Abstract
Reduced signaling of the growth hormone (GH)/insulin-like growth factor-1 (IGF-1) pathway is associated with extended life span in several species. Ames dwarf mice are GH-deficient and live >50% longer than wild-type littermates. Previously, we have shown that tissues from Ames mice exhibit elevated levels of antioxidative enzymes, less H2O2 production, and lower oxidative damage suggesting that mitochondrial function may differ between genotypes. To explore the relationship between hormone deficiency and mitochondria in mice with extended longevity, we evaluated activity, protein, and gene expression of oxidative phosphorylation components in dwarf and wild-type mice at varying ages. Liver complex I + III activity was higher in dwarf mice compared to wild-type mice. The activity of I + III decreased between 3 and 20 months of age in both genotypes with greater declines in wild-type mice in liver and skeletal muscle. Complex IV activities in the kidney were elevated in 3- and 20-month-old dwarf mice relative to wild-type mice. In Ames mice, protein levels of the 39 kDa complex I subunit were elevated at 20 months of age when compared to wild-type mouse mitochondria for every tissue examined. Kidney and liver mitochondria from 20-month-old dwarf mice had elevated levels of both mitochondrially-encoded and nuclear-encoded complex IV proteins compared to wild-type mice (p < 0.05). Higher liver ANT1 and PGC-1α mRNA levels were also observed in dwarf mice. Overall, we found that several components of the oxidative phosphorylation (OXPHOS) system were elevated in Ames mice. Mitochondrial to nuclear DNA ratios were not different between genotypes despite the marked increase in PGC-1α levels in dwarf mice. The increased OXPHOS activities, along with lower ROS production in dwarf mice, predict enhanced mitochondrial function and efficiency, two factors likely contributing to long-life in Ames mice.
Keywords: Ames dwarf mice, Hormones, Aging, Mitochondria, Complex enzymes
Introduction
A progressive decline in physiological function occurs with aging. A combination of protein, DNA, and lipid damage accumulates with time and results in the loss of cellular function thus compromising homeostasis (Kwong and Sohal 2000; Salvioli et al. 2001). Mitochondrially-generated free radicals not countered by normal cellular defenses are believed to induce this cellular damage. This oxidative damage is incurred by both the mitochondria (potentially the most adversely affected organelle) and other cellular components. Damaged mitochondria accumulate and may be responsible for some aspects of the aging phenotype via a disturbed energy budget (Cadenas and Davies 2000; López-Torres et al. 2002; Short et al. 2005). However, several questions remain regarding the physiological relevance of altered mitochondrial function and the damage acquired as organisms age.
Mice with hereditary dwarfism (Ames dwarf) live more than a year longer than their wild-type siblings (Brown-Borg et al. 1996). Ames dwarf mice lack growth hormone (GH), prolactin, and thyrotropin resulting in small body size and delayed puberty. The absence of GH secretion results in undetectable levels of plasma insulin-like growth factor 1 (IGF-1) and therefore, downstream processes are not properly targeted. The biological advantage of reduced GH signaling is related to the enhanced antioxidative defenses, reduced oxidative damage, increased insulin sensitivity, and significant longevity enjoyed by these diminutive mice (Brown-Borg and Rakoczy 2000; Brown-Borg et al. 2001a, b; Brown-Borg 2009; Bartke and Brown-Borg 2004). It has been established that reduced signaling through this pathway confers longevity in mammals (Brown-Borg et al. 1996; Flurkey et al. 2001; Coschigano et al. 2000), flies (Tatar et al. 2001; Clancy et al. 2001), worms (Kimura et al. 1997), and yeast (Fabrizio et al. 2001). Endogenous enzymes that counter oxidative stress in the mitochondrial and cellular compartments are elevated in multiple tissues in Ames mice and include catalase, glutathione peroxidase, and superoxide dismutase 2 (manganese SOD; Brown-Borg et al. 1999; 2002; Brown-Borg and Rakoczy 2000; Hauck and Bartke 2001, and 2000). In turn, mitochondrial hydrogen peroxide production is significantly lower in liver tissue from dwarf mice (Brown-Borg et al. 2001a,b). In support of this evidence, the enhanced protein and activity levels of endogenous antioxidant enzymes are functional as they provide resistance to both in vivo (Bartke 2000; Bokov et al. 2009) and in vitro (Salmon et al. 2005) oxidative challenge resulting in Ames dwarf mice and cells derived from these mice, out-surviving their wild-type counterparts.
To further investigate the role of mitochondria and their relationship to hormone deficiency in the extended life span of the Ames mice, the current study determined protein and mRNA expression and activity levels of several components of the oxidative phosphorylation system. The results of this study suggest that mitochondrial components are altered in long-living mice when compared to age-matched wild-type mice and may play a role in the longevities observed.
Materials and methods
Animals Ames dwarf mice were bred and maintained at the University of North Dakota (UND) vivarium facilities under controlled conditions of photoperiod (12 h light/12 h dark) and temperature (22 ± 1°C) with ad libitum access to food (Harlan Teklad, Wilmington, DE, USA; Lab-diet 8640— ≥ 18%crude protein/5% fat) and water (standard laboratory conditions). The Ames dwarf (df/df) mice used in this study were derived from a closed colony with heterogeneous background (over 25 years). Dwarf mice were generated by mating either homozygous (df/df) or heterozygous (df/+) dwarf males with carrier females (df/+). All procedures involving animals were reviewed and approved by the UND Institutional Animal Care and Use Committee. For reference, the average lifespan of the wild-type mice in our colony is 23–24 months (Brown-Borg et al. 1996). All chemicals were obtained from Sigma (St. Louis, MO, USA) unless otherwise noted.
