SUMMARY
The molecular basis for p53-mediated tumor suppression remains unclear. Here, to elucidate mechanisms of p53 tumor suppression, we use knock-in mice expressing an allelic series of p53 transcriptional activation mutants. Microarray analysis reveals that one mutant, p5325,26, is severely compromised for transactivation of most p53 target genes and cannot induce G1-arrest or apoptosis in response to acute DNA damage. Surprisingly, p5325,26 retains robust activity in senescence and tumor suppression, indicating that efficient transactivation of the majority of known p53 targets is dispensable for these pathways. In contrast, the transactivation-dead p5325,26,53,54 mutant cannot induce senescence or inhibit tumorigenesis, like p53-nullizygosity. Thus, p53 transactivation is essential for tumor suppression, but, intriguingly, in association with a small set of novel p53 target genes. Together, our studies distinguish the p53 transcriptional programs involved in acute DNA-damage responses and tumor suppression -- a critical goal for designing therapeutics that block p53-dependent side effects of chemotherapy without compromising p53 tumor suppression.
INTRODUCTION
The facts that over half of all human cancers sustain mutations in the p53 tumor suppressor gene and that p53 null mice display a dramatic early-onset, completely penetrant cancer predisposition together underscore the fundamental importance of p53 for tumor suppression (Kenzelmann Broz and Attardi, 2010; Vogelstein et al., 2000). p53 serves as a cellular stress sentinel, responding to myriad stresses by restricting cellular expansion under unfavorable conditions (Vousden and Prives, 2009). The best-characterized p53 functions are in inducing cell cycle arrest or apoptosis in response to acute DNA damage signals. The ability of p53 to eliminate cells that have encountered acute genotoxic stress is thought to be an ancestral function, as this response is conserved through lower eukaryotes, including D.melanogaster and C.elegans, where it is critical for culling damaged cells to preserve germline and tissue integrity (Lu and Abrams, 2006). In higher eukaryotes, oncogene expression can also activate p53, leading to cellular senescence or apoptosis as safeguards against neoplasia (Vousden and Prives, 2009).
The extent to which the ability of p53 to respond to DNA damage is involved in mediating tumor suppression has been controversial. Analysis of early human neoplastic lesions has revealed molecular marks of activated DNA damage components, leading to a model whereby oncogene-induced hyperproliferation results in replication fork collapse, DNA double-strand break formation, checkpoint kinase activation, and p53 induction to ultimately impose a barrier to tumor development (Halazonetis et al., 2008). However, studies in mouse models of DNA damage-induced lymphomas and fibrosarcomas have suggested that to serve as a tumor suppressor, p53 responds not to acute DNA damage but rather to oncogene-induced expression of the p19ARF tumor suppressor, which directly activates p53 through sequestration and inhibition of its negative regulator Mdm2 (Christophorou et al., 2006; Efeyan et al., 2006). A clearer understanding of the role of DNA damage-triggered, p53-induced cell cycle arrest or apoptosis in tumor suppression would come from elucidating the underlying molecular mechanisms for p53 action in the contexts of acute DNA damage versus tumor suppression. Moreover, illuminating any distinct downstream aspects to these pathways has critical therapeutic implications, as many of the deleterious side effects of genotoxic chemotherapies result from p53-driven apoptosis in radiosensitive tissues, and therefore identifying strategies to mitigate these side effects without compromising tumor suppressor function throughout the organism would be broadly valuable for cancer therapy (Gudkov and Komarova, 2003).
The molecular underpinnings for p53 action in tumor suppression have remained elusive. p53 serves as a transcriptional activator of numerous target genes, but several other biochemical activities have also been ascribed to p53 (Green and Kroemer, 2009; Vousden and Prives, 2009). Moreover, no p53 target gene knockout mouse strain recapitulates the dramatic cancer predisposition of p53 null mice, illustrating our incomplete understanding of p53 networks involved in tumor suppression and suggesting that other pathways could be involved (Lozano and Zambetti, 2005). Defining the role of transactivation in tumor suppression by p53 is complicated by the fact that p53 contains two distinct transcriptional activation domains (comprising residues 1–40 and 40–83, respectively), whose discrete functions and relative contributions to p53 function are not understood (Candau et al., 1997; Venot et al., 1999; Zhu et al., 1998). Parsing out the specific roles of these two domains for p53 function in vivo could reveal distinct transcriptional requirements for acute DNA damage responses and tumor suppression and lead to the discovery of p53 target genes principally important for tumor suppression.
Here, we investigate the mechanism of p53-mediated tumor suppression and its relationship to acute DNA damage responses by deciphering the p53 transactivation requirements for function in these contexts. We generate a series of transactivation domain (TAD) mutant knock-in mouse strains, with alterations in the first, second, and both TADs. Knock-in mice, in which the mutant genes are expressed from the native p53 promoter, uniquely enable the study of both primary cells ex vivo and tumor development in the physiological context of the organism. Intriguingly, our studies reveal that different p53 transcriptional activation requirements, associated with different target gene expression programs, are important in the settings of acute genotoxic stress and oncogenic stimuli. Our findings thereby provide genetic evidence that the mechanisms through which p53 engages responses to these signals are different and lend fundamental new insight into the networks involved in p53-mediated tumor suppression.
RESULTS
Generation of p53 TAD mutant knock-in mouse strains
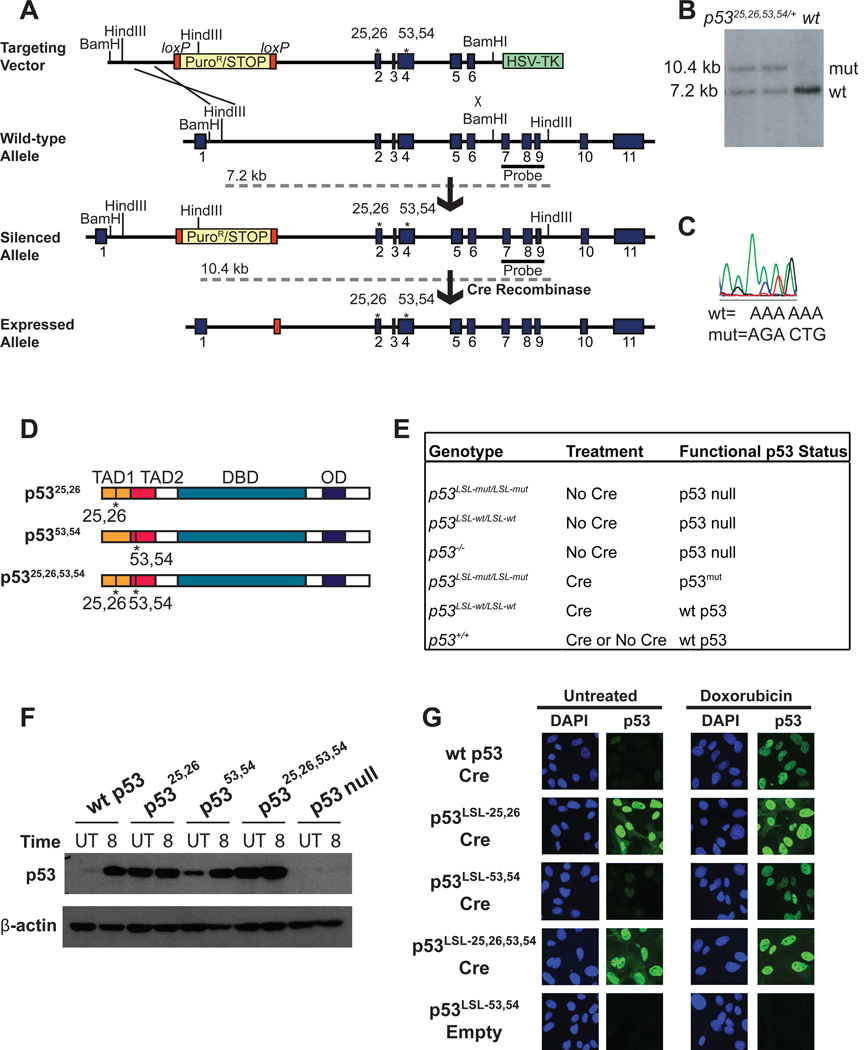
To decipher the discrete roles of the two p53 TADs in DNA damage responses and tumor suppression in vivo, we generated a panel of p53 mutant knock-in mouse strains with alterations in the first (p5325,26), second (p5353,54), or both TADs (p5325,26,53,54). L25Q;W26S knock-in mice were generated previously, and analysis of a small set of p53 target genes in mouse embryo fibroblasts (MEFs) derived from these mice demonstrated compromised transactivation of these genes, except Bax (Johnson et al., 2005). Here we have generated mouse strains bearing either the F53Q;F54S mutations found to incapacitate the second p53 TAD in vitro, or mutations in both TADs (L25Q;W26S;F53Q;F54S; Fig. 1A–D; Candau et al., 1997; Venot et al., 1999; Zhu et al., 1998). All alleles carried a transcriptional stop element flanked by LoxP sites (Lox-Stop-Lox or LSL) in the first p53 intron to allow regulatable p53 expression (Fig. 1A–D).
