Abstract
Background
Plant photoreceptors, phytochromes and cryptochromes, regulate many aspects of development and growth, such as seed germination, stem elongation, seedling de-etiolation, cotyledon opening, flower induction and circadian rhythms. There are several pieces of evidence of interaction between photoreceptors and phyto-hormones in all of these physiological processes, but little is known about molecular and genetic mechanisms underlying hormone-photoreceptor crosstalk.
Methodology/Principal Findings
In this work, we investigated the molecular effects of exogenous phyto-hormones to photoreceptor gene transcripts of tomato wt, as well as transgenic and mutant lines with altered cryptochromes, by monitoring day/night transcript oscillations. GA and auxin alter the diurnal expression level of different photoreceptor genes in tomato, especially in mutants that lack a working form of cryptochrome 1a: in those mutants the expression of some (IAA) or most (GA) photoreceptor genes is down regulated by these hormones.
Conclusions/Significance
Our results highlight the presence of molecular relationships among cryptochrome 1a protein, hormones, and photoreceptors' gene expression in tomato, suggesting that manipulation of cryptochromes could represent a good strategy to understand in greater depth the role of phyto-hormones in the plant photoperceptive mechanism.
Introduction
During evolution, plants have developed accurate mechanisms to integrate internal signal such as hormones and environmental cues like light and temperature, in order to respond as quickly and efficiently as possible to any change. Several growth and developmental processes, such as seed germination, stem elongation, seedling de-etiolation, cotyledon opening, flower induction and circadian rhythms are activated and/or regulated by both light and hormones, suggesting interactions between signalling pathways [1], [2], [3], [4], [5], [6].
Plants have acquired the tools to monitor precisely the changing intensity and spectrum of light, its direction and, in specific cases, its plane of polarization [7], through a number of photoreceptors: the red (R)/far-red(FR) – absorbing phytochromes and the blue/UV-A – absorbing cryptochromes and phototropins [8], [9].
In Arabidopsis, phytochromes are encoded by five different genes, PHYA through PHYE [10], [11], cryptochromes by three genes, CRY1, CRY2 and CRY-DASH [12], [13], [14]. Cryptochromes and phytochromes control several overlapping physiological responses, [15], [16] at all stages of plant development. Although the exact nature of co-action has yet to be well elucidated, it is known that blue light-mediated de-etiolation involves the interaction of both phytochrome and cryptochrome signaling [17], [18], [19].
In tomato (Solanum lycopersicum), four cryptochrome genes have been discovered and analyzed so far: two CRY1-like (CRY1a and CRY1b), one CRY2 and one CRY-DASH gene [20], [21], [22]. The role of the CRY1a gene has been elucidated through the use of antisense [23] and mutant [24] plants. CRY1a controls seedling photomorphogenesis, anthocyanin accumulation, and adult plant development. No effects of CRY1a on flowering time or fruit pigmentation have been observed. The overexpression of tomato CRY2 causes phenotypes similar to but distinct from their Arabidopsis counterparts (hypocotyls and internode shortening under both low and high fluence blue light), but also several novel ones, including a high-pigment phenotype, resulting in overproduction of anthocyanins and chlorophyll in leaves and of flavonoids and lycopene in fruits [25]. Tomato CRY-DASH gene is under the control of circadian machinery with a light-regulated transcription pattern and it is expressed since the earliest phases of tomato development [22].
In tomato, phytochromes are encoded by five genes: PHYA, PHYB1, PHYB2, PHYE and PHYF [26]. Phylogenetic analyses showed orthology between PHYA, PHYE and PHYC/F gene pairs in Arabidopsis and tomato; tomato PHYB1 and PHYB2 were originated by an independent duplication [27]. Roles for PHYA and PHYB1 in the mediation of tomato plant de-etiolation responses to red light (R) have been demonstrated previously [28], [29]. Although the phyAphyB1 double mutant is blind to low-irradiance R, it de-etiolated normally under white light. The phenotype of phyAphyB1phyB2 mutants under natural daylight indicated an important role for PHYB2 in this residual response [30] and it also clear that PHYB2 is also active in R-sensing [31].
Different classes of hormones regulate several aspects of seedling development, often in redundant or antagonistic relationship among them. Gibberellin (GA) and abscisic acid (ABA) are two critical signals with antagonistic effects on seed dormancy and germination [32], [33]. GA and brassinosteroid (BR) are involved in the repression of photomorphogenesis in the dark [34], [35] and with auxin promote hypocotyl elongation [36]. Low levels of auxin induce root growth, whereas high levels have inhibitory effects [37]. Besides, auxin plays an important role in lateral root initiation and growth [38].
The interaction among hormones may be additive, synergistic or antagonistic, making their overall effect more complex (see reviews: [32], [39], [40], [41]). For example, auxin is known to control root growth in part through modulation of the cellular response to GA [42], but it regulates hypocotyl elongation independently of GA [36]. Recent evidence suggests that auxin and BR signaling pathways are overlapping and interdependent: expression of several AUX/IAA genes (SAUR and GH3 homologs) are regulated by both auxin and BR [43], [44], [45].
A few downstream genes are known to modulate or integrate different hormonal signals. For example, the Arabidopsis sax mutant provides strong evidence for interaction among multiple hormones related to BR levels [46], [47]. Finally, SPY gene was recently demonstrated to have a role as a coordinator in cross-talk between GA and cytokinin [48].
Phyto-hormones also play important roles in regulating vegetative and reproductive development. Mutants with a decreased response to GA, BR or auxin are usually characterized by dwarfism, reduced apical dominance, dark-green foliage, and reduced fertility [32], [49], [50], [51]. GA also regulates flowering time and flower organ development [52], [53].
There are several pieces of evidence of interactions between photoreceptors and hormones during plant development. Many studies have suggested that phytochromes and cryptochromes influence the activities of auxin in order to regulate plant growth. Indeed, PHYA, PHYB and CRY1 promote light-dependent effects of the auxin transport inhibitor 1-N-naphthylphthalamic acid on both hypocotyls and root elongation in Arabidopsis [54], [55]. Other reports indicate that cryptochromes regulate the transcription of AUX/IAA genes [56] and that AUX/IAAs are phosphorylated by PHYA [57].