OXPHOS protein activities and expression Levels of oxidative phosphorylation (OXPHOS) proteins and activity were evaluated in mitochondrial preparations to determine potential locations of altered function in dwarf tissues. Liver, kidney, and hind-limb skeletal muscle of dwarf and corresponding wild-type animals at 3, 12, and 20 months of age (n = 8/age/genotype) were collected. Body weights as well as liver and brain weights were recorded the same day as tissue collection.The activity of complex IV was measured and linked assays (described below) were used to measure electron flow through complexes I, II, and III. Liver, muscle, and kidney tissue mitochondria were isolated using standard techniques as described by Trounce et al. (1996). Briefly, fresh tissue was homogenized in buffer (0.25 M sucrose and 10 mM HEPES, pH 7.4) and centrifuged for 1,500×g for 5 min at 4°C (2×). Centrifugation was repeated at 10,000×g for 15 min at 4°C to pellet mitochondria and followed by one wash. Ten to 50 μg of mitochondrial protein were used in each assay depending on the enzyme examined. Protein concentrations were determined using the Bradford (1976) assay.Complex I + III (NADH-cytochrome c oxidoreductase) and complex II + III (succinate-cytochrome c reductase) activities were measured according to procedures described by Kuznetsov et al. (1996). For complex I + III activity, a reaction mixture containing 50 μM ferricytochrome c, 1 mM KCN, and 100 mM K2HPO4 maintained at 30°C was added to the cuvettes. The reaction was initiated by adding 10 mM NADH. For complex II + III activity, a reaction mixture containing 50 μM ferricytochrome c, 1 mM KCN, and 100 mM K2HPO4 maintained at 30°C was added to the cuvettes. The reaction was initiated by adding 20 mM succinate after which the cuvette contents were mixed and the change in absorbance was monitored at a wavelength of 550 nm for 3 min to determine the amount of reduced cytochrome c formed (calculated using a molar extinction coefficient of 19,600 M−1 cm−1). For all enzyme assays, the appropriate controls and blanks were utilized.Complex IV (cytochrome c oxidase) activity was measured by following the oxidation of reduced cytochrome c at 550 nm as described by Cooperstein and Lazorow (1951). Ferrocytochrome c was prepared by dissolving cytochrome c in 10 mM K2HPO4 and adding ascorbate to reduce the cytochrome. The mixture was loaded onto a Sephadex G-25 column and fractions collected. The reduced cytochrome was added to 20 mM in 10 mM K2HPO4. The sample was added and the rate of cytochrome c oxidation was measured by monitoring the decrease in absorbance for 1 min at 550 nm at 30°C. The amount of reduced cytochrome c oxidized was calculated using a molar extinction coefficient of 19,600 M−1 cm−1.Protein expression was determined using previously developed immunoblotting procedures (Brown-Borg and Rakoczy 2000) that employed primary antibodies to specific subunits of complexes I, II, III, IV, and V in tissue mitochondria. The following antibodies from Molecular Probes (Eugene, OR, USA) were used: 39 kDa subunit of complex I (NADH dehydrogenase; clone 20C11), 70 kDa subunit of complex II (succinate dehydrogenase; Clone 2E3), core 2 subunit of complex III (50 kDa; cytochrome bc1; clone 13G12), the 57 kDa mitochondrially-encoded subunit I of complex IV (cytochrome c oxidase; clone 1D6), the 20 kDa nuclear-encoded subunit IV of complex IV (clone 20E8) and the α-subunit of complex V (55 kDa; ATP synthase; Clone 7H10). Differences in levels of OXPHOS proteins were determined densitometrically (UVP Bioimaging Systems, Upland, CA, USA). Ponceau-S staining of membranes was used to evaluate equal loading of protein.
Gene expression Gene expression was evaluated in liver, whole brain, and hind-limb skeletal muscle tissue of 12-month-old dwarf and wild-type mice using real-time reverse transcription polymerase chain reaction (RT-PCR) techniques. Total RNA was extracted from tissues using Ultraspec RNA (Biotecx, Houston, TX, USA; based on method by Chomczynski and Sacchi, 1987). Two micrograms of total RNA were utilized to synthesize cDNA and perform real time quantitative PCR using a QuantiTect SYBR Green RT-PCR kit (Qiagen; Valencia, CA, USA) according to the manufacturer’s protocol. The reaction mixtures contained SYBR green, forward, and reverse primers (Table 1), and QuantiTect RT mix and were assayed using a SmartCycler instrument (Cepheid, Sunnyvale, CA, USA). The reverse transcription reaction ran for 30 min at 50°C. The initial PCR activation step included a 15-min incubation at 95°C followed by 35 cycles of 94°C (15 s), 60°C (30 s), and 72°C (30 s). An annealing temperature of 60°C was used for all primer sets except β2-microglobulin (B2M) (62°C; Table 1). Gene expression was quantitated by using the comparative cycle threshold (CT) method (Heid et al., 1996). CT, the threshold cycle, is the number of cycles it takes for a sample to reach the level where the rate of amplification is the greatest during the exponential phase. ΔCT was obtained for each sample/gene by the following calculation: ΔCT = CT,X − CT,R, where CT,X is the threshold number for target gene amplification and CT,R is the threshold number for reference gene (B2M) amplification. The amount of target (in Ames mice), normalized to an endogenous reference (B2M) and relative to the control group (wild-type mice), is given by the formula 2 ΔΔCT; ΔΔCT = ΔCT,q − ΔCT,cb, where ΔCT, q = ΔCT for an individual sample and ΔCT,cb = mean ΔCT for control group. Thus, the value of 2 ΔΔCT for the amount of target in the wild-type was 1. For the amount of target in the Ames mice, a onefold change indicates no change, greater than onefold change indicates upregulation, and less than onefold change indicates downregulation. Real-time PCR procedures were also utilized to determine the relative amounts of nuclear and mitochondrial DNA levels for comparison between genotypes (n = 11–12/age/genotype). The methods of Wong and Cortopassi (2002) were followed with slight modifications. Briefly, DNA was isolated using a Qiagen DNA extraction kit according to manufacturers instruction. The genes targeted were nuclear cystic fibrosis and mitochondrial nicotinamide adenine dinucleotide dehydrogenase-5. The mouse-specific primer pairs are listed in Table 1. For quantification of nuclear DNA, 10 ng of DNA was used as a template while 0.1 ng DNA was used for mitochondrial DNA quantification. The following cycling parameters were utilized: 50°C for 2 min followed by 95°C for 10 min, then 40 cycles of 95°C (15 s) and 60°C (60 s) then 95°C (15 s), 60°C (15 s), and 95°C (15 s). The linearity of the amplification curve was analyzed using the Cepheid software and the mean cycle time of the linear portion of the curve was designated Ct. The relative mitochondrial copy number to nuclear copy number was assessed by the comparative Ct method described above.
Table 1.