Fig 1. Generation of p53 TAD mutant knock-in mice.
(A) Targeting scheme used to generate knock-in mice, with the p5325,26,53,54 mutant as an example. Mutant p53 expression from targeted alleles is silenced until Cre introduction allows for excision of the stop element. Dotted grey lines indicate the sizes of the fragments generated from each allele upon HindIII digestion. (B) Southern blot showing 2 correctly targeted, heterozygous ES cell clones compared to a wild-type cell line. Genomic DNA was digested with HindIII and the Southern blot probed with the 3’ external probe indicated in (A). (C) Sequencing analysis of the reverse complement confirms the presence of point mutations in properly targeted cell lines. (D) Schematic of p5325,26, p5353,54, and p5325,26,53,54 proteins showing the transactivation domains (TADs), DNA binding domain (DBD), and oligomerization domain (OD). (E) Table summarizing the genotype, treatment, and ultimate functional p53 status of samples used throughout the manuscript. LSL-mut denotes any of the Lox-Stop-Lox p53 TAD mutants. (F) Western blot analysis for p53 in wild-type or homozygous p53LSL-mut MEFs transduced with Ad-Cre or Ad-empty (indicated as p53 null), either left untreated (UT) or treated for 8 hrs with 0.2 µg/ml dox (doxorubicin) (8). β-actin served as a loading control. Comparable, high efficiency (>90% of cells) of Cre recombination was confirmed by immunostaining for p53. (G) Immunofluorescence for p53 in wild-type or homozygous p53LSL-mut MEFs transduced with Ad-Cre or Ad-empty. Cells were left untreated (left) or treated with 0.2 µg/ml dox to stabilize p53 (right). Nuclei were stained with DAPI.
To initially characterize this set of p53 TAD mutant proteins, we cultured homozygous p53LSL-mut MEFs, infected them with adenoviruses expressing Cre recombinase (Ad-Cre), and assayed p53 protein levels and localization (Fig 1E–G). Using this approach we typically observed over 90% p53 positivity, and in all experiments, we verified widespread p53 expression in the population being examined. Furthermore, MEFs expressed p53 only after Cre introduction, indicating effective silencing of the locus by the LSL cassette (Fig. 1G), and allowing us to use MEFs infected with empty adenoviruses (Ad-empty) as convenient p53 null controls. Although basal p5325,26 and p5325,26,53,54 protein levels were elevated relative to wild-type (wt) p53 levels because mutation of residues 25/26 inhibits binding of the Mdm2 ubiquitin ligase (Lin et al., 1994), protein levels were in a physiological range, accumulating to levels only slightly higher than those of wild-type p53 after DNA damage (Fig. 1F, G). Additionally, p5353,54 basal levels were slightly increased relative to wild-type p53, consistent with the reported contribution of residues 53/54 to the p53-Mdm2 interaction (Chi et al., 2005). All mutant proteins displayed clear nuclear localization (Fig. 1G).
p53 mutants display a range of transcriptional activation capacities
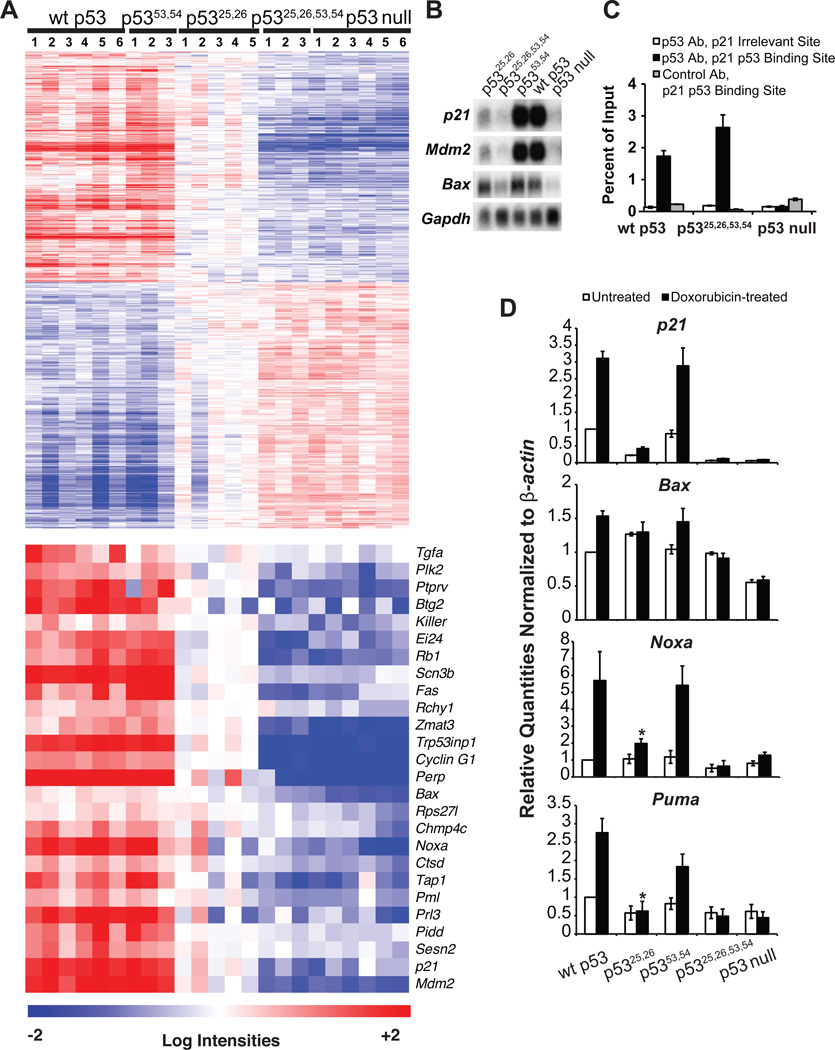
To delineate the relative contributions of the respective TADs to overall p53 transactivation function, we examined the activity of the p53 mutants both on a genome-wide scale and quantitatively at select target genes. Initially, we performed gene expression profiling experiments using a model for oncogenic Hras (HrasV12)-driven, p53-dependent senescence in MEFs (Serrano et al., 1997). By comparing HrasV12;p53 wild-type and HrasV12;p53 null MEFs using Significance Analysis of Microarrays (Tusher et al., 2001), we defined a group of p53-dependent genes, including numerous established p53 targets such as p21 and Mdm2 (Fig. 2A). Expression of these genes in the mutant MEFs was compared by heat map analysis and northern blotting to reveal notable differences in the activities of the three mutants. First, expression profiles of p5353,54/53,54 cells resembled those of cells expressing wild-type p53, suggesting that p5353,54 retains p53 transactivation function (Fig. 2A, B). In contrast, analysis of p5325,26/25,26 MEFs revealed gene expression profiles intermediate between those observed in wild-type and p53 null cells (Fig. 2A). To elaborate on this pattern, we focused on individual established p53-inducible genes, to ensure analysis of direct p53 targets. This analysis demonstrated that p5325,26 merely drives extremely low-level expression of p21, Noxa, and Puma but induces efficient expression of Bax comparable to wild-type p53, suggesting that p5325,26 is severely impaired for transactivation of most but not all p53 target genes (Fig. 2B, D). Finally, the expression profile of p5325,26,53,54/25,26,53,54 MEFs closely resembled that of p53 null MEFs, suggesting that mutation of both TADs abolishes transactivation activity (Fig. 2A, B). Chromatin immunoprecipitation (ChIP) demonstrated that p53 TAD mutants bind p53 target gene promoters in cells, indicating that these mutations selectively disrupt transactivation function but not chromatin association (Fig. 2C, Fig. S1A). qRT-PCR on DNA damage-treated MEFs echoed the microarray results, but emphasized how minimally p5325,26 activated targets relative to p53 null MEFs and further revealed that p5325,26,53,54 retained a slight capacity to activate Bax (Fig. 2D; Fig. S1B). Together, our analyses identify an allelic series of p53 transactivation mutants that can be used to elucidate the contribution of different extents of transactivation to p53 biological function downstream of acute genotoxic and oncogenic stress.
Fig. 2. Analysis of the transcriptional activation potential of the p53 TAD mutants.