Gibberellins are known to be a component of light signalling [58]; phytochromes and GAs act in coordination to regulate multiple aspects of Arabidopsis development such as flowering and hypocotyls elongation [59], [60], [61]. Phytochromes affect GA levels, by regulating expression of the GA2ox and GA3ox genes [62], and may also regulate GA responsiveness [63], [64], [65]. It has been recently shown that PHYA and PHYB mediate light stabilization of the DELLA proteins, which may, at least partially, result from the phytochrome-dependent regulation of GA homeostasis [66].
Light and GA play an antagonistic role during photomorphogenesis [34]. It has been reported that light inhibits the ability of Phytochrome Interacting Factors (PIFs) to promote dark-type growth (elongation of hypocotyl and repression of chloroplast development), through a stabilizing action of PIF proteins in the dark, rather than the destabilization, mediated by activated phytochromes, that occurs in the light. On the other hand, PIF responses are restored by the destabilizing action of GA over DELLA [4], [67].
Phytochromes and GAs are also involved (together with auxins and ethylene) in regulating shade-avoidance responses, that maximize light capture by positioning the leaves out of the shade [68].
In comparison to the phytochrome-regulated responses, the relationship between cryptochromes and GA in the blue light responses is less clear in Arabidopsis. It has been found in pea that CRY1 and PHYA redundantly regulate GA2ox and GA3ox expression and GA signaling [65], [69]. A recent report demonstrated that cryptochromes mediate blue light regulation of GA catabolic/metabolic genes, which affect GA levels and hypocotyl elongation [5].
Furthermore cytokinins in Arabidopsis are involved in the regulation of the circadian clock mechanism [6], in which both cryptochromes and phytochromes are also involved. Besides Vandenbussche and collegues [70] concluded that HY5, a positive regulator of photomorphogenesis induced by CRY1 and CRY2 [71], represents a point of convergence between cryptochrome and cytokinin signalling pathways.
Several other examples of hormone-over-photoreceptor interaction could be reported; however there is little or no information about effects of phyto-hormones on photoreceptors and possible alteration of their gene transcript accumulation.
We decided to investigate the effects of the addition of exogenous phyto-hormones to the photoreceptor system of tomato wt and transgenic lines with altered crypthochromes, by monitoring the day/night transcript oscillations. We demonstrated that exogenous GA and auxin are able to modify the tomato photoreceptor diurnal expression patterns, especially in cry1a mutants, suggesting the presence of a molecular network among cryptochrome 1a, hormones, and photoreceptor genes in tomato.
Results and Discussion
To investigate whether phyto-hormones influence the diurnal expression pattern of the tomato cryptochrome (CRY1a, CRY1b, CRY2 AND CRY-DASH) and phytochrome (PHYA, PHYB1, PHYB2, PHYE AND PHYF) genes, we have exogenously added citokinin (t-zeatin), gibberellic acid (GA3), auxin (IAA) and abscisic acid (ABA) phyto-hormones to wt tomato, to a mutant genotype with a non functional CRY1a (cry1a-) [24] and to a transgenic line overexpressing the cryptochrome 2 (CRY2OX) [25]. All tomato plants were grown hydroponically under a light cycle of 16 h light/8 h darkness (LD), as described in Methods. Two hours before the presumptive dawn (ZT-2) a specific phyto-hormone (t-zeatin, GA3, IAA or ABA) was added (for details, see Methods). Aerial components of the hormone-added plants and control plants (without hormone) were sampled at distinct time points over a diurnal cycle (ZT0, ZT6, ZT12, ZT16 and ZT20) and subjected to cryptochrome and phytochrome gene expression assays, by Q-RT PCR. We further analyzed the diurnal transcription pattern of two genes for which the transcription is strictly light-regulated: GIGANTEA (GI), involved in the regulation of the plants' circadian rhythm [72] and CAB4, a member of the large family of Chlorophyll a/b-binding proteins [73].
The effects of cryptochrome alterations on the photoreceptors' transcription pattern, without hormone treatment, are relatively minor, with the obvious exception of the fact that CRY2 transcripts are constantly up-regulated in CRY2OX genotype. Furthermore, GI and CAB4 transcripts show the widest day/night oscillation and a sharp peak at 12 h and 6 h after dawn, respectively; the different genotypes influence the peak amplitude rather than the phase of the cycling transcripts (Fig S1). Transcript alteration patterns similar to the above mentioned ones have already been observed in our previous work carried out using soil grown plants in LD [74]. However, in hydroponically grown plants we don't have strong effects on other CRYs and PHYs transcripts except for significant down-regulation of some photoreceptor transcripts (CRY1a, CRY1b, CRY-DASH, PHYA, PHYB1, PHYE, PHYF) at several time points in CRY2 over-expressing tomatoes (Fig. S1).
The reciprocal interaction between light and phyto-hormones is a well-known physiological process: light was found to regulate directly the biosynthesis of active gibberellins [75], ethylene [76] and ABA, as well [58]. The molecular mechanisms that regulate this interaction during plant development and life remain unclear, although they are starting to be unraveled [77]; here we provide evidence of a remarkable level of control of gibberellin and auxin on cryptochrome and phytochrome gene expression in tomato. Our results show that this control varies according to the analyzed genotype (Fig. 1). In general, the genotype with non functional cryptochrome 1a, cry1a-, appears to be much more sensitive to exogenous hormones than wt (Fig. 1). The data regarding CRY2 expression in CRY2OX genotype showed that the presence of an overexpression construct driven by a constitutive promoter is presumably able to dilute any hormonal effects on the transcription of this cryptochrome (Fig. 2A, 3A, 4A, 5A).
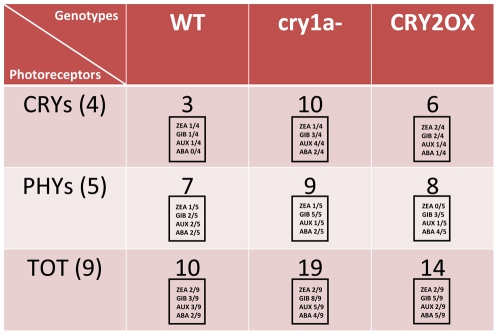
Figure 1. Number of transcription patterns altered in at least three points per cycle, by ZEA, GIB, AUX and ABA phyto-hormones in wt, cry1a- and CRY2OX genotypes.
We considered four cryptochrome (CRYs (4)) and five phytochrome (PHYs (5)) gene transcripts. In the squares is indicated the number of altered patterns for each hormone.