Primer sets (5′-3′) used for real-time PCR analysis of gene expression
| Gene of interest | Forward primer | Reverse primer |
|---|---|---|
| Cystic fibrosis | tgttgtgaagacgagctgatg | tgcattaaaagagagcatgtgttg |
| Nicotinamide adenine dinucleotide dehydrogenase-5 | tggatgatggtacggacgaa | tgcggttatagaggattgcttgt |
| Complex I (ND1) mitochondrially-encoded | caggatgagcctcaaactcc | ggtcaggctggcagaagtaa |
| Complex I (ND2) | agggatcccactgcacatag | cctatgtgggcaattgatga |
| Complex II (SDHC) nuclear-encoded | acaaatggtctcttcctatggca | cccctccactcaaggctattc |
| Complex III (cytb) nuclear-encoded | acgtccttccatgaggacaa | gaggtgaacgattgctaggg |
| Complex IV (cox1a) mitochondrially-encoded | cttttatcctcccaggatttgg | gctaaatactttgacaccgg |
| Complex IV (cox5a) nuclear-encoded | ctttaaatgaattgggaatctccac | gcccatcgaagggagtttaca |
| Complex V (atp6) | aattacaggcttccgacacaaac | tggaattagtgaaattggagttcct |
| Peroxisome proliferator-activated receptor γ coactivator 1 alpha | caatgaatgcagcggtctta | gtgtgaggagggtcatcgtt |
| Insulin-like growth factor 1 | ctgagctggtggatgctctt | cactcatccacaatgcctgt |
| Adenine nucleotide transporter 1 | acttcgccttcaaagacaagtaca | gcgccagaactgcttatgg |
| β2-microglobulin | aagtatactcacgccaccca | aagaccagtccttgctgaag |
Statistical analysis In each experiment, differences between means were assessed utilizing Prism (Graphpad, San Diego, CA, USA). For activity assays, two-way analyses of variance and when appropriate, Bonferroni post hoc testing was used to determine significant differences among means. For comparison of protein levels within age, based on densitometric analysis, Students t tests were employed. Genotype differences in gene expression were compared using Student’s t tests. Data are reported as mean ± SEM.
Results
Relative levels of mitochondrial oxidative phosphorylation enzyme activities and protein were determined in long-living Ames dwarf mice and their wild-type littermates. Three ages of mice were evaluated to ascertain whether a change in activity with age was detectable. Potential differences in gene expression were also examined in 12-month old dwarf and wild-type tissue samples. As reported previously, body weights of 3, 12, and 12-month-old dwarf mice were lower than wild-type mice at each age (3-month dwarf, 10.15 ± 0.26 vs. wild type 30.29 ± 1.35 g; 12-month dwarf, 15.02 ± 1.84 vs. wild type 32.44 ± 1.20; 20-month dwarf, 16.06 ± 1.01 vs. wild-type 36.65 ± 0.87 g; p < 0.0001 age; p < 0.0001 genotype). Liver weights were also lower in dwarf mice in comparison to wild-type mice at each age (3-month dwarf, 0.44 ± 0.02 vs. wild-type 1.75 ± 0.12; 12-month dwarf, 0.61 ± 0.05 vs. wild type 1.48 ± 0.08; 20-month dwarf, 0.70 ± 0.06 vs. wild type 1.87 ± 0.10 g; p = 0.0075 age; p < 0.0001 genotype; p = 0.018 interaction). Whole brain weights were lower in dwarf mice but when expressed on a body weight basis, the brain represented a higher percentage of body weight in comparison to wild-type mice (3-month dwarf 3.6% vs. 3-month wild type 1.5%; 3-month dwarf, 0.36 ± 0.01 vs. wild type 0.46 ± 0.01; 12-month dwarf, 0.37 ± 0.02 vs. wild type 0.41 ± 0.02; 20-month dwarf, 0.38 ± 0.01 vs. wild type 0.44 ± 0.01 g; p < 0.0001 genotype).
To ensure that the levels of protein and activities of OXPHOS enzymes were not due to differences in mitochondrial numbers, the ratio of mitochondrial DNA to nuclear DNA was determined in two representative tissues. Neither liver nor brain tissues exhibited differences in this ratio at 12 and 24 months of age (data not shown). The activities of OXPHOS enzymes were measured in mitochondria of young, adult and old dwarf and wild-type mice. Complex I + III activity in liver mitochondria was significantly influenced by genotype (p = 0.01) and was elevated in dwarf compared to wild-type mitochondria at all ages (Table 2). However, genotype did not influence complex I + III activity in kidney or skeletal muscle mitochondria. Age significantly affected complex I + III activity in liver (p < 0.0001) and skeletal muscle (p < 0.0001) mitochondria. Complex I + III activity in liver mitochondria decreased 56% and 71% between 3 and 20 months of age in dwarf and wild-type mice, respectively. Complex I + III activity in muscle mitochondria decreased 35% and 44% between 3 and 20 months of age in dwarf and wild-type mice, respectively. Although complex I + III activity in kidney mitochondria was not influenced by gender or age, the gender x age interaction was significant (p = 0.018). Comparison of individual means showed that dwarf kidney mitochondria had lower I + III activity at 3 months of age relative to wild-type mitochondria at the same age (p = 0.0022), but at 12 and 20 months of age, no differences were observed. In contrast, kidney activity levels in the dwarf increased 29% between 3 and 20 months of age while in wild-type animals, the activity levels exhibited an 18% decrease. Thus, in all wild-type tissues and all but kidney in the dwarf, complex I + III levels declined with age. However, the age-related declines in complex I + III activity were greater in the wild-type mice.
Table 2.