(A) (Upper) Heat map defining transactivation capacity of p53 TAD mutants using a p53-dependent gene set identified by microarray analysis through comparison of 6 HrasV12;p53 wild-type to 6 HrasV12;p53 null MEF samples. Columns indicate independent MEF lines. (Lower) Heat map examining p53 mutant activity on confirmed, direct p53 target genes (Brady and Attardi, 2010; Riley et al., 2008). (B) Northern blot using RNA samples from (A). (C) ChIP showing that p5325,26,53,54 binds to the p53 response element in the p21 promoter and not to an irrelevant region 3 kb downstream. p16 antibody serves as a negative control. (D) qRT-PCR of RNA from MEFs either untreated (white bars) or treated with 0.2 µg/ml dox for 8 hrs (black bars). Graphs indicate averages +/− SEM of quantities normalized first to β-actin and then to wild-type untreated sample values from 3 independent MEF lines. * indicates non-significant difference of p > 0.05 v. p53 null. See also Figure S1.
DNA damage-induced cell cycle arrest and apoptosis require full p53 transcriptional activity
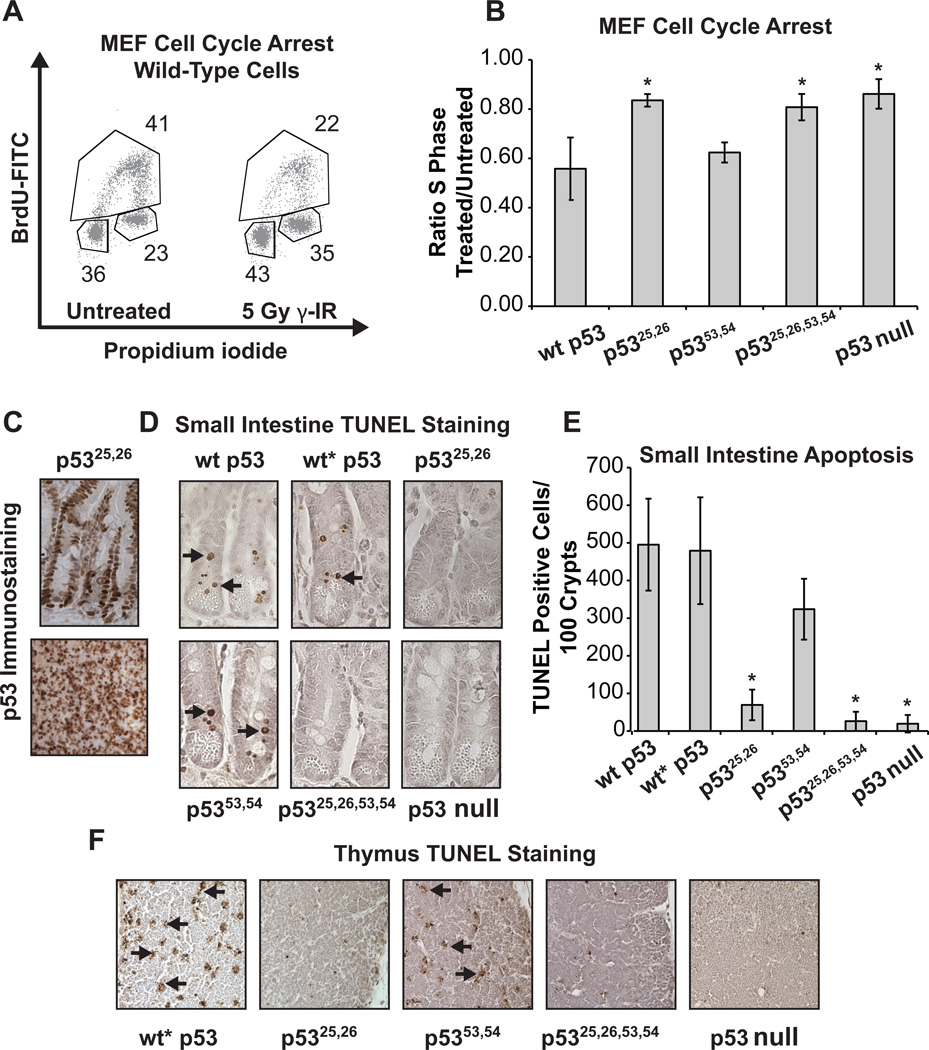
In the face of acute genotoxic stress, p53 triggers either cell cycle arrest or apoptosis to limit the propagation of damaged cells. We first defined transactivation requirements for p53-dependent G1 cell cycle arrest using a classical MEF model in which cell cycle profiles are examined 18 hrs after 5 Gy γ-irradiation (Kastan et al., 1992). While wild-type and p5353,54/53,54 cells arrested efficiently, both p5325,26/25,26 and p5325,26,53,54/25,26,53,54 cells failed to arrest, indicating that activity of the first, but not the second, p53 TAD is essential for DNA damage-induced G1 arrest (Fig. 3A, B). To assess p53 transactivation function in DNA damage-triggered apoptosis, we examined p53-dependent apoptosis in radiosensitive tissues in vivo upon exposure to ionizing radiation (Lowe et al., 1993; Merritt et al., 1994). Using Rosa26CreERT2 mice allowed widespread p53 expression in the small intestine and less efficient expression in the thymus upon tamoxifen administration (Fig. 3C; Ventura et al., 2007). Mice homozygous for each p53 allele were exposed to 5 Gy of whole-body ionizing radiation, and apoptosis in the small intestine and thymus was examined 6 hrs later. We found that p5325,26 failed to promote apoptosis in vivo, consistent with our previous analyses examining doxorubicin-induced apoptosis in E1A-MEFs and studies in cultured lymphocytes (Chao et al., 2000; Johnson et al., 2005; Gaidarenko and Xu 2009; Fig. 3D–F). Moreover, compound mutation of both TADs did not alter this phenotype, as p5325,26,53,54 also failed to induce apoptosis. In contrast, mutation of the second TAD alone did not greatly affect p53-dependent apoptosis. Together, these experiments underscore the vital roles of the first TAD and robust transactivation for p53 to promote responses to acute DNA damage and further reveal that the second TAD plays little to no role in these responses.
Fig. 3. The first p53 TAD plays the predominant role in acute DNA damage responses.
(A) Representative propidium iodide and BrdU flow cytometry data from asynchronous wild-type MEFs showing G1 arrest response after γ-irradiation. (B) Average S-phase ratio of γ-irradiation-treated/untreated MEFs. Wild-type, p53LSL-25,26/LSL-25,26, p53LSL-53,54/LSL-53,54, and p53LSL-25,26,53,54/LSL-25,26,53,54 cells were infected with Ad-Cre or Ad-empty and irradiated. Averages of 3–5 experiments +/− SD are graphed. * indicates a significant difference of p < 0.001 v. p53 wt, one-way ANOVA with Bonferroni post tests. (C) p53 immunostaining in p5325,26-expressing small intestine (top) and thymus (bottom) from Rosa26CreER;p5325,26/25,26 mice treated with tamoxifen. (D) Representative TUNEL-staining images showing apoptotic cells (arrows) in small intestines. Wild-type p53, p53LSL-wt/LSL-wt (wt*), p53LSL-25,26/LSL-25,26, p53LSL-53,54/LSL-53,54, and p53LSL-25,26,53,54/LSL-25,26,53,54 mice carrying the Rosa26CreERT2 allele were treated with tamoxifen and irradiated, then tissues were collected 6 hrs later. Cre-negative mice treated with tamoxifen served as p53 null controls. (E) Average number of TUNEL positive cells per 100 crypts of the small intestine +/− SD from at least 4 mice per genotype. * indicates a significant difference of p < 0.001 v. p53 wt, one-way ANOVA with Bonferroni post tests. (F) Representative TUNEL staining of thymi, with arrows indicating apoptosis.