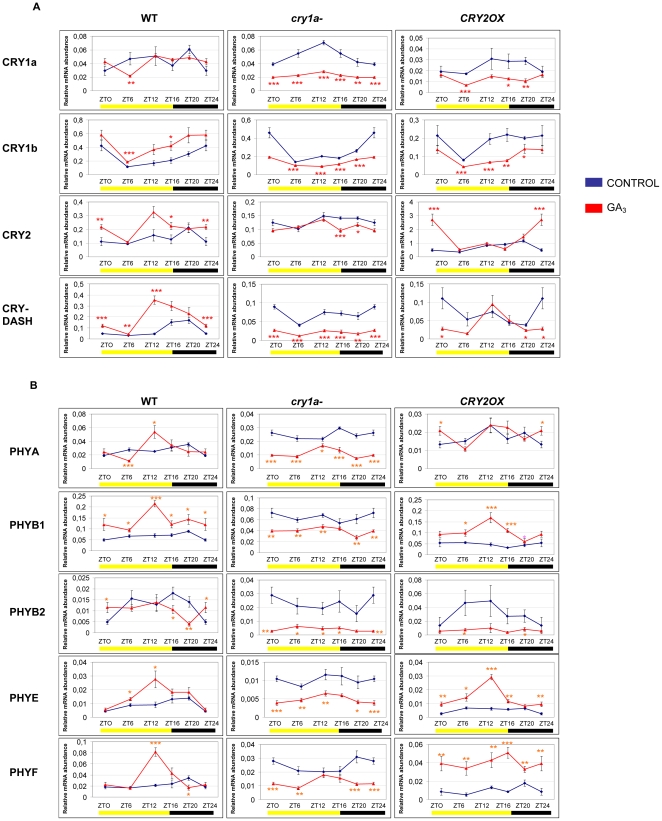
Figure 2. Diurnal expression pattern of Cryptochrome (A) and Phytochrome (B) transcripts analyzed by QRT-PCR in wt, cry1a- and CRY2OX GA3-treated tomato plants.
Results are presented as a ratio after normalization with β-actin. Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. The control data, of gene expression in the absence of hormone applications, are reproduced, for clarity, from those in Figure S1. Data shown are the average of two biological replicates, with error bars representing SEM. Hormone-treated plant transcripts significantly different from the corresponding ones of control plants are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001).
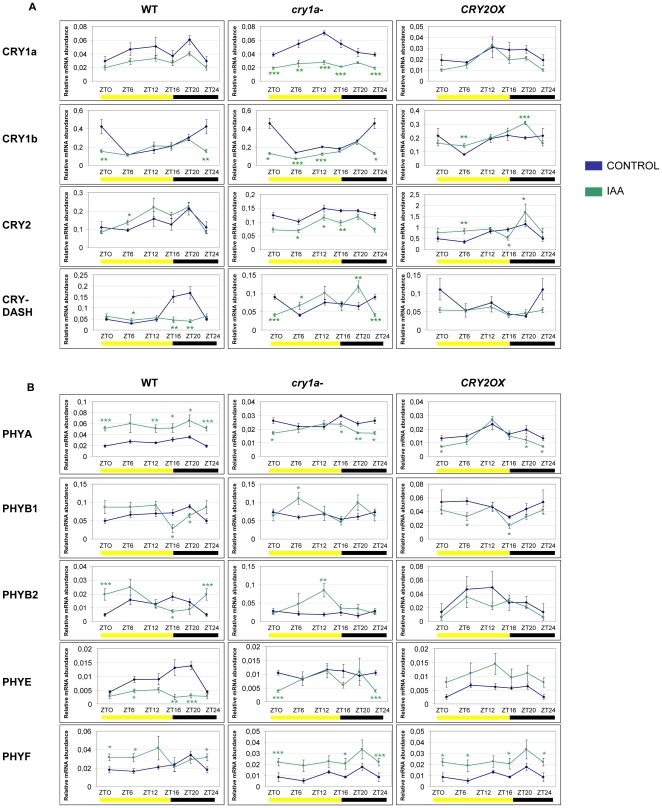
Figure 3. Diurnal expression pattern of Cryptochrome (A) and Phytochrome (B) transcripts analyzed by QRT-PCR in wt, cry1a- and CRY2OX IAA-treated tomato plants.
Results are presented as a ratio after normalization with β-actin. Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. The control data, of gene expression in the absence of hormone applications, are reproduced, for clarity, from those in Figure S1. Data shown are the average of two biological replicates, with error bars representing SEM. Hormone-treated plant transcripts significantly different from the corresponding ones of control plants are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001). Data from control plants are replotted from Figure 2.
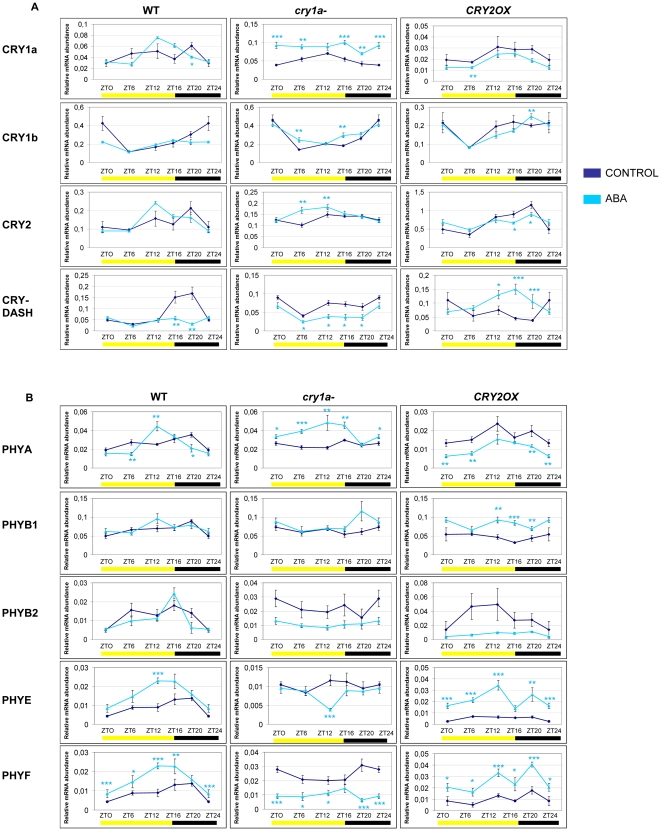
Figure 4. Diurnal expression pattern of Cryptochrome (A) and Phytochrome (B) transcripts analyzed by QRT-PCR in wt, cry1a- and CRY2OX ABA-treated tomato plants.