Complex I + III activity (nmol/min × mg) in tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice
| Genotype | Dwarf | Wild-type | Dwarf | Wild-type | Dwarf | Wild-type | P value | ||
|---|---|---|---|---|---|---|---|---|---|
| Age | 3 Months | 3 Months | 12 Months | 12 Months | 20 Months | 20 Months | Genotype | Age | Interaction |
| Liver | 387.9 ± 43.5 | 306.6 ± 33.9 | 166.2 ± 8.6 | 142.5 ± 17.3 | 170.8 ± 28.9 | 87.7 ± 15.6 | 0.0100 | <0.0001 | 0.5054 |
| Kidney | 367.1 ± 9.9 | 540.1 ± 45.3 | 418.5 ± 55.4 | 401.7 ± 45.9 | 473.7 ± 23.0 | 440.6 ± 29.0 | 0.1988 | 0.4006 | 0.0175 |
| Skeletal muscle | 306.2 ± 32.1 | 353.8 ± 36.4 | 387.5 ± 47.8 | 373.9 ± 49.5 | 199.0 ± 10.6 | 198.7 ± 8.7 | 0.7083 | <0.0001 | 0.6680 |
Values represent mean ± SEM. P values represent results of a two-way ANOVA. n = 7–8/genotype/age
The activity of complex II + III in liver mitochondria was not influenced by genotype, age, or genotype × age interaction (Table 3). Genotype significantly influenced complex II + III activity in skeletal muscle mitochondria (p = 0.0012). Overall, the activity was lower in dwarf compared to wild-type mice at all ages. Complex II + III activity in kidney mitochondria was influenced by a significant genotype × age interaction (p = 0.0204). This interaction arose because the activity in dwarf mitochondria was 43% higher than the activity in wild-type mitochondria at 20 months of age (p = 0.004, Bonferroni contrast). In contrast to the age-related pattern in complex I + III activity where activities primarily declined with age, the activities of complex II + III were either not affected by age as found in liver or the age-related differences were confounded by significant genotype × age interactions as found in kidney. Mitochondria from dwarf mice tended to show increases in kidney and muscle activity between 3 and 20 months of age while in wild-type mice, activity levels in muscle appeared to decline between 3 and 20 months of age. In skeletal muscle, activity levels at 12 months of age tended to be higher than that observed in younger mice.
Table 3.
Complex II + III activity (nmol/min × mg) in tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice
| Genotype | Dwarf | Wild-type | Dwarf | Wild-type | Dwarf | Wild-type | P value | ||
|---|---|---|---|---|---|---|---|---|---|
| Age | 3 Months | 3 Months | 12 Months | 12 Months | 20 Months | 20 Months | Genotype | Age | Interaction |
| Liver | 107.2 ± 8.2 | 105.2 ± 5.6 | 111.5 ± 11.8 | 85.6 ± 15.1 | 102.7 ± 13.9 | 113.9 ± 12.3 | 0.5735 | 0.6862 | 0.2893 |
| Kidney | 194.2 ± 6.1 | 188.1 ± 17.7 | 155.7 ± 10.0 | 145.2 ± 11.9 | 254.6 ± 17.3 | 178.6 ± 12.1 | 0.0066 | <0.0001 | 0.0204 |
| Skeletal muscle | 65.0 ± 4.8 | 139.8 ± 19.5 | 105.9 ± 11.5 | 142.4 ± 22.3 | 84.5 ± 8.2 | 98.1 ± 10.2 | 0.0012 | 0.0861 | 0.1204 |
Values represent mean ± SEM. P values represent results of a two-way ANOVA. n = 7–8/genotype/age
Genotype and age played significant roles in the activities of complex IV in several tissues. Complex IV activity in liver mitochondria was highest in 20-month-old mice (p = 0.0006 for effect of age) but was not affected by genotype (Table 4). The activity of complex IV in kidney mitochondria was highest in 3- and 20-month-old mice (p < 0.0001 for effect of age) and was higher, overall, in dwarf mice (p = 0.0252 for effect of genotype). In muscle mitochondria, complex IV activity was lowest in 20-month-old mice (P = 0.0005 for effect of age) but was lower, overall, in dwarf mice.
Table 4.
Complex IV activity (nmol/min × mg) in tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice
| Genotype | Dwarf | Wild-type | Dwarf | Wild-type | Dwarf | Wild-type | P value | ||
|---|---|---|---|---|---|---|---|---|---|
| Age | 3 Months | 3 Months | 12 Months | 12 Months | 20 Months | 20 Months | Genotype | Age | Interaction |
| Liver | 1,369 ± 103 | 1,238 ± 124 | 1,252 ± 65 | 1,313 ± 82 | 1,755 ± 106 | 1,528 ± 64 | 0.2067 | 0.0006 | 0.3100 |
| Kidney | 2,307 ± 131 | 1,986 ± 36 | 1,609 ± 30 | 1,580 ± 71 | 2,630 ± 100 | 2,048 ± 204 | 0.0252 | <0.0001 | 0.2151 |
| Skeletal muscle | 1,587 ± 96 | 1,920 ± 219 | 1,557 ± 167 | 1,952 ± 276 | 961 ± 71 | 1,256 ± 102 | 0.0213 | 0.0005 | 0.9595 |
Values represent mean ± SEM. P values represent results of a two-way ANOVA. n = 7–8/genotype/age
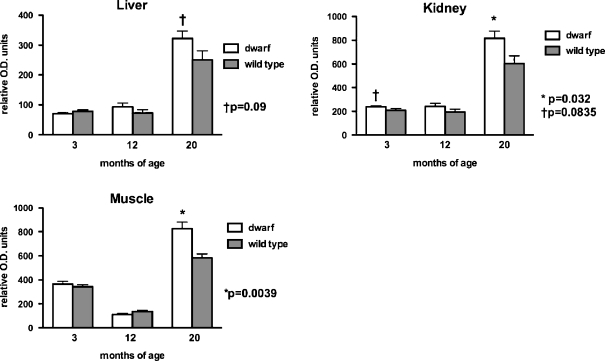
To complement activity levels, several OXPHOS proteins were also measured in tissue mitochondrial samples using standard immunoblotting techniques. Levels of complex I (evaluated using an antibody that detects the 39 kDa subunit) were significantly increased in kidney and muscle mitochondria from 20-month-old dwarf mice when compared to age-matched wild-type mice (Fig. 1) and tended to be elevated in liver mitochondria of 20-month-old dwarf mice. In addition, mitochondrial complex I proteins tended to be elevated in kidney mitochondria of 3-month-old dwarf mice. No differences in protein levels of complex I were observed at 12 months of age in any tissue. Protein levels between ages were not compared as each age was assayed independently.
Fig. 1.