Full p53 transactivation function is dispensable for cellular senescence
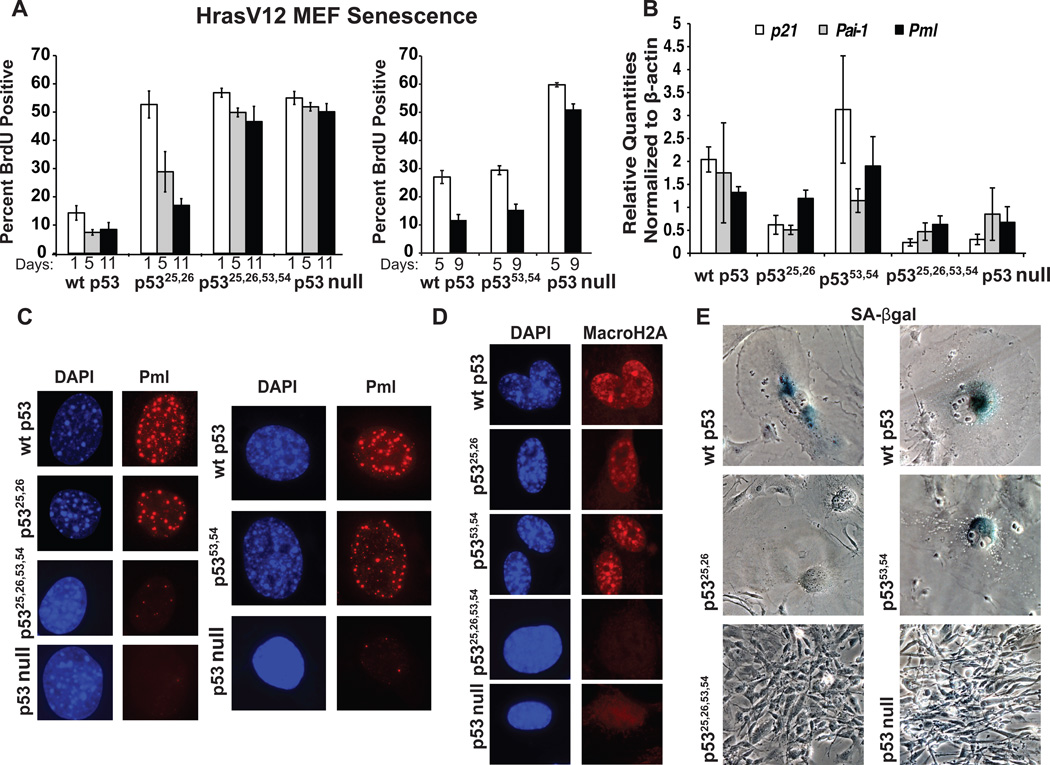
To elucidate p53 transactivation requirements in the context of oncogenic signals, we examined the allelic series of p53 mutants in HrasV12-induced cellular senescence (Serrano et al., 1997). HrasV12 was expressed in MEFs, and cell cycle arrest was assessed by BrdU labeling. As expected, few cells with wild-type p53 incorporated BrdU, whereas those lacking p53 proliferated efficiently upon HrasV12 expression (Fig. 4A). Interestingly, both HrasV12;p5325,26/25,26 and HrasV12;p5353,54/53,54 MEFs underwent a proliferative arrest with time and displayed hallmarks of cellular senescence, including flattened, enlarged morphologies and both transcriptional induction of Pml and widespread Pml nuclear body accumulation (de Stanchina et al., 2004; Ferbeyre et al., 2000; Fig. 4A–C, E, Fig. S2A, B). Additionally, all arresting cells displayed strong nuclear staining for the histone variant macroH2A, another indicator of senescence (Kennedy et al., 2010; Fig. 4D). Unlike their wild-type and p5353,54/53,54 counterparts, however, senescent p5325,26/25,26 cells were negative for SA-β-galactosidase, another common senescence marker (Dimri et al., 1995), suggesting that full p53 transactivation potential is essential for this activity (Fig. 4E). Thus, despite being highly impaired for transactivation of most p53 target genes, including the senescence-relevant targets p21 and Pai-1 (Brown et al., 1997; Kortlever et al., 2006; Fig. 4B), p5325,26 can induce senescence in the context of oncogenic signals. This surprising finding indicates that full p53 transactivation potential is dispensable for p53 to promote senescence. While either single TAD mutant alone was able to induce senescence, mutation of both domains completely reversed this phenotype, as indicated by the high level of proliferation throughout the time course, the paucity of Pml bodies, and the absence of macroH2A staining in HrasV12;p5325,26,53,54/25,26,53,54 cells, mimicking HrasV12;p53−/− MEFs (Fig. 4A, C, D). The observation that p5325,26,53,54 cannot engage the senescence program indicates that, although the full p53 transactivation program is dispensable, some limited level of p53 transactivation is required for promoting senescence. Together, our findings suggest fundamentally distinct requirements for transcriptional activation between the acute DNA damage and senescence responses: full p53 transactivation is paramount for proper p53 action downstream of genotoxic stress, in both G1 checkpoint function and apoptosis, whereas more selective p53 transactivation function suffices for senescence.
Fig. 4. p5325,26, but not p5325,26,53,54, induces cellular senescence in HrasV12 MEFs.
(A) Average percentages of BrdU-positive cells over time. Left: HrasV12-expressing p53+/+, p53LSL-25,26/LSL-25,26, and p53LSL-25,26,53,54/LSL-25,26,53,54 MEFs were infected with Ad-Cre and cultured. Total percentages of p53-positive cells displaying BrdU positivity are shown. Right: HrasV12-expressing p53+/+, p5353,54/53,54, and p53LSL-53,54/LSL-53,54 (p53 null) MEFs were cultured and analyzed for BrdU incorporation. The timelines under the graphs denote either days post-Ad-Cre (performed 4 days after HrasV12 transduction; left) or days after HrasV12 transduction (right, where Ad-Cre is not used). Averages +/− SEM of at least 3 cell lines per genotype are graphed. (B) qRT-PCR analysis of the expression of the senescence-related target genes p21, Pai-1, and Pml in HrasV12 MEFs (C) Pml immunostaining of HrasV12 MEFs at the final timepoints of senescence assays. DAPI stains nuclei. (D) MacroH2A staining of HrasV12 MEFs at the final timepoints of senescence assays. DAPI stains nuclei. (E) Phase contrast images of SA-β-galactosidase staining and morphology of HrasV12 MEFs at the final timepoints of senescence assays. See also Figure S2.
p5325,26 and p5353,54 are potent tumor suppressors
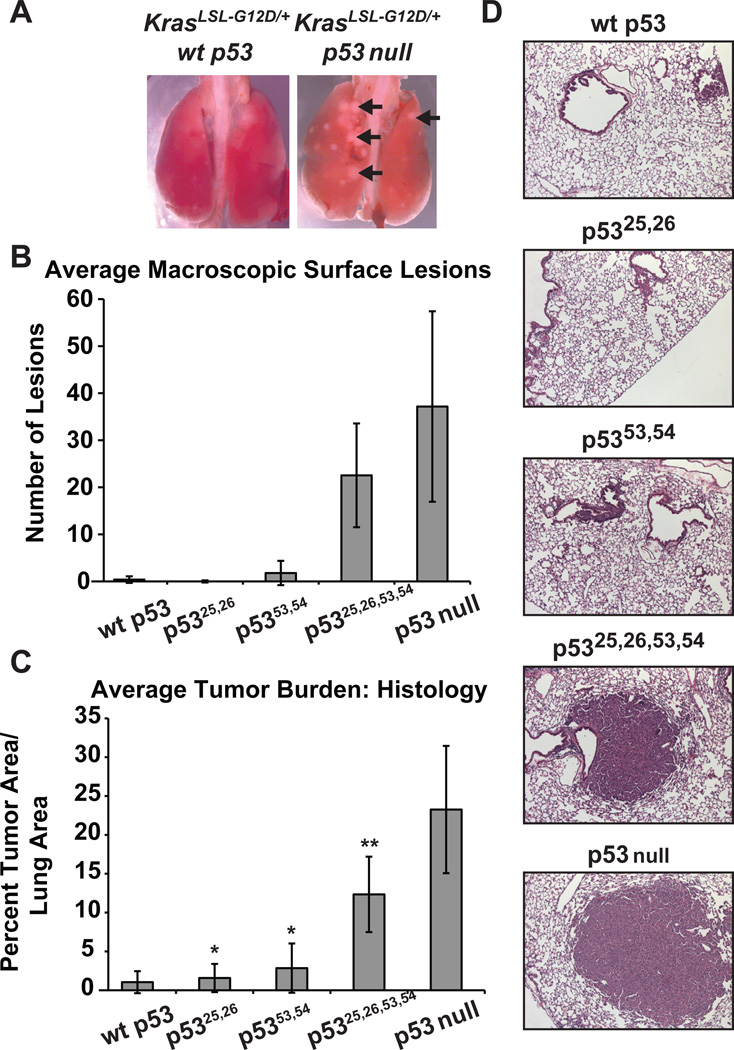
To identify the mechanisms of p53 action downstream of oncogenic signals in vivo, we interrogated the contribution of p53 transactivation to tumor suppression. We employed a model for human non-small cell lung cancer (NSCLC) driven by expression of oncogenic KrasG12D from its endogenous promoter after Cre-mediated excision of an upstream Lox-Stop-Lox element (Jackson et al., 2001). p53 loss promotes the progression of these Kras-driven lung tumors to more advanced lesions, making this an optimal system to query the mechanism of p53 action (Jackson et al., 2001; Johnson et al., 2001). Cohorts of KrasLSL-G12D/+ mice homozygous for the different p53 alleles were subjected to intranasal Ad-Cre instillation, and tumor burden was assessed 12 weeks later. KrasG12D/+;p53−/− mice exhibited visible tumors studding the lung surfaces, while lungs from KrasG12D/+;p53+/+ mice appeared grossly normal (Fig. 5A, B). Additionally, the average tumor burden was dramatically increased in KrasG12D/+;p53−/− mice compared to KrasG12D/+;p53+/+ mice (Fig. 5C, D). Strikingly, KrasG12D/+;p5325,26/25,26 and KrasG12D/+;p5353,54/53,54 lungs were macroscopically normal with minimal tumor burden, comparable to KrasG12D/+;p53+/+ lungs (Fig. 5B–D). In marked contrast, KrasG12D/+;p5325,26,53,54/25,26,53,54 lungs exhibited many macroscopic lesions and significantly greater tumor burdens than KrasG12D/+;p53+/+ lungs, indicative of a failure to suppress tumor growth. p5325,26,53,54 did, however, display minimal tumor suppressor activity relative to p53-deficiency, potentially reflecting residual transactivation of genes such as Bax or another p53 activity. Thus, p5325,26 and p5353,54 retain the ability to suppress KrasG12D-induced lung tumor growth, but combined mutation of both transcriptional activation domains largely reverses tumor suppressor activity. The remarkable observation that p5325,26 can suppress tumor growth, despite an inability to efficiently activate most known p53 target genes, including p21, Puma, and Noxa, suggests that full transactivation is dispensable for p53 tumor suppressor function - a notion bolstered by our studies in several tumor models of different lineages (data not shown). Collectively, our studies unveil an unexpected contribution of both p53 TADs to tumor suppression and provide an unequivocal demonstration that transcriptional activation function is critical for p53 tumor suppression in vivo, albeit in a selective fashion.