Results are presented as a ratio after normalization with β-actin. Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. The control data, of gene expression in the absence of hormone applications, are reproduced, for clarity, from those in Figure S1. Data shown are the average of two biological replicates, with error bars representing SEM. Hormone-treated plant transcripts significantly different from the corresponding ones of control plants are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001). Data from control plants are replotted from Figure 2.
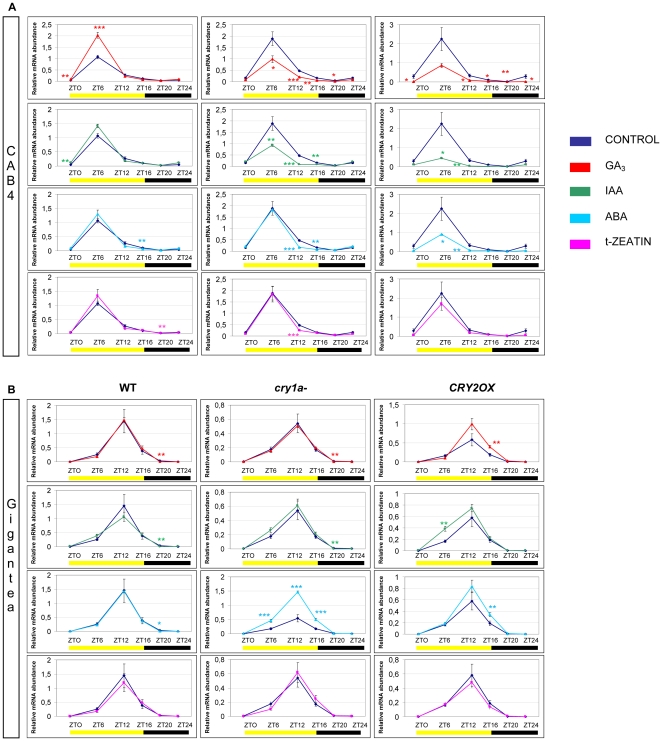
Figure 5. Diurnal expression pattern of CAB4 (A) and GIGANTEA (B) transcripts analyzed by QRT-PCR in wt, cry1a- and CRY2OX hormone-treated tomato plants.
Results are presented as a ratio after normalization with β-actin. Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. The control data, of gene expression in the absence of hormone applications, are reproduced, for clarity, from those in Figure S1. Data shown are the average of two biological replicates, with error bars representing SEM. Hormone-treated plant transcripts significantly different from the corresponding ones of control plants are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001).
Effects of phyto-hormones on tomato photoreceptor diurnal transcription
The modification of cryptochrome and phytochrome transcription pattern following addition of GA3 is remarkable, especially in cry1a- plants (Fig. 1 and Fig. 2AB). In this genotype, GA3 produces strong downregulation of both cryptochrome and phytochrome transcripts, with the only exception of CRY2, at all time points (Fig. 2A). The lack of a functional CRY1a protein produces a generic and strong signal of downregulation of the photo-perceptive apparatus of tomato in GA3 treated plants with regard to the untreated ones, suggesting a pivotal role for CRY1a in mediating light and gibberellin stimuli. Analyzing in greater detail the behavior of cryptochrome transcripts following GA3 treatment in wt tomato plants, it is evident that cryptochromes are quite unaffected by rapid change of hormone concentration in the culture medium, the only exception being the upregulation of CRY-DASH (Fig. 2A). On the other hand, in CRY2OX and cry1a- genotypes cryptochrome 1 transcripts are mostly downregulated (Fig. 2A). This hints that CRY1a and CRY2 play an antagonistic role in CRY1a and CRY1b transcriptional regulation, when gibberellin is added.
The transcription pattern of the phytochrome gene family following treatment with GA3, evidenced an opposite response in cry1a- plants with respect to wt and CRY2OX tomatoes (Fig. 2B). Indeed, when a functional form of CRY1a protein is absent, all five phytochromes are constantly downregulated (Fig. 2B); on the contrary, when CRY1a works normally (in wt and CRY2OX plants) the same genes, but PHYB2, appear to be mostly upregulated, especially at ZT12 (Fig. 2B). These results demonstrate that the presence of a CRY1a working protein is a decisive factor for transcript regulation of phytochrome genes. This effect is particularly evident in PHYB1 transcription (Fig. 2B), suggesting a possible role of PHYB1 in regulating the molecular network among hormones, photoreceptors and light in tomato, as an element downstream of CRY1a.
The photoreceptor response to auxin (IAA) treatment is lower than to that of gibberellin (Fig. 3AB). Once again, the most sensitive genotype to exogenous hormone is clearly cry1a-, especially when focusing on the cryptochrome mRNA transcripts: CRY1a, CRY1b and CRY2 are downregulated in at least three time points analyzed (Fig. 3A). In wt and CRY2OX plants no clear pattern of up or downregulation of cryptochrome transcripts was observed (Fig. 3A). CRY1a may play a crucial role in the regulation of chryptochrome expression also under auxin stimulus; however, this role seems to be absent for phytochromes, which are almost totally unaffected in cry1a- plants (Fig. 3B). Therefore, the action of CRY1a over tomato photoreceptor gene transcripts changes according to different hormonal stimuli. Within the phytochrome family only PHYA appears to be sensitive to auxin treatment: indeed, in wt plants this gene is up regulated across the day; this effect is not visible in cry1a- and CRY2OX plants (Fig. 3B), suggesting that CRY1a and CRY2 can play a positive and a negative role, respectively, in the auxin induced alteration of PHYA transcripts.
Generally, ABA does not cause dramatic effects on transcription of cryptochrome genes. Nevertheless some very interesting exceptions must be remarked: the strong upregulation of CRY1a and the downregulation of CRY-DASH in cry1a- plants, as well as, the upregulation of CRY-DASH in CRY2OX tomatoes (Fig. 4A). It is interesting to note that the transcription of CRY-DASH, whose function as photoreceptor has been heavily discussed [78], [79], is influenced by the other two main tomato cryptochromes, at least under hormonal stimulus.