Complex I (39 kDa subunit) protein levels in tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice. Values represent mean ± SEM. N = 7–8/age/genotype. Protein levels are expressed as relative optical density (OD) units. Separate gels were utilized per age group; therefore, statistical comparisons were performed between genotypes within an age group and not across age groups
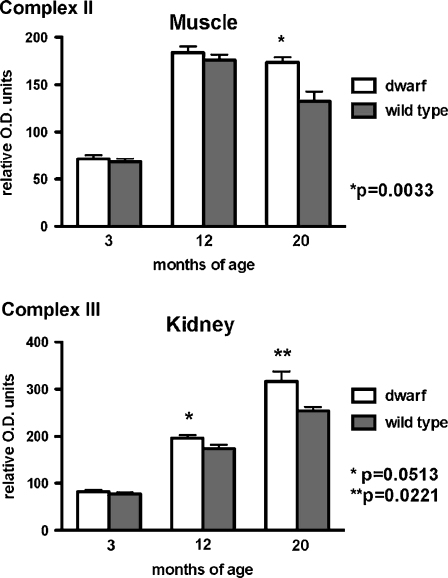
Fewer differences between dwarf and wild-type mitochondrial expression were detected when complex II protein levels were evaluated using antibodies that detect the 70 kDa subunit (Fig. 2). Three-month-old dwarf liver mitochondria expressed 15% less protein compared to wild-type mice (dwarf 753.6 ± 39.8 vs. wild type 882.4 ± 55.1 relative O.D. units; p = 0.087). Kidney mitochondria did not exhibit genotype differences in levels of complex II. The content of complex II was higher in hindlimb muscle mitochondria from 20-month-old dwarf mice compared to age-matched wild-type mice (p = 0.0033; Fig. 2).
Fig. 2.
Complex II (70 kDa subunit) and complex III (core 2 subunit) protein expression in tissue mitochondria from dwarf and wild-type mice at 3, 12, and 20 months of age. Values represent mean ± SEM. N = 5–8/age/genotype. Protein levels are expressed as relative optical density (OD) units. Separate gels were utilized per age group; therefore, statistical comparisons were performed between genotypes within an age group and not across age groups
Significantly greater expression of the core 2 subunit of complex III protein was observed only in kidney tissue mitochondria from dwarf mice at 12 (p = 0.0513) and 20 (p = 0.0221) months of age compared to age-matched wild-type mice (Fig. 2). No genotype differences in complex III protein were observed in mitochondria from liver or skeletal muscle at any age examined.
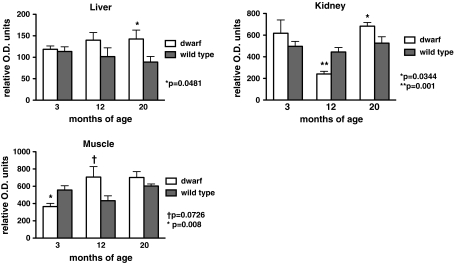
Protein levels from two different subunits of complex IV were determined in mitochondria from both genotypes. Subunit I of complex IV (IV-I) is mitochondrially-encoded while subunit IV of complex IV (IV-IV) is a nuclear-encoded protein. Complex IV-I content (Fig. 3) in liver mitochondria was greater in dwarf mice compared to wild-type mice at 20 months of age (p = 0.0481). Similarly, kidney mitochondria from 20-month-old dwarf mice exhibited higher levels of complex IV-I when compared to age-matched wild-type mice (p = 0.0344). However, at 12 months of age, dwarf kidney mitochondria contained less IV-I protein than wild-type mice (p = 0.001). At 12 months of age, complex IV-I content in skeletal muscle tended to be higher in dwarf mice (p = 0.0726) while the content at 3 months of age was significantly lower in dwarf mice (p = 0.008).
Fig. 3.
Complex IV (subunit I) protein levels in liver, kidney, and skeletal muscle tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice. Values represent mean ± SEM. N = 7–8/age/genotype. Protein levels are expressed as relative optical density (OD) units. Separate gels were utilized per age group; therefore, statistical comparisons were performed between genotypes within an age group and not across age groups
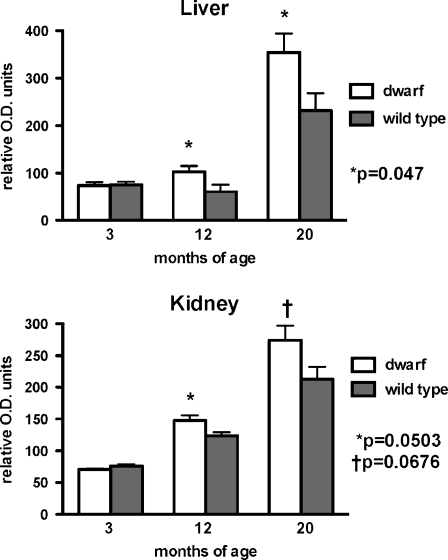
The expression of the nuclear-encoded complex IV-IV was not altered in mitochondria of 3-month-old animals from any tissue or genotype tested. However, the content of complex IV-IV was higher in both liver (p = 0.047) and kidney (p = 0.0503) mitochondria from dwarf mice compared to age-matched wild-type mice at 12 months of age (Fig. 4). The complex IV-IV content was also higher in liver and kidney mitochondria from 20-month-old dwarf mice compared to age-matched wild-type mice. Muscle tissue mitochondria from dwarf and wild-type mice did not differ significantly in the levels of this protein.
Fig. 4.
Complex IV (subunit IV) protein levels in liver and kidney tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice. Values represent mean ± SEM. N = 6–8/age/genotype. Levels are expressed as relative optical density (OD) units. Separate gels were utilized per age group; therefore, statistical comparisons were performed between genotypes within an age group and not across age groups
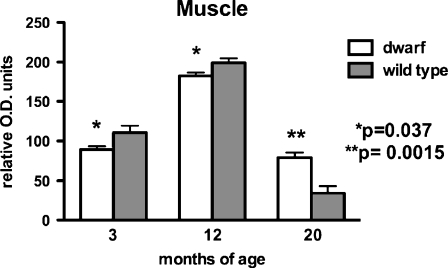
Complex V protein expression was measured using antibodies that detect the α-subunit. No significant differences in expression were detected between dwarf and wild-type mitochondria in the proliferative type tissues, liver, and kidney. The level of this complex was lower in muscle mitochondria from 3- and 12-month-old dwarfs when compared to age-matched wild-type mice (p = 0.037; Fig. 5). However, at 20 months of age, complex V content was higher in muscle mitochondria of dwarf mice (p = 0.0015).
Fig. 5.