Fig. 5. p5325,26 and p5353,54, but not p5325,26,53,54, are potent tumor suppressors.
KrasLSL-G12D/+ mice with p53+/+ (n=9), p53LSL-25,26/LSL-25,26 (n=6), p53LSL-53,54/LSL-53,54 (n=12), p53LSL-25,26,53,54/LSL-25,26,53,54 (n=13), or p53−/− (n=6) status were infected intranasally with Ad-Cre at 6–8 weeks, and lungs were collected 12 weeks later. (A) Whole mount images of lungs from KrasG12D;p53+/+ and KrasG12D;p53−/− mice, with arrows indicating tumors. (B) Average number of macroscopic lung tumors +/− SD in mice of all genotypes. (C) Average tumor burden, calculated as the ratio of total tumor area to total lung area on H&E-stained sections, +/− SD in mice of all genotypes. * indicates no significant difference, p > 0.05 v. p53 wild-type, ** indicates significant difference of p < 0.001 v. p53 wt, one-way ANOVA Bonferroni post tests. (D) Representative histological sections from lungs of each genotype.
Analysis of p53 TAD mutants uncovers target genes associated with human cancer
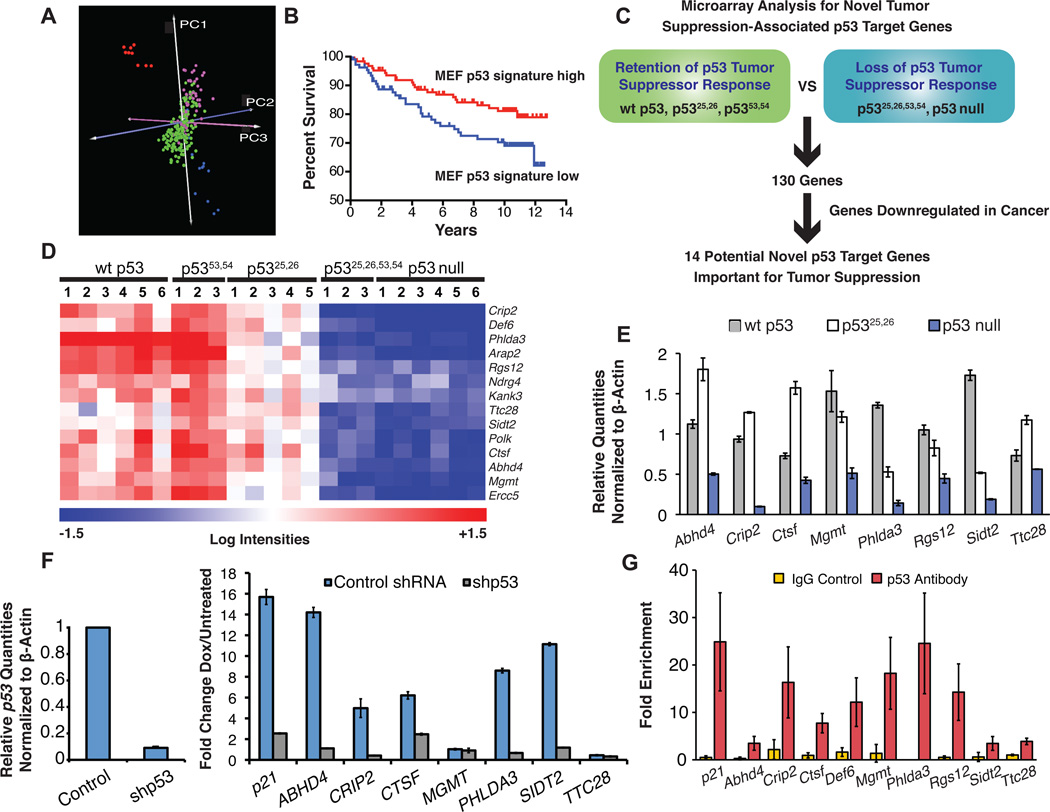
The striking finding that p5325,26 can efficiently activate only a small subset of all p53 target genes yet it retains full biological activity in tumor suppression suggests that target genes effectively induced by p5325,26 may encode components important for mediating p53 function in tumor suppression. Using our gene expression data (Figure 2A), we sought to identify genes robustly activated by p5325,26 that could account for its biological activity. Toward this end, we identified genes efficiently induced in cells with a wild-type p53 tumor suppressor phenotype (wt p53, p5325,26, and p5353,54) relative to cells failing to display a tumor suppressor phenotype (p5325,26,53,54 and p53 null) and obtained a list of 130 genes, the vast majority of which are not known p53 target genes. We first sought to determine if these p53-dependent genes had significance in human cancer by using this signature to predict the p53 status of a set of human breast cancer samples of known p53 sequence (Miller et al., 2005). Strikingly, the MEF-derived gene list was able to distinguish p53 wild-type from p53 mutant human breast cancers with high efficiency (80% properly segregated using the k-nearest neighbor algorithm; Fig. 6A). Moreover, our p53 signature also effectively stratified these breast cancer samples according to tumor grade and patient survival, revealing a strong correlation between our expression profile reflecting loss of p53 tumor suppressor activity and both higher-grade tumors and reduced survival (data not shown; Fig. 6B). Together, these findings underscore the relevance of our gene set for human carcinogenesis.
Fig. 6. Microarray analysis to identify p53 targets associated with tumor suppression.
(A) Gene expression profiles of HrasV12 MEFs from p53 genotypes that retain wild-type p53 tumor suppressor activity (wt p53, p5353,54/53,54 and p5325,26/25,26) were compared to those lacking p53 tumor suppressor activity (p5325,26,53,54/25,26,53,54 and p53 null). Those genes expressed at least 2-fold and 1.5 standard deviations higher in the wild-type group relative to the p53 null group were used as a p53 signature to predict p53 status of human breast cancer samples by PCA (Miller et al., 2005; Blue= wt p53, p5353,54 and p5325,26 MEFs, red= p5325,26,53,54 and p53 null MEFs, green=p53wt human breast cancers, pink=p53mut human breast cancers). (B) Kaplan-Meier curves demonstrate the effectiveness of MEF p53 signature in stratifying human breast cancer samples by patient survival; p=0.0422, log rank test. (C) Schematic showing approach to identify p53 target genes potentially involved in tumor suppression. (D) Heat map displaying the 14 genes meeting the filtering criteria described in (C). (E) qRT-PCR validation showing the average expression levels +/− SD of target genes in HrasV12 MEFs homozygous for wt p53, p5325,26, or p53 null alleles, after normalization to β-actin. (F) Induction of target genes by DNA damage is p53-dependent in GM00011 human fibroblasts. qRT-PCR demonstrates the efficacy of p53 knockdown with p53 shRNA compared to scrambled control shRNA (left) and the p53-dependence of target gene induction after 24 hrs of treatment with 0.2 µg/ml dox (right). Values are the averages of 3 replicates +/− SD. (G) ChIP for p53 binding to consensus sites in target genes in wild-type MEFs treated with 0.2 µg/ml dox for 6 hrs. IgG antibody serves as a negative control. Values represent the fold enrichment of binding to the p53 consensus site compared to binding to an irrelevant gene desert site and are the average of 3 replicates +/− SD. See also Figure S3.
