Recent work of several laboratories has identified FACT (facilitates chromatin transcription) protein complex as an emerging target for development of anticancer drugs and suggested a mechanism of FACT action during transcription and cell differentiation.
FACT is the transcription and replication factor [1], which can stimulate transcript elongation through chromatin in vitro [2, 3]. Human FACT is a heterodimer composed of two subunits Spt16 and SSRP1 and has a histone chaperone activity [3, 4]. In vivo, FACT and RNA polymerase II (Pol II) co-localize and display similar kinetics of recruitment and chromosome tracking [5, 6]. FACT also maintains the nucleosome integrity during transcription initiation and transcript elongation by Pol II [6-9]. Currently, two different models have been proposed to explain the functions of FACT to nucleosomes. On one hand, it has been suggested that FACT-assisted histone H2A/H2B dimer dissociation from nucleosome facilitates transcription through chromatin [3] and perhaps chromatin re-assembly. On the other hand, the studies of yeast FACT suggested that it can globally affect the nucleosome structure to allow access of regulatory proteins to histone-covered DNA [10, 11].
In a recent careful biochemical study from laboratory of K. Luger [12] interactions between FACT and its various potential interaction targets in chromatin (histones H2A-H2B and H3-H4, histone tails, nucleosomal and linker DNA) were quantified. It was shown that FACT preferentially binds to H2A-H2B dimer and this high-affinity binding is largely mediated by the acidic C-terminal domain of the Spt16 subunit. The observation that FACT competes with DNA for a shared interaction interface on H2A-H2B dimer suggests a possible mechanism of FACT-facilitated DNA displacement from the histone octamer and possibly subsequent nucleosome recovery. Indeed, as various processive DNA-targeted enzymes (e.g. polymerases and ATP-dependent chromatin remodelers) encounter nucleosomes and attempt to displace histones from DNA, progression of the enzymes should be greatly facilitated by the FACT-dimer interactions. The recovery of chromatin structure could be facilitated by chaperone activity of FACT though facilitated re-binding of the dimers to chromatin or their more efficient survival during transcription, as was suggested recently [13].
Analysis of FACT expression in various tissues using immunohistochemical methods and NCBI Gene Expression Omnibus data analysis (laboratory of K. Gurova) have shown that the SSRP1 and SPT16 subunits are expressed in a coordinated manner, and the levels of expression are highly variable within the tissues [14]. The levels of expression are higher in non-differentiated and cancer cells, and do not correlate with expression of the Ki67 proliferation marker, suggesting that that FACT expression is more related to differentiation than to proliferation. Induction of oncogenes in different cells in most cases results in an increased expression of FACT subunits. Furthermore, tumor cells are sensitive to FACT downregulation, suggesting that FACT could be an important new target for development of anti-cancer drugs.
The latter line of research has been extended in another work from Gurova group [15]. It was shown that cancer cells, like undifferentiated cells, contain higher levels of FACT. Small DNA-intercalating molecules, curaxins, have a strong anticancer activity, activate p53 and inhibit NF-kB without causing detectable genotoxicity. The effects of curaxins on p53 and NF-kB, as well as their toxicity to cancer cells, result from “chromatin trapping” of FACT (depletion of soluble, functionally active FACT), presumably through interaction with DNA-bound curaxins that is particularly detrimental for cancer cells. Trapping of FACT leads to phosphorylation of the p53 by casein kinase 2 (CK2) and inhibition of NF-kB-dependent genes that require FACT activity during transcript elongation stage.
How can the changes in FACT content during cell differentiation and carcinogenesis be rationalized in the framework of the current models of FACT action? Since FACT works primarily during transcript elongation step (likely via interactions with H2A/H2B dimers, Fig. 1), it is required in large amounts in cells where gene expression occurs at a higher level, such as cancer or undifferentiated cells. Therefore these cells are more sensitive to inhibitors affecting expression or activity of FACT. Indeed, recent proteomic studies of Xiong et al. [16] suggest that proteins involved in DNA replication, chromatin remodeling, and Pol II-dependent transcription are down-regulated in differentiated cells. Future analysis of the molecular mechanisms of FACT action and its interaction with anticancer drugs should reveal whether FACT expression plays a regulatory role in differentiation and carcinogenesis, and should result in development of more efficient cancer therapies.
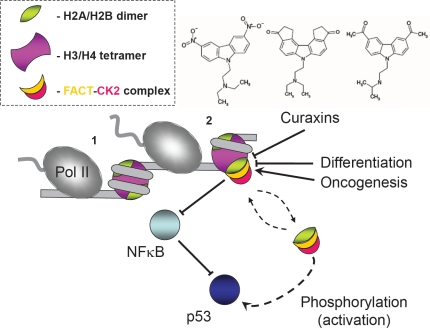
Figure 1. FACT as a regulatory hub in cell differentiation and carcinogenesis.
As RNA polymerase II (Pol II) encounters nucleosomes (1) and uncoils nucleosomal DNA from core histones (2), FACT interacts with the H2A/H2B dimer and facilitates transcription through chromatin. Binding of FACT results in cell growth inhibition through inhibition of NK-κB and activation of p53 by CK2 phosphorylation. Curaxins intercalate into the DNA, induce a conformational change in the DNA and interfere with recruitment of FACT.
Acknowledgments
This work was supported by NIH GM58650 and Government of the Russian Federation (order #220) grants to V.M.S.
REFERENCES
- 1.Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 3.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 4.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 5.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 6.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 10.McCullough L, Rawlins R, Olsen A, Xin H, Stillman DJ, Formosa T. Insight Into the Mechanism of Nucleosome Reorganization From Histone Mutants That Suppress Defects in the FACT Histone Chaperone. Genetics. 2011;188:835–846. doi: 10.1534/genetics.111.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler DD, Muthurajan UM, Hieb AR, Luger K. The histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011 doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia H, Fleyshman D, Kolesnikova K, Safina A, Commane M, Paszkiewicz G, et al. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget. 2011 doi: 10.18632/oncotarget.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasparian AV, Burkhart CA, Purmal AA, Brodsky L, Pal M, Saranadasa M, et al. Curaxins: anticancer compounds that simultaneously suppress NF-kappaB and activate p53 by targeting FACT. Sci Transl Med. 2011;3:95ra74. doi: 10.1126/scitranslmed.3002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong L, Darwanto A, Sharma S, Herring J, Hu S, Filippova M, et al. Mass spectrometric studies on epigenetic interaction networks in cell differentiation. J Biol Chem. 2011;286:13657–13668. doi: 10.1074/jbc.M110.204800. [DOI] [PMC free article] [PubMed] [Google Scholar]