Complex V (α-subunit) protein levels in skeletal muscle tissue mitochondria from 3-, 12-, and 20-month old Ames dwarf and wild-type mice. Values represent mean ± SEM. N = 7–8/age/genotype. Levels are expressed as relative optical density (OD) units. Separate gels were utilized per age group; therefore, statistical comparisons were performed between genotypes within an age group and not across age groups
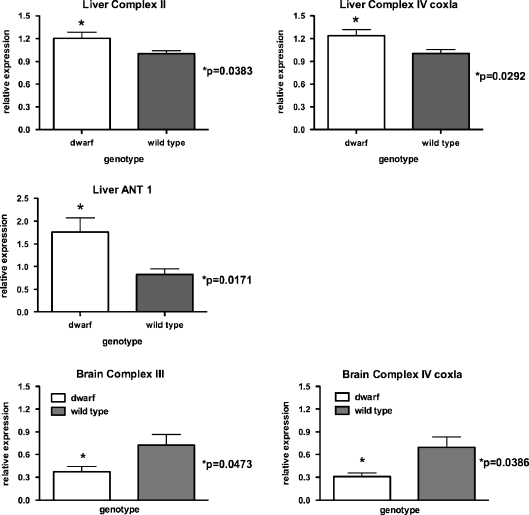
Representative OXPHOS genes from each complex were measured in liver, brain, and muscle tissues from 12-month-old dwarf and wild-type mice. In liver (Fig. 6), mRNA expression of complex II (p = 0.0383), complex IV (cox1a; p = 0.0292) and ANT 1 (p = 0.0171) were higher in dwarf compared to wild-type mice. No genotype differences were detected in complexes I (ND1, ND2), III, IV (cox5a), or V mRNA in liver tissue. The variability in mRNA expression in muscle tissue was high and thus none of the OXPHOS genes were observed to differ between genotypes (data not shown). In brain (Fig. 6), mRNA expression for complex III (p = 0.0473) and complex IV (cox1a; p = 0.0386) was lower in dwarf mice compared to wild type. Genotype did not affect expression of mRNA for complexes I, II, IV (cox5a), V, or ANT in brain.
Fig. 6.
OXPHOS gene expression in 12-month-old dwarf and wild-type mouse liver and brain tissue. Real-time PCR analysis of mRNA normalized to β2-microglobulin. Values represent mean ± SEM. N = 5–8/age/genotype. mRNA levels are expressed as relative expression to wild-type mice
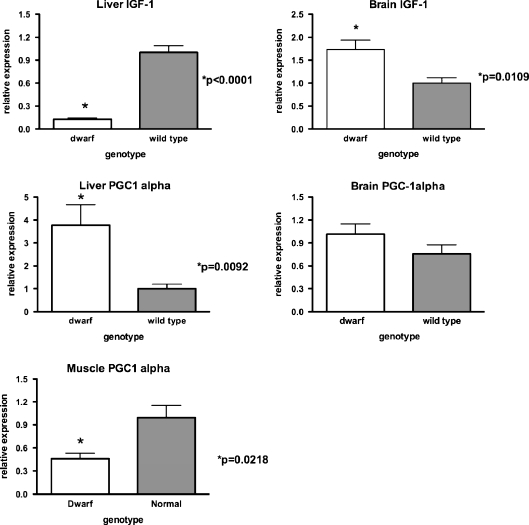
Dwarf IGF1 gene expression was also examined in 12-month-old mice as a control. The expression of this gene was markedly lower in liver from dwarf compared to wild-type mice as expected (p < 0.0001; Fig. 7). However, brain IGF-1 mRNA levels were higher in dwarf compared to wild-type mice (p = 0.0109). Liver PGC-1α mRNA expression was greater in dwarf compared to wild-type mice (p = 0.0092). However, muscle PGC-1α mRNA expression was significantly lower (p = 0.0218) in dwarf relative to wild-type mice.
Fig. 7.
mRNA expression levels of IGF-1 and PGC-1α in liver, brain, and muscle tissues of 12-month-old dwarf and wild-type mice. Values represent mean ± SEM. N = 7–8/age/genotype. mRNA levels are expressed as relative to wild-type mice
Discussion
Major life span extension is observed in GH deficient Ames dwarf mice relative to their wild-type littermates with Ames mice living >50% longer than normal mice. Previous studies have focused on antioxidative defense, oxidative damage, and pathways leading to and including glutathione metabolism. We believe that the reduced GH signaling in the Ames mouse contributes to the enhanced antioxidative capacity, lower damage to proteins and DNA, and an increase in functioning of the methionine and glutathione metabolic pathways (Brown-Borg and Rakoczy 2003; 2005; Uthus and Brown-Borg 2003). An integral player in each of these observations is the mitochondria, organelles that drive energy metabolism, and possibly cellular aging according to several hypotheses of aging. The delayed aging phenotype of the Ames dwarf is remarkable and has not been examined in terms of the oxidative phosphorylation system. The enhanced antioxidant status in the Ames dwarf coupled with their longevity prompted further study of OXPHOS components. Understanding this system in Ames mice will provide information regarding the role of hormone deficiency and mitochondrial function in aging and longevity.
Complex I of the OXPHOS system is comprised of multiple proteins and is the largest of the ETC protein complexes. Seven of the ∼46 peptides are mitochondrially encoded. This multimeric protein complex contributes significantly to the generation of free radicals, the rate of respiration, and overall control of the electron transport chain in mammals (Fearnley et al. 2001; Chomyn et al. 1986). Complex I activity is the major regulator of OXPHOS, even as little as 25% inhibition of this enzyme, severely impairs energy metabolism. In addition, antioxidant status has been shown to be important in the maintenance of mitochondrial energy thresholds (Davey et al. 1998). If glutathione is depleted in mitochondria, the threshold is eliminated. Thus, the expression of this particular mitochondrial enzyme was of great interest in the Ames mouse knowing that they exhibit an elevated oxidative defense capacity.
In the current study, increased liver complex I protein levels in old dwarf mice were accompanied by a significant increase in electron flow between complex I and complex III when compared to age-matched wild-type mice. Complexes I and II exert the greatest control over the respiratory chain flux (Capaldi et al. 1988). It is also well accepted that defects in the electron transport chain contribute to the etiology of several disease states (reviewed in Wallace and Fan 2010). Similarly, alterations in this system resulting in less efficient energy production are thought to occur with aging (reviewed in Navarro and Boveris 2007; Short et al. 2005; Conley et al. 2000; McCarter 1995). The increased complex I activity and protein observed in 20-month-old dwarf mice suggests that mitochondrial function may be better preserved in the dwarf at older ages.