To focus functionally on those genes most relevant to tumor suppression, we then filtered this p53-dependent 130 gene set for those genes downregulated in human and mouse cancers of a variety of types using EBI’s Gene Atlas database (EFO_0000311 ontology term, downregulated in any organism; Kapushesky et al., 2009; Fig 6C). In this manner, we derived a list of 14 genes (Fig. 6D). We validated that these genes displayed p53-dependent expression, which was either completely or partially maintained in cells expressing the p5325,26 mutant (Fig. 6E). The targets identified include genes involved in cell signaling (Abhd4, Phlda3, and Rgs12), cytoskeletal function (Crip2, Def6, Kank3, Arap2), and DNA repair (Polk, Mgmt, and Ercc5; Bugni et al., 2009; Kawase et al., 2009; Liu et al., 2008; van Ham et al., 2003). Additional genes included Sidt2, an orthologue of a C. elegans dsRNA transporter that encodes a lysosomal protein in human cells (Duxbury et al., 2005; Jialin et al., 2010) and Ttc28, whose encoded protein interacts with the BRCA1-containing BRCC complex (Sowa et al., 2009). Notably, Phlda3 was recently described as a p53 target gene encoding an AKT inhibitor and as frequently deleted in lung tumors, and Mgmt and Ndrg4 have been reported as tumor suppressor genes (Kawase et al., 2009; Bugni et al., 2009; Melotte et al., 2009). To further examine the significance of these genes as potential p53 tumor suppressor target genes in humans, we tested whether a subset was induced by p53 in primary human fibroblasts. Indeed, we observed robust, p53-dependent upregulation of most genes, except MGMT or TTC28, upon doxorubicin treatment (Fig. 6F). MGMT and TTC28 may be induced by p53 in other human cell settings, however, as suggested by the demonstrated binding of human p53 to sites near these genes in ChIP-PET experiments (Wei et al., 2006). We also investigated whether these genes represent direct p53 effectors using ChIP. We found that p53 in fact binds to specific consensus binding sites in the regulatory regions of these genes, demonstrating that their induction is proximal to p53 activation (Fig. 6G, Fig. S3A, B). Together, our studies have identified a cadre of tumor suppression-associated target genes activated by p53 in both mouse and human cells.
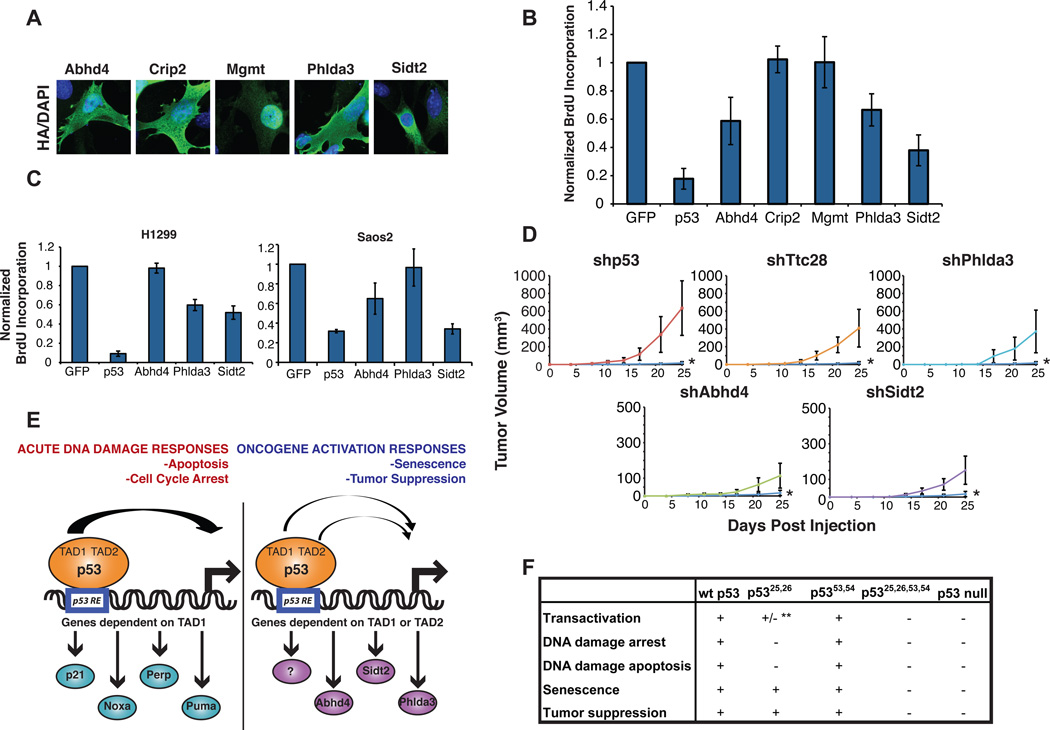
Novel p53 target gene products display tumor suppressor activity
We next sought to assess the biological contribution of several of these gene products, representing various functional categories, to p53 function. We first examined the function of these gene products in a senescence model through overexpression in HrasV12;p53−/− MEFs. Consistent with a role in DNA methylation, Mgmt was localized in the nucleus, while Abhd4, Phlda3, Crip2, and Sidt2 were found in the cytoplasm, in keeping with functions in signaling, modulation of the cytoskeleton, or lysosome biology (Fig. 7A). We then focused on effects on cell cycle progression through analysis of BrdU incorporation, with p53 and GFP serving as positive and negative controls, respectively, for cell cycle arrest. Interestingly, we found that the different targets showed varying capacities to limit proliferation: Phlda3, Abhd4, and Sidt2 efficiently inhibited cell cycling, while Mgmt and Crip2 did not (Fig. 7B, Fig. S4). These gene products could similarly restrict cell cycling in p53-deficient human cancer cells, although their activity depended on the cell types examined (Fig. 7C). These findings suggest that the activity of these targets is conserved in humans, but in a cell-type-dependent fashion, likely because of the specific pathways deregulated in these particular cancer cells. The fact that the observed arrest responses were more moderate than that seen with p53 is in alignment with the notion that p53 regulates programs of genes that coordinately affect a particular p53 response (Lozano and Zambetti, 2005). In addition, it is not surprising that some target gene products fail to show a phenotype in this assay, as they likely affect cycle-cycle-independent aspects of tumor suppression. For example, Mgmt plays a key role in DNA repair (Bugni et al., 2009), while Crip2 localizes to actin-rich structures, suggesting a role in regulating actin dynamics or cell migration (van Ham et al., 2003). Together, our data suggest that p53 may trigger multiple sub-programs that cooperate to promote tumor suppression.
Fig. 7. Novel p53 target gene products display tumor suppressor activity.
(A) Immunofluorescence indicating localization of HA-tagged proteins expressed in HrasV12;p53 null MEFs. (B) Quantification of BrdU labeling in HrasV12;p53 null MEFs expressing different target gene products and BrdU-pulsed 24 hrs post-transfection. The BrdU labeling index of HA-positive cells was assessed by immunofluorescence. Values are normalized to the proliferative index of cells expressing HA-GFP, and p53 is used as a positive control for arrest. Averages +/− SD are shown. (C) Quantification of BrdU labeling in H1299 and Saos2 cells expressing p53 or different target gene products, as described in (B). Averages +/− SEM are shown. (D) Average tumor volumes +/− SEM, as a function of time, in Scid mice injected with E1A-HRasV12 transformed MEFs with knockdown of various genes. shGFP control tumors are indicated by the blue line in each graph and labeled *. (E) Models for p53 action in response to acute DNA damage versus oncogenic signals. p53 responses to acute DNA damage, including apoptosis and cell cycle arrest, rely on the activity of the first p53 TAD and robust transactivation of canonical p53 target genes such as p21, Noxa, Puma, and Perp. In contrast, p53 responses downstream of oncogenic signaling in senescence and tumor suppression can be driven by either p53 TAD. A more limited p53 transactivation program, such as through efficient activation of novel p53 target genes regulated by either the first or second p53 TAD, including Phlda3, Abhd4, and Sidt2, can account for p53 function in these contexts. (F) Table summarizing findings described in this entire study. **indicates minimal transactivation of most, but not all, p53 target genes. See also Figure S4.
To evaluate the functional contribution of these new p53 targets to tumor suppression, we investigated whether inhibiting their expression promotes cancer. Using MEFs transformed with the oncoproteins E1A and HrasV12, followed by RNA interference to knock-down select individual targets, we examined tumor growth in immunocompromised mice. shRNAs directed against GFP or p53 served as negative or positive controls for tumor growth, respectively. Intriguingly, we observed that knockdown of Phlda3 and Ttc28 resulted in efficient tumor growth, while knockdown of Sidt2 and Abhd4 resulted in more moderate tumor growth, although still enhanced relative to GFP shRNA controls (Fig. 7D). The enhanced tumor growth observed with diminished expression of any of several of these novel p53 target genes demonstrates that these genes have tumor suppressor activity. Thus, collectively these studies define a critical network of direct p53 target genes that are mediators of tumor suppression.
