on RHAU. Interestingly a potential RGG box motif (47-49 aa), which has been reported previously to be involved in the RNA interaction of several proteins, is present in the G-rich region of RHAU. For example, two RGG box sequences identified at the C terminus of the FMRP protein were shown to be necessary for an RNA and G-quadruplex interaction of FMRP and also for the induction of SG formation by EGFP-FMRP overexpression (42, 43). Moreover Caprin-1, an SG-associated protein, contains three RGG box motifs in its C terminus that are also important for its specific RNA interaction and SG induction (41). Our analyses of the RHAU N terminus shows that the deletion of the G-rich region together with the RGG box motif reduces RNA binding activity of RHAU and its accumulation in SGs (42%) suggesting that the RGG box motif may be involved in the RNA interaction and RHAU localization in SGs. However, the G-rich region with the RGG box motif alone was not sufficient for the recruitment of a fused β-galactosidase reporter protein to SGs (see Fig. 7B). Only the complete RNA-binding domain enables RHAU localization in SGs and RNA interaction, indicating that a domain other than the G-rich region takes part in RHAU recruitment to SGs. The detailed nature of the cooperation and the underlying mechanism between the G-rich region and the RSM remain interesting questions for the future.
According to the classification of putative RNA helicases by Abdelhaleem et al. (20), RHAU belongs to the DEAH box family containing a highly conserved C terminus next to the conserved helicase core region. Studies of DEAH proteins in yeast have shown that the specific function of each member is determined mainly by its N terminus and that helicase and ATPase activities are essential for their function during splicing. For example, Prp16, the ortholog of mammalian DHX38, localizes in the nucleus and recognizes the spliceosome through its N terminus (25). Its C terminus enhances spliceosome binding, whereas the helicase core region drives the process by binding and hydrolyzing ATP. Similarly Prp22 (DHX8), another splicing factor, requires the N-terminal domain for its ATPase and helicase function (24, 45). Our results show that the RHAU helicase follows the same principle because its specific localization in SGs and RNA interaction is governed by its N-terminal domain where the RNA-binding domain is located.
Importantly CLIP analysis confirmed that RHAU is an RNA-binding protein, which was suggested by our previous work when RHAU was first identified as an ARE-associated protein (26) but was not formally tested. Although we do not have direct evidence that RHAU-associated RNAs are mRNAs, the correlation between RHAU RNA binding activity and RHAU accumulation in SGs, which are aggregates of protein-mRNA complexes, strongly suggests this to be the case. Furthermore results showing that the abrogation of RHAU ATPase activity increases the amount of associated RNA (see Fig. 8A) are in good agreement with the accepted model of the closed and open conformation of DEX(H/D) box helicases. The structural and fluorescence resonance energy transfer information of complete DEAD box helicases has shown that the cooperative binding of ATP and RNA induces a closed conformation of helicases that is relaxed upon ATP hydrolysis, leading to the release of bound RNA (46-49). A similar effect has been reported for the Prp22 splicing factor that releases bound RNA upon ATP hydrolysis, thereby reducing its apparent RNA binding affinity (34). In this context, it is noteworthy that the ATPase-deficient mutant is excluded from the nucleus despite its strong RNA interaction indicating that it may associate mostly with mRNA and that the presence of RHAU in the nucleus is not essential for its localization to SGs. The nuclear exclusion of the ATPase-deficient mutant DAIH is not a consequence of it being trapped in the cytoplasm by a strong, irreversible interaction with RNA as we proposed previously (30). The N terminus-deleted ATPase-deficient mutant DAIH(105-1008), which showed markedly reduced RNA interaction and localization in SGs, remained mainly in the cytoplasm (Fig. 8D). Therefore, the nuclear-cytoplasmic translocation of RHAU is most probably regulated by its ATPase activity. A similar role for ATPase activity in nuclear export via the CRM1 protein was reported previously for the DEAD box helicase An3 (27).