Analyzing phytochrome responses to exogenous ABA in CRY2OX treated-plants, we observed strong upregulation during the day for PHYB1, PHYE and PHYF; conversely, PHYA shows downregulation (Fig. 4B).
In ABA treated cry1a- plants the scenario is completely inverted: PHYA is upregulated at all time points, but ZT20 (presumptive night); contrarily, PHYF is constantly downregulated, with the sole exception of ZT16 (Fig. 4B). In CRY2OX genotype PHYA appears to be downregulated with the exception of ZT12 and ZT16, whereas PHYE and PHYF are up regulated during almost all cycle (Fig. 4B). In general, cryptochrome 1–2 type proteins seem to play a role in phytochrome responses to ABA treatment, in accordance with what was already discussed for gibberellin treatment.
The effect of exogenous t-zeatin on photoreceptor gene expression is very weak and quite independent from the genotype (Fig. S2 and Fig. 1).
Effects of phyto-hormones on tomato CAB4 and GIGANTEA diurnal transcription
Transcription of the photosynthetic gene CAB4 is unaffected by the addition of exogenous t-zeatin in all three genotypes under study (Fig. 5A). On the contrary, GA3-treatment generates significant upregulation in wt plants and a downregulation in both cry1a- and CRY2OX genotypes, especially during the light phase of the day (Fig. 5A). It is surprising that in wt genotype gibberellin can stimulate the expression of a gene like CAB4, implicated in the perception of light stimuli, when Arabidopsis spy mutant, that is hypersensitive to GA, presents a pale phenotype, very similar to photoreceptors mutants [80]. Our data suggest that the upregulation of CAB4 is probably driven by CRY1a and antagonized by CRY2, since they are downregulated in both mutant and overexpressor genotypes after GA3-treatment (Fig. 5A).
A similar situation is evident in auxin-treated plants except that the addition of IAA does not interfere with CAB4 transcription in wt genotype, providing evidence that addition of auxin can alter CAB4 transcription only as a consequence of the abnormal presence of functional cryptochromes (Fig. 5A). Furthermore, downregulation of CAB4 is also evident after ABA treatment but limited to CRY2OX plants (Fig. 5A), evidencing a specific dose-effect of the cryptochrome 2 over ABA induced transcript alterations.
It is known that the expression of the circadian and flowering gene GI is (at least partially) under the control of cryptochromes in Arabidopsis [81], and, more specifically, under the control of CRY1a in tomato [74]. Our results here reveal that GI transcripts are not affected by exogenous adding of t-zeatin, gibberellin and auxin in all the three genotypes observed (Fig. 5B); on the contrary, GI is very sensitive to ABA, but only in cry1a- plants, where its transcripts are dramatically upregulated during the part of the day in which GI is more expressed (from ZT6 to ZT16) (Fig. 5B). In a recent work [74], we have already demonstrated that the lack of an active form of CRY1a causes downregulation of GI; these new experiments highlight that in cry1a- plants CRY1a and ABA signaling components are redundant in maintaining optimal GI expression, resulting, most likely, in fine modulation of numerous important physiological processes in tomatoes.
Concluding remarks
The main finding of this work is that without a functional cryptochrome 1a, both GA3 and IAA can perturb the diurnal expression pattern of tomato photoreceptors: GA3 downregulates both cryptochrome and phytochrome expression pattern, whereas IAA is able to downregulate cryptochrome diurnal transcription.
Data presented here reveal a substantial degree of control of cryptochromes (especially CRY1a) over the regulatory networks formed by phytohormones, light and photoreceptors. We demonstrated that cryptochromes have a main role in the regulation of the diurnal expression pattern of both cryptochrome and phytochrome genes under hormonal stimulus. Particularly, the absence of a working CRY1a protein makes “the tomato system” more sensitive to changes of phyto-hormone concentration in the growing medium.
In cry1a- tomatoes, most photoreceptors, especially phytochromes, become repressible by GA addition. The loss of photoperception via CRY1a is able to compound the skotomorphogenic phenotype caused by gibberellin action, as in that combined situation the transcription of most other photoreceptors is also repressed; CRY2 overexpression can, in some cases (PHYB1, PHYE),antagonize this action.
Moreover, under the given treatments, cryptochrome 1a, and in a milder manner cryptochrome 2, can regulate not only the expression of photoreceptor gene transcripts, but also the transcription pattern of genes involved in photosynthetic processes and circadian rhythm, as CAB4 and GI. This hints a major involvement of phyto-hormones in mediating the physiological response of plants to light stimuli by an interaction with photoreceptors.
Materials ands Methods
Standard molecular biology protocols were followed as described in Sambrook and colleagues [82].
Plant material
All experiments were carried out in Solanum lycopersicum (cv Moneymaker) background, cry1a- (80B mutant) and transgenic CRY2OX seeds [24], [25]. Tomato seeds were germinated in standard paper towels. After germination, uniform seedlings were placed into transparent plastic boxes (14 seedlings of the same genotype per box) and grown hydroponically for 28 days in a growth chamber in LD conditions (16 h light/8 h dark-25°C) without humidity control. Light intensity of about 50 µmol m−2 s−1 was provided by Osram (Munich) 11–860 daylight lamps. The composition of the full nutrient solution used during the plant growth was: 1 mM MgSO4, 2.5 mM Ca(NO3)2, 2 mM KNO3, 0.1 mM K2HPO4, 10 µM Fe-EDDHA, 10 µM B, 2 µM Mn, 1 µM Zn, 0.5 µM Cu, 0.2 µM Mo, 0.2 µM Co, 0.2 µM Ni and 25 µM Cl [83]. Nutrient solution was replaced in each box every 2 days. The solution pH was maintained at 7.5 with CaCO3. At ZT -2 (ZT- Zeitgeber time = number of hours after the onset of illumination) [84] of the 29th day of growth, 20 µM phyto-hormones were added to nutrient solution of test-plants (this hormone concentration is within experimental ranges commonly used pharmacologically for a given phyto-hormone, and is within a physiologic range); control-plants were let in the standard nutrient solution. The aerial parts of 10–14 plants for each genotype (wt, cry1a- and CRY2OX) both for treated and control plants were harvested at the times shown.