DISCUSSION
Here, using an allelic series of p53 transactivation domain mutant knock-in mice, we reveal distinct mechanisms for p53 action in the contexts of acute DNA damage and oncogenic signals. Our findings suggest that the transcriptional activation function of p53 is fundamental for both pathways, but that there are qualitative differences in the requirements for transactivation. p53 responses to acute DNA damage in primary cells rely on an intact first TAD of p53 as the L25Q;W26S mutations within this domain severely impair transactivation of most classical p53 targets including p21, Noxa, and Puma, and abrogate activity in both DNA damage-induced G1 arrest and apoptosis (Fig. 7E, F). These observations suggest that full p53 transcriptional activation is essential for responses downstream of acute DNA damage, a notion consistent with the known requirements of p21 as well as Puma and Noxa, genes robustly induced by DNA damage, in G1 arrest and apoptosis, respectively (Lozano and Zambetti, 2005).
In contrast, we find that p53 responses downstream of oncogenic signals require more selective p53 transactivation function governed by both TADs, as both the p5325,26 and p5353,54 single TAD mutants retain the ability to drive senescence and tumor suppression, whereas p5325,26,53,54 lacks these activities (Fig. 7E, F). Similarly, human p5325,26 can induce apoptosis in some transformed cell contexts (Baptiste-Okoh et al., 2008). It is very surprising that p5325,26 retains tumor suppressor function because of its severely impaired ability to transactivate most p53 target genes, and these observations suggest that potent transactivation of a complete set of p53 target genes is dispensable for p53-mediated tumor suppression. However, our finding that p5325,26,53,54 lacks both transactivation capacity and activity downstream of oncogenic signals shows definitively that transcriptional activation is critical for tumor suppression. Analysis of the p5325,26 mutant suggests that tumor suppressor function could reflect minimal transactivation of canonical p53 target genes such as p21 and Puma (Efeyan et al., 2007; Hemann et al., 2004), normal transactivation of the small subset of genes properly activated by p5325,26, or collaborative action of both classes of genes. Indeed, the p53R172P mutant requires p21 to suppress tumor development, and Puma is critical for suppressing lymphomagenesis in Eu-myc transgenic mice (Barboza et al., 2006; Hemann et al., 2004). Our identification of a new set of direct p53 target genes efficiently induced by p5325,26 and associated with p53 tumor suppression in human cancers suggests compelling additional candidates involved in mediating p53 tumor suppressor function. Accordingly, our studies of cell cycle arrest upon overexpression of target genes in cultured cells and of tumor growth upon knockdown of target genes in a fibrosarcoma model together support roles for Abhd4, Phlda3, and Sidt2 in tumor suppression. Collectively, our studies thus define novel p53 networks critically linked to tumor suppression. Additionally, while we have focused on defining the contribution of transcriptional activation by p53 to its function in different settings, our studies do not preclude other p53 activities being relevant for p53 function in vivo (Green and Kroemer, 2009).
Of significance, the functional redundancy of the first and second p53 TADs can explain the lack of TAD mutations in human cancer. The majority of reported p53 mutations in cancer target the p53 DNA binding domain, where one amino acid alteration can ablate both p53 DNA binding and transactivation activity (Brady and Attardi, 2010). Although mutating both TADs can also severely cripple p53 tumor suppression function, it is less probable that the four amino acid substitutions required for loss of function would occur during carcinogenesis.
Because the p53 TADs are inextricably linked to the regions required for negative regulation by Mdm2, mutating the TADs – particularly residues 25/26 – results in p53 stabilization (Lin et al., 1994). This coupling of transcriptional activation to protein stability is a common theme amongst transcription factors (Kodadek et al., 2006). However, the findings that the p5325,26 protein retains many activities of wild-type p53, but p5325,26,53,54 does not, despite both mutants exhibiting similarly enhanced stability, indicate that the observed phenotypes relate specifically to transcriptional activation potential rather than simple stabilization.
These studies open the door for detailed analyses of the mechanisms of p53 target gene induction during responses to acute DNA damage and oncogenic signaling. Mutations in key p53 TAD residues likely compromise critical interactions between p53 and important cofactors. This notion is supported by in vitro studies showing that mutation of the first TAD disrupts interactions with TBP, Taf9, Taf6, CBP and the TRAP80 component of the mediator, mutation of the second TAD perturbs interaction with the p62 component of TFIIH, and mutation of both TADs impedes binding to p300 and STAGA (Gamper and Roeder, 2008 and references therein; Teufel et al., 2007). Our studies demonstrate that the domains defined in vitro are indeed essential for transcriptional activation by p53 in vivo and further expand our understanding by revealing that TADs display selectivity according to context. It may be that activation of genes important for DNA damage responses relies on specific cofactor contacts with the first TAD of p53, whereas activation of genes involved in tumor suppression requires an alternate cofactor(s) interacting with both TADs. Future investigation of cofactor recruitment to different classes of p53 target genes using ChIP analyses may augment our knowledge of the mechanisms of p53 transcriptional activation.
Through analysis of p5325,26, a mutant unable to elicit responses to acute DNA damage, our results indicate that p53 tumor suppressor activity in different tissue types, including epithelia (this study, unpublished observations), does not rely on its ability to respond to acute genotoxic insults. These findings are consistent with previous studies indicating that p53-mediated tumor suppression in DNA damage-induced lymphomas and fibrosarcomas is independent of p53’s ability to trigger a response to acute DNA damage (Christophorou et al., 2006; Efeyan et al., 2006). However, our findings do not rule out the possibility that in incipient tumors, p53 may react to low-level, chronic DNA damage, caused by such factors as replication stress or telomere attrition, through a different mechanism (Halazonetis et al., 2008). The idea that the DNA damage pathway downstream of chronic genotoxic stress may be mechanistically different from that downstream of acute genotoxic injury provides a potential resolution to the controversy regarding the role of DNA damage signaling in activating p53 in nascent tumors and warrants further investigation.
Our studies elaborating a mechanistic distinction between p53 pathways downstream of acute DNA damage and oncogene expression also have significant implications for improving cancer therapy. Radiation and chemotherapies can be highly beneficial for cancer treatment, but at the cost of inducing p53-dependent deleterious side effects in radiosensitive tissues (Gudkov and Komarova, 2003). Therefore, identifying a strategy to selectively inhibit some p53 functions to mitigate the deleterious side effects of genotoxic therapeutics, without incurring increased risk of new cancers, would be broadly valuable for cancer therapy. An inhibitor of the first p53 TAD, administered during radiation or chemotherapy treatment of p53 mutant tumors, could potentially minimize associated pathologies without compromising p53 tumor suppressor function throughout the organism. An additional therapeutic application of our studies is the identification of strategies to restore p53 function in tumors via activation of p53 targets uniquely important for tumor suppression, providing a promising new route for cancer therapy.
EXPERIMENTAL PROCEDURES
Generation and Analysis of Mice
The generation of knock-in mice is described in the Supplemental Information. p53 mutant strains were crossed to Rosa26CreERT2 and KrasLSL-G12D mice (Tuveson et al., 2004; Ventura et al., 2007). For the apoptosis experiments, 5 mg tamoxifen (Sigma) dissolved in 2% ethanol in corn oil (v/v) was administered for 2 days by oral gavage, and then mice were X-ray-irradiated 72 hrs after the last tamoxifen dose. For the tumor study, mice were intranasally infected with 4 × 107 PFU of Ad-Cre (University of Iowa GTVR) as described (Jackson et al., 2005) and sacrificed 12 weeks later. All animal work was done in accordance with Stanford University APLAC.
Cell Culture
Adenoviral infections were performed at an MOI of ~100 using adenoviral Cre or empty (Ad5 Cre, denoted “Ad-Cre” or Ad5 empty, denoted “Ad-empty”, University of Iowa GTVR) for 24 hrs. Efficient p53 expression (>90% of cells) was confirmed by immunofluorescence. The DNA damage G1 arrest assays were performed in MEFs infected with Ad-Cre or Ad-empty and irradiated 48h later with 5 Gy of γ-radiation using a 137Cs source. 14 hrs later, cells were pulsed with BrdU and prepared for flow cytometry as described (Attardi et al., 2004). For senescence experiments, retroviral transduction of MEFs with pWZL-HrasV12 (Serrano et al., 1997) was followed by adenovirus infection 4 days later. For overexpression experiments, MEFs and human cells were transfected using Fugene6 (Roche) and Lipofectamine 2000 (Invitrogen), respectively, per manufacturer’s instructions. In all experiments, 4 hr BrdU pulses (3 µg/ml) were performed at the indicated timepoints.