Many SG-associated proteins are RNA-binding proteins involved in different intracellular processes mediated by or acting upon mRNA. Several of these proteins, including TIA-1, TTP, heterogeneous nuclear RNP A1, PCBP-2, MLN51, and G3BP (6, 9, 11-13), have been shown by FRAP analysis to rapidly shuttle into and out of SGs. This suggests that SGs are not static storage centers for untranslated mRNA but rather dynamic structures that sort individual transcripts for storage, reinitiation, or decay (4, 10). FRAP analysis also revealed that proteins behave with differing kinetics. For instance the PABP-1 protein exhibits a slower and only partial recovery (60%) 30 s after bleaching compared with TIA-1 (6, 11, 13). Like G3BP, wild-type RHAU shows very rapid shuttling into and out of SGs. In striking contrast, the kinetics of the ATPase-deficient mutant were different from RHAU and G3BP: green fluorescent protein signal recovery in SGs was as slow as for PABP-1. Thus, ATP hydrolysis affects the shuttling kinetics of RHAU into SGs. Furthermore our analysis showed that the FMRP protein is not a member of the group of rapidly shuttling RNA-binding proteins. Rather it exhibits slow shuttling like the Fas-activated serine/threonine kinase protein and, thus, may play a scaffolding role in SGs (6). Although RHAU exhibits the rapid mobility of G3BP, TIA-1, and TTP, it may be involved in RNP remodeling or their recruitment to SGs as suggested for TIA-1 and TTP (13).
With the detection of RHAU in SGs, we are the first to show the association of a DEAH box RNA helicase with SGs. Our FRAP analysis shows the striking difference in SG shuttling kinetics between fully active RHAU protein and its ATPase-deficient mutant, tendering the hypothesis that ATPase activity takes part in the dynamic remodeling of RNPs in SGs. Furthermore the sorting and remodeling of RNPs in SGs presumably requires energy, which may be provided by RNA helicases that utilize energy from ATP hydrolysis to rearrange inter- or intramolecular RNA structures or to dissociate protein-RNA complexes. To our knowledge, three DEAD box RNA helicases, rck/p54 (DDX6), DDX3, and eIF4A, have been shown to localize in SGs (32, 50, 51). Considering that various remodeling processes take place in these foci, it will be no surprise if the number of RNA helicases found associated with SGs is revealed to be much higher.
Supplementary Material
Acknowledgments
We thank J. Tanaka (University of the Ryukyus, Nishihara, Japan), D. Schmitz Rohmer, and B. A. Hemmings (Friedrich Miescher Institute, Basel, Switzerland) for kindly providing valuable materials and P. King, C. Du Roure (Friedrich Miescher Institute), P. Svoboda (Institute of Molecular Genetics AS CR, Prague, Czech Republic), and G. Stoecklin (German Cancer Research Center, Heidelberg, Germany) for critical comments on the manuscript. We also thank S. Thiry and S. Pauli (Friedrich Miescher Institute) and M. Yazdani and L. Jaskiewicz (Biozentrum, Basel, Switzerland) for excellent technical assistance and advice. This work was supported in part by CREST grant (to Y. Fujiki) from the Science and Technology Agency of Japan.
This work was supported in part by CREST grant (to Y. F.) from The Science and Technology Agency of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: SG, stress granule; RNP, ribonucleoprotein; FRAP, fluorescence recovery after photobleaching; CCCP, carbonyl cyanide m-chlorophenylhydrazone; CLIP, cross-linking immunoprecipitation; ARE, AU-rich element; RSM, RHAU-specific motif; uPA, urokinase-type plasminogen activator; PABP-1, poly(A)-binding protein 1; ZBP1, zipcode-binding protein 1; aa, amino acids; TTP, tristetraprolin; HuR, ELAV-like protein 1 (Hu-antigen R); FMRP, Fragile X mental retardation; PBS, phosphate-buffered saline; PIPES, 1,4-piperazinediethanesulfonic acid; DAPI, 4′,6-diamidino-2-phenylindole; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; GST, glutathione S-transferase; RISP, RNA-Interaction Site Prediction; ROI, regions of interest; EGFP, enhanced green fluorescent protein; Nter, N terminus; HCR, helicase core region; Cter, C terminus; WT, full-length RHAU (wild type); β-gal, β-galactosidase.