Quantitative RT-PCR
Total RNA (1 µg) was reverse-transcribed with oligo-dT and Superscript III (Invitrogen), according to the manufacturer's instructions. First strand cDNA (5 ng) was used as template for QRT-PCR. QRT-PCR assays were carried out with gene-specific primers, using an ABI PRISM 7900HT (Applied Biosystems) and the Platinum SYBR Green master mix (Invitrogen), according to manufacturer's instructions. The primer sequences are:
CRY1a TCCTTGCTAACTTTTTGTTAGTATCTGTG; TACGATCTTTTGTTAGCCTGCCT
CRY1b: ATATCGATGTAATGCAAGAACTATGGA; TCTGGTACAGAGAAGTAGAGGCATCA
CRY2: CAAAGGGTGCCATCAATGC; GCTTGTTATCATTGAGCTTCTTTGTT
CRY-DASH: GACACTCTCCTGGAATGATG; CACCAGTCTTCTTGGTATATCC
PHYA: GAATCGAAGGTGACTATAGAGCGATT; GAACACCAGCCAAATTGATCAG
PHYB1: GGGCTTCCTCCTGAATTGG; GCTCAGTCCTAGGCCTTCCTG
PHYB2: TGATTTCTTACAGATTATGGCAAGCT; TTGGTCGAAGATGGACTTCTACC
PHYE: TTGCTTAGTGTAGTGCACCATGC; GTTTCAAACCAGGTAACACCTTGA
PHYF: TTGAGCAAGGATCAAAGGCA; GTGTCGTCAATGATCTTGGCTAGT
GI: GCAACCATTGGAAAACAAAG; CAGACAGAAGCAAGGACATAAG
CAB: GAAGGGTCCAATTGAGAAC; GTACAAAGTTTGTCCCGTAAG
ACTIN: AGGTATTGTGTTGGACTCTGGTGAT; ACGGAGAATGGCATGTGGAA.
PCR conditions were: 5 min at 95°C, followed by 45 cycles at 95°C for 15 sec, and at 58°C for 60 sec. At the end of the PCR, the thermocycler has been programmed to generate a thermal denaturation curve of the amplified DNA and to measure the melting temperature of the PCR product(s). The shape of the melting curve indicates whether the amplified products are homogeneous and the melting temperature provides confirmation that the correct product has been specifically amplified. Relative template abundance was quantified using the relative standard curve method described in the ABI PRISM 7900HT manual and the data were normalized for the quantity of the β-actin transcript [85], an housekeeping gene whose transcripts do not oscillate during the day (data not shown). A serial dilution of 10-, 100-, 1000-,10000-, and 100000-fold of each studied gene fragment was used to determine the amplification efficiency of each target and housekeeping gene. At least three PCR runs were carried out for each cDNA to serve as technical replicates and two independent experiments were carried out by using two biological replicates for each genotype. Means from two independent experiments were subjected to SEM calculation, Student's t test using PAST.
Supporting Information
Effect of CRY1a loss-of-function and CRY2 over-expression on diurnal expression of tomato cryptochrome (A), phytochrome (B) and GIGANTEA / CAB4 (C) genes. Wt, cry1a- and CRY2OX tomato plants were grown hydroponically under LD conditions. The abundance of the mRNAs was measured by QRT-PCR. Results are presented as a proportion of the highest value after normalization with β-actin. Yellow-black box along the horizontal axis represents light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. Data shown are the average of two biological replicates, with error bars representing SEM. Time points of CRY2OX and cry1a- genotypes, significantly different from the corresponding ones in wt genotype are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001).
(DOCX)
Diurnal expression pattern of Cryptochrome (A) and Phytochrome (B) transcripts analyzed by QRT-PCR in wt , cry1a - and CRY2OX t-ZEATIN-treated tomato plants. Results are presented as a ratio after normalization with β-actin. Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. The control data, of gene expression in the absence of hormone applications, are reproduced, for clarity, from those in Figure S1. Data shown are the average of two biological replicates, with error bars representing SEM. Hormone-treated plant transcripts significantly different from the corresponding ones of control plants are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001). Data from control plants are replotted from Figure 2.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Work supported by the Italian Ministry of Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, et al. Gibberellin biosynthesis and response during Arabidopsis seed germination (vol 15, pg 1591, 2003). Plant Cell. 2004;16:783–783. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Penfield S, Hall A. A Role for Multiple Circadian Clock Genes in the Response to Signals That Break Seed Dormancy in Arabidopsis. Plant Cell. 2009;21:1722–1732. doi: 10.1105/tpc.108.064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai T, Nagashima A, Uehara Y. The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-Naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant and Cell Physiology. 2008;49:1250–1255. doi: 10.1093/pcp/pcn092. [DOI] [PubMed] [Google Scholar]
- 4.Deng XW, Feng SH, Martinez C, Gusmaroli G, Wang Y, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–U479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Yu X, Foo E, Symons GM, Lopez J, et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007;145:106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis SJ, Hanano S, Domagalska MA, Nagy F. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes to Cells. 2006;11:1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 7.Kendrick R, Kronenberg G. Photomorphogenesis in plants. 2nd edn. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- 8.Casal JJ. Phytochromes, cryptochromes, phototropin: Photoreceptor interactions in plants. Photochemistry and Photobiology. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Quail PH. Photosensory perception and signalling in plant cells: new paradigms? Current Opinion in Cell Biology. 2002;14:180–188. doi: 10.1016/s0955-0674(02)00309-5. [DOI] [PubMed] [Google Scholar]
- 10.Sharrock RA, Quail PH. Novel Phytochrome Sequences in Arabidopsis-Thaliana - Structure, Evolution, and Differential Expression of a Plant Regulatory Photoreceptor Family. Genes & Development. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- 11.Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, et al. Phytochromes - Photosensory Perception and Signal-Transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad M, Cashmore AR. Hy4 Gene of a-Thaliana Encodes a Protein with Characteristics of a Blue-Light Photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Ahmad M, Chan J, Cashmore A. CRY2, a second member of the Arabidopsis cryptochrome gene family. Plant Physiology. 1996;110:1047. [Google Scholar]
- 14.Kleine T, Lockhart P, Batschauer A. An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant Journal. 2003;35:93–103. doi: 10.1046/j.1365-313x.2003.01787.x. [DOI] [PubMed] [Google Scholar]