A significant effect of age was also observed in complex I + III activity in several tissues in both genotypes. The enzyme activity in mitochondria from liver and skeletal muscle was higher in young mice compared to mice at 20 months of age. In addition, a greater decline in activity with age was observed in wild-type mice versus Ames mice in all tissues examined. There are many reports confirming that complex I activities decline with aging and that this decline contributes to the reduction in mitochondrial respiration with aging (Castelluccio et al. 1994; Ventura et al. 2002; Cooper et al. 1992; Sugiyama et al. 1993). It has been proposed that complex I therefore, may become more rate controlling with age (Ventura et al. 2002). Key evidence in the current study demonstrates that the percent decline was lower in long-living Ames dwarf mice compared to their normal counterparts and that the Ames mice maintained higher activities at 20 months of age. Therefore, if complex I is indeed a major controller, and that a decline over time contributes to aging processes, then Ames mice may exhibit an advantage in many tissues over wild-type mice that may contribute to the delayed aging phenotype in dwarf mice. Mitochondrial complex I + III activities from the kidney of dwarf mice increased with age while a decline was observed in wild-type mice. This upregulation of activity in the dwarf kidney was complemented by significant increases in complexes I and III protein in Ames mice. Overall, these increases in the kidney and the lower rate of decline in other tissues as compared to wild-type mice, likely play a role in preservation of mitochondrial function.
The OXPHOS capacity varies among tissues and is dependent on tissue function and mitochondrial content. The liver is an extremely important tissue as it orchestrates the supply of energy substrates to other tissues. A pattern of declining complex II + III activity in some tissues of older wild-type mice was apparent. Although 3-month-old dwarf muscle mitochondria had lower II + III activity levels than wild-type mice, at 20 months of age, these long-living mice had 30% greater activity than was observed at 3 months. At the same time, II + III activity declined 30% in wild-type skeletal muscle tissue. In contrast to the other tissues, dwarf kidney II + III activity was elevated in old Ames mice positively correlating with the increased levels of complex III proteins in these older mice a finding similar to that observed in complex I + III activity in dwarf kidney. These elevated activities likely result from the higher levels of both complexes I and III proteins observed in dwarf kidney. If complex I is indeed the master regulator governing overall electron transport chain function, then the differences observed in both complex I activities and proteins in dwarf mice may underlie the elevated levels of other downstream enzymes in this chain (i.e., III, IV, and V).
In the proliferative tissues, liver, and kidney, mitochondria from old dwarf mice expressed higher complex IV activities in comparison to age-matched wild-type mice, activities that matched the greater abundance of both nuclear-encoded and mitochondrially-encoded complex IV subunit proteins. Additionally, activity of this enzyme complex increased with age in dwarf mitochondria (liver, kidney) and wild-type liver but wild-type muscle activities declined with age.
The five multimeric enzyme complexes that drive OXPHOS and cellular respiration are contained within the inner mitochondrial membrane. Factors that regulate the synthesis and activities of the OXPHOS complexes are largely unknown (Trounce 2000). Growth hormone in particular, has not been shown to directly regulate the activities of these enzyme complexes although we believe that this anabolic hormone affects mitochondrial metabolism. Westbrook et al. (2009) showed that the decreased GH signaling in Ames mice and GH receptor knockout mice (GHRKO) was associated with increased metabolism. These long-living mice exhibited increased oxygen consumption (VO2), decreased respiratory quotients and increased heat generated per gram of body weight when compared to wild-type mice. The fact that the GHRKO mice were similar to the Ames in terms of metabolism suggests that the reduced GH signaling is likely the culprit as opposed to thyroid hormone as the GHRKO mice are actually slightly hyperthyroid (elevated T3 and T3/T4 ratio compared to wild-type mice; Westbrook et al. 2009). In addition, in mice that express highly elevated levels of circulating GH and exhibit significantly shortened life spans, the GH transgenics, have in fact lower oxygen consumption, and heat produced per gram of body weight, and an increased respiratory quotient (Westbrook et al. 2009) compared to normal mice. Approximately 90% of mammalian VO2 is mitochondrial, thus the increased VO2 levels in the Ames mice indicates that these mice exhibit mitochondrial oxidative metabolism (Rolfe and Brown 1997). Several peroxisome proliferator-activated receptor α-regulated genes and gene products including those involved in β-oxidation of fatty acids are elevated in dwarf mice (including Ames, Snell, and GHRKO mice; Stauber et al. 2005).
Complex V, also known as ATP synthase, catalyzes the synthesis of ATP during OXPHOS. Old dwarf skeletal muscle mitochondria exhibited elevated levels of ATP synthase compared to wild-type mice. Other studies have shown that the activity of this protein declines with aging and that some of the subunits are subjected to oxidative protein modification over time, thus affecting their function (Choksi et al. 2007). Moreover, ATP synthase is not coupled to electron transport chain processes so the higher levels in Ames mouse tissues are not likely a result of a general upregulation in ETC complexes. The adenine nucleotide translocator (ANT) is also an important component of both OXPHOS and the mitochondrial permeability transition pore, and is known to exhibit oxidative damage during aging (Yan and Sohal 1998; Feng et al. 2008; Halestrap 2009). The high levels of ANT mRNA in the dwarf liver relative to wild-type mice suggest that Ames mice may maintain more efficient mitochondrial functioning well into old age.
Ames dwarf mice do not produce pituitary GH and thus liver IGF-1 is not properly targeted resulting in negligible levels of plasma IGF-1. The gene expression data of liver IGF-1 in this study reflects the lack of GH stimulation and thus the lack of anabolic activity that may stimulate mitochondrial metabolism. We have preliminary data showing that GH treatment in Ames mice suppresses expression of complex I protein suggesting that this hormone may influence OXPHOS (Brown-Borg, unpublished observations). Extremely low plasma thyroid hormone concentrations in dwarf mice also preclude the assertion that enhanced metabolic activity may be responsible for the enhanced OXPHOS output in older dwarf mice. Other studies also suggest that glucocorticoids promote OXPHOS and enhance ATP synthesis via increases in ANT and ATP synthase (Arvier et al. 2007). Ames dwarf mice exhibit significantly elevated plasma corticosteroid levels that may also contribute to the enhancement of the OXPHOS system observed in this study (Borg et al. 1995).