F. Iwamoto and Y. Fujiki, unpublished data.
References
- 1.Garneau, N. L., Wilusz, J., and Wilusz, C. J. (2007) Nat. Rev. Mol. Cell Biol. 8 113-126 [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., and Kedersha, N. (2002) J. Cell Sci. 115 3227-3234 [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P., and Kedersha, N. (2002) Cell Stress Chaperones 7 213-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, P., and Kedersha, N. (2006) J. Cell Biol. 172 803-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., and Anderson, P. (2002) Mol. Biol. Cell 13 195-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., Scheuner, D., Kaufman, R. J., Golan, D. E., and Anderson, P. (2005) J. Cell Biol. 169 871-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stohr, N., Lederer, M., Reinke, C., Meyer, S., Hatzfeld, M., Singer, R. H., and Huttelmaier, S. (2006) J. Cell Biol. 175 527-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999) J. Cell Biol. 147 1431-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guil, S., Long, J. C., and Caceres, J. F. (2006) Mol. Cell. Biol. 26 5744-5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson, P., and Kedersha, N. (2008) Trends Biochem. Sci. 33 141-150 [DOI] [PubMed] [Google Scholar]
- 11.Baguet, A., Degot, S., Cougot, N., Bertrand, E., Chenard, M. P., Wendling, C., Kessler, P., Le Hir, H., Rio, M. C., and Tomasetto, C. (2007) J. Cell Sci. 120 2774-2784 [DOI] [PubMed] [Google Scholar]
- 12.Fujimura, K., Kano, F., and Murata, M. (2008) RNA 14 425-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., Golan, D. E., and Anderson, P. (2000) J. Cell Biol. 151 1257-1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimura, K., Kano, F., and Murata, M. (2008) Exp. Cell Res. 314 543-553 [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss, G., Kim, V. N., and Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3 195-205 [DOI] [PubMed] [Google Scholar]
- 16.Linder, P. (2006) Nucleic Acids Res. 34 4168-4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jankowsky, E., and Jankowsky, A. (2000) Nucleic Acids Res. 28 333-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jankowsky, E., Gross, C. H., Shuman, S., and Pyle, A. M. (2001) Science 291 121-125 [DOI] [PubMed] [Google Scholar]
- 19.Bleichert, F., and Baserga, S. J. (2007) Mol. Cell 27 339-352 [DOI] [PubMed] [Google Scholar]
- 20.Abdelhaleem, M., Maltais, L., and Wain, H. (2003) Genomics 81 618-622 [DOI] [PubMed] [Google Scholar]
- 21.Jankowsky, E., and Bowers, H. (2006) Nucleic Acids Res. 34 4181-4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohr, G., Del Campo, M., Mohr, S., Yang, Q., Jia, H., Jankowsky, E., and Lambowitz, A. M. (2008) J. Mol. Biol. 375 1344-1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valgardsdottir, R., and Prydz, H. (2003) J. Biol. Chem. 278 21146-21154 [DOI] [PubMed] [Google Scholar]
- 24.Schneider, S., and Schwer, B. (2001) J. Biol. Chem. 276 21184-21191 [DOI] [PubMed] [Google Scholar]
- 25.Wang, Y., and Guthrie, C. (1998) RNA 4 1216-1229 [PMC free article] [PubMed] [Google Scholar]
- 26.Tran, H., Schilling, M., Wirbelauer, C., Hess, D., and Nagamine, Y. (2004) Mol. Cell 13 101-111 [DOI] [PubMed] [Google Scholar]
- 27.Askjaer, P., Rosendahl, R., and Kjems, J. (2000) J. Biol. Chem. 275 11561-11568 [DOI] [PubMed] [Google Scholar]