- 15.Quail PH. Phytochrome photosensory signalling networks. Nature Reviews Molecular Cell Biology. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 16.Lin CT, Shalitin D. Cryptochrome structure and signal transduction. Annual Review of Plant Biology. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad M, Cashmore AR. The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant Journal. 1997;11:421–427. doi: 10.1046/j.1365-313x.1997.11030421.x. [DOI] [PubMed] [Google Scholar]
- 18.Casal JJ, Mazzella MA. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiology. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mas P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 20.Perrotta G, Ninu L, Flamma F, Weller JL, Kendrick RE, et al. Tomato contains homologues of Arabidopsis cryptochromes 1 and 2. Plant Mol Biol. 2000;42:765–773. doi: 10.1023/a:1006371130043. [DOI] [PubMed] [Google Scholar]
- 21.Perrotta G, Yahoubyan G, Nebuloso E, Renzi L, Giuliano G. Tomato and barley contain duplicated copies of cryptochrome 1. Plant Cell and Environment. 2001;24:991–997. [Google Scholar]
- 22.Facella P, Lopez L, Chiappetta A, Bitonti MB, Giuliano G, et al. CRY-DASH gene expression is under the control of the circadian clock machinery in tomato. FEBS Lett. 2006;580:4618–4624. doi: 10.1016/j.febslet.2006.07.044. [DOI] [PubMed] [Google Scholar]
- 23.Ninu L, Ahmad M, Miarelli C, Cashmore AR, Giuliano G. Cryptochrome 1 controls tomato development in response to blue light. Plant Journal. 1999;18:551–556. doi: 10.1046/j.1365-313x.1999.00466.x. [DOI] [PubMed] [Google Scholar]
- 24.Weller JL, Perrotta G, Schreuder MEL, van Tuinen A, Koornneef M, et al. Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant Journal. 2001;25:427–440. doi: 10.1046/j.1365-313x.2001.00978.x. [DOI] [PubMed] [Google Scholar]
- 25.Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, et al. Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 2005;137:199–208. doi: 10.1104/pp.104.051987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauser BA, CordonnierPratt MM, DanielVedele F, Pratt LH. The phytochrome gene family in tomato includes a novel subfamily. Plant Molecular Biology. 1995;29:1143–1155. doi: 10.1007/BF00020458. [DOI] [PubMed] [Google Scholar]
- 27.Pratt LH, Cordonnierpratt MM, Hauser B, Caboche M. Tomato Contains 2 Differentially Expressed Genes Encoding B-Type Phytochromes, Neither of Which Can Be Considered an Ortholog of Arabidopsis Phytochrome-B. Planta. 1995;197:203–206. doi: 10.1007/BF00239958. [DOI] [PubMed] [Google Scholar]
- 28.Vantuinen A, Kerckhoffs LHJ, Nagatani A, Kendrick RE, Koornneef M. Far-Red Light-Insensitive, Phytochrome a-Deficient Mutants of Tomato. Molecular & General Genetics. 1995;246:133–141. doi: 10.1007/BF00294675. [DOI] [PubMed] [Google Scholar]
- 29.Vantuinen A, Kerckhoffs LHJ, Nagatani A, Kendrick RE, Koornneef M. A Temporarily Red Light-Insensitive Mutant of Tomato Lacks a Light-Stable, B-Like Phytochrome. Plant Physiology. 1995;108:939–947. doi: 10.1104/pp.108.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerckhoffs LHJ, Kelmenson PM, Schreuder MEL, Kendrick CI, Kendrick RE, et al. Characterization of the gene encoding the apoprotein of phytochrome B2 in tomato, and identification of molecular lesions in two mutant alleles. Molecular and General Genetics. 1999;261:901–907. doi: 10.1007/s004380051037. [DOI] [PubMed] [Google Scholar]
- 31.Weller JL, Schreuder MEL, Smith H, Koornneef M, Kendrick RE. Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato. Plant Journal. 2000;24:345–356. doi: 10.1046/j.1365-313x.2000.00879.x. [DOI] [PubMed] [Google Scholar]
- 32.Gazzarrini S, McCourt P. Cross-talk in plant hormone signalling: What arabidopsis mutants are telling us. Annals of Botany. 2003;91:605–612. doi: 10.1093/aob/mcg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steber CM, Cooney SE, McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;134:1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 36.Collett CE, Harberd NP, Leyser O. Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiology. 2000;124:553–561. doi: 10.1104/pp.124.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahman A, Amakawa T, Goto N, Tsurumi S. Auxin is a positive regulator for ethylene-mediated response in the growth of arabidopsis roots. Plant and Cell Physiology. 2001;42:301–307. doi: 10.1093/pcp/pce035. [DOI] [PubMed] [Google Scholar]
- 38.Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, et al. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant Journal. 2002;29:325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- 39.Neff MM, Fankhauser C, Chory J. Light: an indicator of time and place. Genes & Development. 2000;14:257–271. [PubMed] [Google Scholar]
- 40.Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends in Plant Science. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- 41.Swarup R, Parry G, Graham N, Allen T, Bennett M. Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol. 2002;49:411–426. doi: 10.1007/978-94-010-0377-3_12. [DOI] [PubMed] [Google Scholar]
- 42.Fu XD, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- 43.Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, et al. Comprehensive comparison brassinosteroid-regulated of auxin-regulated and brassinosteroid-regulated genes in arabidopsis. Plant Physiology. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, et al. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiology. 2003;133:1843–1853. doi: 10.1104/pp.103.030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. Plos Biology. 2004;2:1460–1471. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, et al. The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant Journal. 1999;18:315–320. doi: 10.1046/j.1365-313x.1999.00455.x. [DOI] [PubMed] [Google Scholar]
- 47.Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant Journal. 1999;18:303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- 48.Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, et al. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop GJ, Koncz C. Brassinosteroids and plant steroid hormone signaling. Plant Cell. 2002;14(Suppl):S97–110. doi: 10.1105/tpc.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- 52.Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10:791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Ito T, Zhao Y, Peng J, Kumar P, et al. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci U S A. 2004;101:7827–7832. doi: 10.1073/pnas.0402377101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, et al. Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta. 2006;224:995–1003. doi: 10.1007/s00425-006-0280-6. [DOI] [PubMed] [Google Scholar]
- 55.Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiology. 1998;116:455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP. Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 2003;36:203–214. doi: 10.1046/j.1365-313x.2003.01870.x. [DOI] [PubMed] [Google Scholar]