Microarray Analysis
Total RNA was isolated from HrasV12 MEFs 48 hrs after Ad-Cre or Ad-empty infection and processed for analysis by Affymetrix Mouse Genome 430 2.0 expression arrays per manufacturer’s instructions. Probe-level data were processed using BRB-ArrayTools (Biometric Research Branch of the National Cancer Institute), with Robust Multichip Average for background adjustment, normalization, and expression summarization. All subsequent analysis was performed using MeV (Saeed et al., 2006).
qRT-PCR, Northern Blotting, and Western Blotting
RNA was isolated using Trizol (Invitrogen) and reverse transcribed using MMLV reverse transcriptase (Invitrogen) and random primers. PCR was performed in triplicate using SYBR green (SA-Biosciences) and a 7900HT Fast Real-Time PCR machine (Applied Biosystems), and results were computed relative to a standard curve made with cDNA pooled from all samples. Northern blotting was performed as described (Johnson et al, 2005). p53 western blotting was performed using anti-p53 antibodies (CM5; Vector Labs, 1:1000) and anti-actin antibodies (Santa Cruz, 1:1000), using standard protocols.
Chromatin Immunoprecipitation
Cells were crosslinked with 1% formaldehyde and neutralized with 0.125 M glycine. Purified chromatin was sonicated to ~500 bp. Immunoprecipitations were performed with anti-p53 antibodies (CM5; Vector Labs) or IgG or p16 antibodies as controls (Santa Cruz) and washed 5X with LiCl wash buffer (100 mM Tris pH 7.5, 500 mM LiCl, 1% NP-40, 1% Na-deoxycholate) and 1X with TE. The ChIP’ed DNA was eluted for 1 hr at 65°C in elution buffer (1% SDS, 0.1 M NaHCO3). After reverse crosslinking the samples, qPCR was performed in triplicate as described above and compared to a standard curve of immunoprecipitation input.
Cell and Tissue Staining
Immunofluorescence was performed using standard protocols and the following antibodies: anti-human p53 (rabbit FL-393; 1:250; Santa Cruz), anti-HA (rabbit; 1:200; Invitrogen), anti-mouse p53 (rabbit CM5; 1:150, Vector Labs), anti-PML (1:100, gift of S. Lowe), and anti-BrdU (mouse; 1:50, BD Biosciences). Where relevant, cells were co-stained with p53 and the antibody of interest and only p53-positive cells were used in analysis. TUNEL staining on mouse tissues was performed as described (Ihrie et al., 2003). Hematoxylin and eosin (H&E) staining of mouse lungs was performed using standard protocols on paraffin-embedded lung. Lungs were sliced into several coronal pieces prior to embedding, and H&E sections were analyzed using Bioquant Osteo imaging software (Bioquant), where tumors were manually circled and quantified to determine area and then divided by the total lung area, excluding airspace. For SA-β galactosidase staining, cells were fixed in 2% formaldehyde/0.2% glutaraldehyde and incubated with X-gal at pH 6.0 for 48 hrs as described (Dimri et al., 1995).
Supplementary Material
ACKNOWLEDGMENTS
Microarray analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. We thank T. Jacks for providing the Rosa26CreERT2 mice, S. Lowe for the pWZL-HrasV12 plasmid and Pml antibody, S. Artandi for the 3xHA expression plasmid, and W. Hahn and A. Schinzel and the Dana Farber RNAi facility for the lentiviral shRNA constructs; D. Burkhart, A. Krieg, J. Sage, and L. Sayles for technical assistance; and S. Artandi, A. Brunet, A. Giaccia, and J. Sage for critical reading of the manuscript. This work was supported by a Smith Stanford Graduate Fellowship to C.A.B, a Lucille P. Markey Biomedical Research Stanford Graduate Fellowship to T.M.J., National Science Foundation Graduate Research Fellowships to C.A.B. and T.M.J., a Gerald Lieberman Dissertation Fellowship to C.A.B., a C.A.P.E.S Fellowship to S.S.M, a Swiss National Science Foundation Fellowship to D.K.B., and funding from the American Cancer Society, the Leukemia and Lymphoma Society, and the National Institutes of Health to L.D.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession number GSE27901.
REFERENCES
- Attardi LD, de Vries A, Jacks T. Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene. 2004;23:973–980. doi: 10.1038/sj.onc.1207026. [DOI] [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1937–1942. doi: 10.1073/pnas.0711800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science (New York, NY. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- Bugni JM, Meira LB, Samson LD. Alkylation-induced colon tumorigenesis in mice deficient in the Mgmt and Msh6 proteins. Oncogene. 2009;28:734–741. doi: 10.1038/onc.2008.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Scolnick DM, Darpino P, Ying CY, Halazonetis TD, Berger SL. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- Chao C, Saito S, Kang J, Anderson CW, Appella E, Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. The EMBO journal. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Lee SH, Kim DH, Ahn MJ, Kim JS, Woo JY, Torizawa T, Kainosho M, Han KH. Structural details on mdm2-p53 interaction. The Journal of biological chemistry. 2005;280:38795–38802. doi: 10.1074/jbc.M508578200. [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury MS, Ashley SW, Whang EE. RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem Biophys Res Commun. 2005;331:459–463. doi: 10.1016/j.bbrc.2005.03.199. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Collado M, Velasco-Miguel S, Serrano M. Genetic dissection of the role of p21Cip1/Waf1 in p53-mediated tumour suppression. Oncogene. 2007;26:1645–1649. doi: 10.1038/sj.onc.1209972. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Garcia-Cao I, Herranz D, Velasco-Miguel S, Serrano M. Tumour biology: Policing of oncogene activity by p53. Nature. 2006;443:159. doi: 10.1038/443159a. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes & development. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- Gaidarenko O, Xu Y. Transcription activity is required for p53-dependent tumor suppression. Oncogene. 2009 doi: 10.1038/onc.2009.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Roeder RG. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Mol Cell Biol. 2008;28:2517–2527. doi: 10.1128/MCB.01461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3:117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science (New York, NY. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. Suppression of tumorigenesis by the p53 target PUMA. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, Reczek E, Horner JS, Khachatrian L, Sage J, Jacks T, Attardi LD. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr Biol. 2003;13:1985–1990. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer research. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jialin G, Xuefan G, Huiwen Z. SID1 transmembrane family, member 2 (Sidt2): a novel lysosomal membrane protein. Biochem Biophys Res Commun. 2010;402:588–594. doi: 10.1016/j.bbrc.2010.09.133. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Johnson TM, Hammond EM, Giaccia A, Attardi LD. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nature genetics. 2005;37:145–152. doi: 10.1038/ng1498. [DOI] [PubMed] [Google Scholar]
- Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, Rustici G, Williams E, Parkinson H, Brazma A. Gene expression atlas at the European bioinformatics institute. Nucleic acids research. 2009;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kawase T, Ohki R, Shibata T, Tsutsumi S, Kamimura N, Inazawa J, Ohta T, Ichikawa H, Aburatani H, Tashiro F, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–550. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Kennedy AL, McBryan T, Enders GH, Johnson FB, Zhang R, Adams PD. Senescent mouse cells fail to overtly regulate the HIRA histone chaperone and do not form robust Senescence Associated Heterochromatin Foci. Cell Div. 2010;5:16. doi: 10.1186/1747-1028-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Attardi LD. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis. 2010 doi: 10.1093/carcin/bgp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodadek T, Sikder D, Nalley K. Keeping transcriptional activators under control. Cell. 2006;127:261–264. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes & development. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang L, Harvey-White J, Huang BX, Kim HY, Luquet S, Palmiter RD, Krystal G, Rai R, Mahadevan A, et al. Multiple pathways involved in the biosynthesis of anandamide. Neuropharmacology. 2008;54:1–7. doi: 10.1016/j.neuropharm.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- Lozano G, Zambetti GP. What have animal models taught us about the p53 pathway? J Pathol. 2005;205:206–220. doi: 10.1002/path.1704. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell death and differentiation. 2006;13:909–912. doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- Melotte V, Lentjes MH, van den Bosch SM, Hellebrekers DM, de Hoon JP, Wouters KA, Daenen KL, Partouns-Hendriks IE, Stessels F, Louwagie J, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- Merritt AJ, Potten CS, Kemp CJ, Hickman JA, Balmain A, Lane DP, Hall PA. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer research. 1994;54:614–617. [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras( G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- van Ham M, Croes H, Schepens J, Fransen J, Wieringa B, Hendriks W. Cloning and characterization of mCRIP2, a mouse LIM-only protein that interacts with PDZ domain IV of PTP-BL. Genes Cells. 2003;8:631–644. doi: 10.1046/j.1365-2443.2003.00660.x. [DOI] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene. 1999;18:2405–2410. doi: 10.1038/sj.onc.1202539. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. The Journal of biological chemistry. 1998;273:13030–13036. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.