- 28.Schmitt, C., von Kobbe, C., Bachi, A., Pante, N., Rodrigues, J. P., Boscheron, C., Rigaut, G., Wilm, M., Seraphin, B., Carmo-Fonseca, M., and Izaurralde, E. (1999) EMBO J. 18 4332-4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner, J. D., Jankowsky, E., Company, M., Pyle, A. M., and Abelson, J. N. (1998) EMBO J. 17 2926-2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto, F., Stadler, M., Chalupnikova, K., Oakeley, E., and Nagamine, Y. (2008) Exp. Cell Res. 314 1378-1391 [DOI] [PubMed] [Google Scholar]
- 31.Vaughn, J. P., Creacy, S. D., Routh, E. D., Joyner-Butt, C., Jenkins, G. S., Pauli, S., Nagamine, Y., and Akman, S. A. (2005) J. Biol. Chem. 280 38117-38120 [DOI] [PubMed] [Google Scholar]
- 32.Mazroui, R., Sukarieh, R., Bordeleau, M. E., Kaufman, R. J., Northcote, P., Tanaka, J., Gallouzi, I., and Pelletier, J. (2006) Mol. Biol. Cell 17 4212-4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ule, J., Jensen, K., Mele, A., and Darnell, R. B. (2005) Methods 37 376-386 [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, N., and Schwer, B. (2005) Biochemistry 44 9795-9803 [DOI] [PubMed] [Google Scholar]
- 35.Terribilini, M., Sander, J. D., Lee, J. H., Zaback, P., Jernigan, R. L., Honavar, V., and Dobbs, D. (2007) Nucleic Acids Res. 35 W578-W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong, J., Jiang, P., and Lu, Z. H. (2008) Comput. Methods Programs Biomed. 90 148-153 [DOI] [PubMed] [Google Scholar]
- 37.Jeong, E., Chung, I. F., and Miyano, S. (2004) Genome Inform. 15 105-116 [PubMed] [Google Scholar]
- 38.Do, C. B., Mahabhashyam, M. S., Brudno, M., and Batzoglou, S. (2005) Genome Res. 15 330-340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecklin, G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W. F., Blackwell, T. K., and Anderson, P. (2004) EMBO J. 23 1313-1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallouzi, I. E., Brennan, C. M., Stenberg, M. G., Swanson, M. S., Eversole, A., Maizels, N., and Steitz, J. A. (2000) Proc. Natl. Acad. Sci. U. S. A 97 3073-3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon, S., Xu, Y., Wang, B., David, M. D., Schubert, P., Kennedy, D., and Schrader, J. W. (2007) Mol. Cell. Biol. 27 2324-2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos, A., Hollingworth, D., and Pastore, A. (2003) RNA 9 1198-1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazroui, R., Huot, M. E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E. W. (2002) Hum. Mol. Genet. 11 3007-3017 [DOI] [PubMed] [Google Scholar]
- 44.Tourriere, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J. M., Bertrand, E., and Tazi, J. (2003) J. Cell Biol. 160 823-831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Schneider, S., Hotz, H. R., and Schwer, B. (2002) J. Biol. Chem. 277 15452-15458 [DOI] [PubMed] [Google Scholar]
- 46.Theissen, B., Karow, A. R., Kohler, J., Gubaev, A., and Klostermeier, D. (2008) Proc. Natl. Acad. Sci. U. S. A 105 548-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caruthers, J. M., Johnson, E. R., and McKay, D. B. (2000) Proc. Natl. Acad. Sci. U. S. A 97 13080-13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng, Z., Coller, J., Parker, R., and Song, H. (2005) RNA 11 1258-1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi, H., Cordin, O., Minder, C. M., Linder, P., and Xu, R. M. (2004) Proc. Natl. Acad. Sci. U. S. A 101 17628-17633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilczynska, A., Aigueperse, C., Kress, M., Dautry, F., and Weil, D. (2005) J. Cell Sci. 118 981-992 [DOI] [PubMed] [Google Scholar]
- 51.Lai, M. C., Lee, Y. H., and Tarn, W. Y. (2008) Mol. Biol. Cell 19 3847-3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.