- 57.Colon-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124:1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novakova M, Motyka V, Dobrev PI, Malbeck J, Gaudinova A, et al. Diurnal variation of cytokinin, auxin and abscisic acid levels in tobacco leaves. Journal of Experimental Botany. 2005;56:2877–2883. doi: 10.1093/jxb/eri282. [DOI] [PubMed] [Google Scholar]
- 59.Blazquez MA, Weigel D. Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 1999;120:1025–1032. doi: 10.1104/pp.120.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamiya Y, Garcia-Martinez JL. Regulation of gibberellin biosynthesis by light. Current Opinion in Plant Biology. 1999;2:398–403. doi: 10.1016/s1369-5266(99)00012-6. [DOI] [PubMed] [Google Scholar]
- 61.Vandenbussche F, Verbelen JP, Van Der Straeten D. Of light and length: regulation of hypocotyl growth in Arabidopsis. Bioessays. 2005;27:275–284. doi: 10.1002/bies.20199. [DOI] [PubMed] [Google Scholar]
- 62.Reid JB, Botwright NA, Smith JJ, O'Neill DP, Kerckhoffs LH. Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol. 2002;128:734–741. doi: 10.1104/pp.010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reed JW, Foster KR, Morgan PW, Chory J. Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 1996;112:337–342. doi: 10.1104/pp.112.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao D, Hussain A, Cheng H, Peng J. Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta. 2005;223:105–113. doi: 10.1007/s00425-005-0057-3. [DOI] [PubMed] [Google Scholar]
- 65.Foo E, Ross JJ, Davies NW, Reid JB, Weller JL. A role for ethylene in the phytochrome-mediated control of vegetative development. Plant J. 2006;46:911–921. doi: 10.1111/j.1365-313X.2006.02754.x. [DOI] [PubMed] [Google Scholar]
- 66.Achard P, Liao L, Jiang C, Desnos T, Bartlett J, et al. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007;143:1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–U411. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 68.Van der Straeten D, Vandenbussche F, Pierik R, Millenaar FF, Voesenek LA. Reaching out of the shade. Current Opinion in Plant Biology. 2005;8:462–468. doi: 10.1016/j.pbi.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Symons GM, Reid JB. Hormone levels and response during de-etiolation in pea. Planta. 2003;216:422–431. doi: 10.1007/s00425-002-0860-z. [DOI] [PubMed] [Google Scholar]
- 70.Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, et al. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant Journal. 2007;49:428–441. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 72.Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA Acts in Blue Light Signaling and Has Biochemically Separable Roles in Circadian Clock and Flowering Time Regulation. (vol 143, pg 473, 2007). Plant Physiology. 2010;153:364–364. doi: 10.1104/pp.106.088757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giuliano G, Hoffman NE, Ko K, Scolnik PA, Cashmore AR. A Light-Entrained Circadian Clock Controls Transcription of Several Plant Genes. Embo Journal. 1988;7:3635–3642. doi: 10.1002/j.1460-2075.1988.tb03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Facella P, Lopez L, Carbone F, Galbraith DW, Giuliano G, et al. Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS One. 2008;3:e2798. doi: 10.1371/journal.pone.0002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Foster KR, Morgan PW. Genetic-Regulation of Development in Sorghum-Bicolor .9. The Ma(3)(R) Allele Disrupts Diurnal Control of Gibberellin Biosynthesis. Plant Physiology. 1995;108:337–343. doi: 10.1104/pp.108.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jasoni RL, Cothren JT, Morgan PW, Sohan DE. Circadian ethylene production in cotton. Plant Growth Regulation. 2002;36:127–133. [Google Scholar]
- 77.Blazquez M, Alabadi D. Molecular interactions between light and hormone signaling to control plant growth. Plant Molecular Biology. 2009;69:409–417. doi: 10.1007/s11103-008-9400-y. [DOI] [PubMed] [Google Scholar]
- 78.Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deisenhofer J, Huang YH, Baxter R, Smith BS, Partch CL, et al. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17701–17706. doi: 10.1073/pnas.0608554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacobsen SE, Olszewski NE. Mutations at the Spindly Locus of Arabidopsis Alter Gibberellin Signal-Transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Samach A, Paltiel J, Amin R, Gover A, Ori N. Novel roles for GIGANTEA revealed under environmental conditions that modify its expression in Arabidopsis and Medicago truncatula. Planta. 2006;224:1255–1268. doi: 10.1007/s00425-006-0305-1. [DOI] [PubMed] [Google Scholar]
- 82.Sambrook J, Fritsch E, Maniatis T, editors. Molecular cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 83.Buckhout TJ, Bell PF, Luster DG, Chaney RL. Iron-Stress Induced Redox Activity in Tomato (Lycopersicum esculentum Mill.) Is Localized on the Plasma Membrane. Plant Physiol. 1989;90:151–156. doi: 10.1104/pp.90.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of CRY1a loss-of-function and CRY2 over-expression on diurnal expression of tomato cryptochrome (A), phytochrome (B) and GIGANTEA / CAB4 (C) genes. Wt, cry1a- and CRY2OX tomato plants were grown hydroponically under LD conditions. The abundance of the mRNAs was measured by QRT-PCR. Results are presented as a proportion of the highest value after normalization with β-actin. Yellow-black box along the horizontal axis represents light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. Data shown are the average of two biological replicates, with error bars representing SEM. Time points of CRY2OX and cry1a- genotypes, significantly different from the corresponding ones in wt genotype are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001).
(DOCX)
Diurnal expression pattern of Cryptochrome (A) and Phytochrome (B) transcripts analyzed by QRT-PCR in wt , cry1a - and CRY2OX t-ZEATIN-treated tomato plants. Results are presented as a ratio after normalization with β-actin. Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Time points are measured in hours from dawn (zeitgeber Time [ZT]); data at ZT24 constitute a replotting of those at ZT0. The control data, of gene expression in the absence of hormone applications, are reproduced, for clarity, from those in Figure S1. Data shown are the average of two biological replicates, with error bars representing SEM. Hormone-treated plant transcripts significantly different from the corresponding ones of control plants are marked with a * (Student's t test, P≤0.05), two ** (Student's t test, P≤0.01) and three *** (Student's t test, P≤0.001). Data from control plants are replotted from Figure 2.
(DOC)