The peroxisome proliferator-activated receptor γ coactivator 1-α is considered a master regulator of mitochondrial biogenesis (Puigserver et al. 1998) and mitochondrial performance (Muoio and Koves 2007). Our lab (this study) as well as others, have reported that Ames mice exhibit greater levels of PGC-1α mRNA in tissues as compared to control animals (Corton and Brown-Borg 2005; Bartke et al. 2008). The high levels of liver PGC-1α mRNA did not result in greater mitochondrial numbers as estimated by the mitochondrial-nuclear DNA ratios. In turn, more mitochondrial mass does not explain the elevated OXPHOS protein and activities observed in Ames mice. Thus, the PGC-1α may be contributing to other metabolic perturbations of nuclear hormone activation in Ames mice that contribute to their longevity including antioxidant defense, β-oxidation, and insulin sensitivity.
Mitochondria from aged individuals exhibit selective diminished activities of complexes I and IV, enzymes that are integral to the inner mitochondrial membrane and key to mitochondrial function (Navarro and Boveris 2007). The results of our experiments demonstrate that mitochondria from older Ames mice exhibit elevated OXPHOS components, specifically complexes I and IV, perhaps enhancing both mitochondrial function and efficiency. To fully address mitochondrial functioning, examination of mitochondrial respiration is required. Based on the current results and preliminary data generated in our laboratory showing that dwarf mouse mitochondria are more uncoupled than those from wild-type mice, we predict that mitochondrial respiratory function will be better preserved in dwarf mice with age (Brown-Borg, unpublished data). These observations, coupled with greater antioxidant defenses and lower ROS production, contribute to the extended life span of Ames dwarf mice.
Acknowledgments
The authors express sincere gratitude to the National Institutes of Health (Grant #01322899) and the UND School of Medicine and Health Sciences Pharmacology, Physiology and Therapeutics Department for supporting this research.
Footnotes
The US Department of Agriculture, Agricultural Research Service, Northern Plains Area is an equal opportunity/affirmative action employer and all agency services are available without discrimination.Mention of a trademark or proprietary product does not constitute a guarantee of warranty of the product by the United States Department of Agriculture and does not imply its approval to the exclusion of other products that may also be suitable.
References
- Arvier M, Lagoutte L, Johnson G, Dumas JF, Sion B, Grizard G, Malthièry Y, Simard G, Ritz P. Adenine nucleotide translocator promotes oxidative phosphorylation and mild uncoupling in mitochondria after dexamethasone treatment. Am J Phys Endo Metab. 2007;293(5):E1320–E1324. doi: 10.1152/ajpendo.00138.2007. [DOI] [PubMed] [Google Scholar]
- Bartke A. Delayed aging in Ames dwarf mice. Relationships to endocrine function and body size. In: Hekimi S, editor. Results and problems in cell differentiation: the molecular genetics of aging. Berlin: Springer; 2000. pp. 181–202. [DOI] [PubMed] [Google Scholar]
- Bartke A, Bonkowski M, Masternak MT. How diet interacts with longevity genes. Hormones (Athens) 2008;7(1):17–23. doi: 10.14310/horm.2002.1111033. [DOI] [PubMed] [Google Scholar]
- Bartke A, Brown-Borg HM (2004) Life extension in the dwarf mouse. In: Schatten GP (ed) Current topics in Developmental Biology Chapter 6, vol 63. Elsevier Academic Press, San Diego, CA, pp 189–225 [DOI] [PubMed]
- Bokov AF, Lindsey ML, Khodr C, Sabia MR, Richardson A. Long-lived Ames dwarf mice are resistant to chemical stressors. J Geron A Biol Sci. 2009;64:819–827. doi: 10.1093/gerona/glp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg KE, Brown-Borg HM, Bartke A. Assessment of the primary adrenal cortical and pancreatic hormone basal levels in relation to plasma glucose and age in the unstressed Ames dwarf mouse. Proc Soc Exp Biol Med. 1995;210(2):126–133. doi: 10.3181/00379727-210-43931. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM. Hormonal control of aging in rodents: the somatotropic axis. Mol Cell Endo. 2009;299:64–71. doi: 10.1016/j.mce.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG. Catalase expression in delayed and premature aging mouse models. Exp Geron. 2000;35:199–212. doi: 10.1016/S0531-5565(00)00079-6. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG. Growth hormone administration to long-living dwarf mice alters multiple components of the antioxidative defense system. Mech Ageing Dev. 2003;124:1013–1024. doi: 10.1016/j.mad.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG. Glutathione metabolism in long-living Ames dwarf mice. Exp Geron. 2005;40:115–120. doi: 10.1016/j.exger.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the aging process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Bode AM, Bartke A. Antioxidative mechanisms and plasma growth hormone levels: potential relationship in the aging process. Endocr. 1999;11:41–48. doi: 10.1385/ENDO:11:1:41. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Johnson WT, Rakoczy SG, Kennedy MA, Romanick MA. Mitochondrial oxidant production and oxidative damage in Ames dwarf mice. J Am Aging Assoc. 2001a;24:85–96. doi: 10.1007/s11357-001-0012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Rakoczy SG, Kennedy MA, Romanick MA (2001b) Relationship between plasma growth hormone, antioxidants and oxidative damage in premature and delayed aging mice. 83rd Annual Meeting of the Endocrine Society p. 237
- Brown-Borg HM, Rakoczy SG, Romanick MA, Kennedy MA. Effects of growth hormone and insulin-like growth factor-1 on hepatocyte antioxidative enzymes. Exp Biol Med. 2002;227:94–104. doi: 10.1177/153537020222700203. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Rad Biol Med. 2000;29(3–4):222–230. doi: 10.1016/S0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Capaldi RA, Halphen DG, Zhang YZ, Yanamura W. Complexity and tissue specificity of the mitochondrial respiratory chain. J Bioenerg Biomembr. 1988;20(3):291–311. doi: 10.1007/BF00769634. [DOI] [PubMed] [Google Scholar]
- Castelluccio C, Baracca A, Fato R, Pallotti F, Maranesi M, Barzanti V, Gorini A, Villa RF, Parenti Castelli G, Marchetti M, et al. Mitochondrial activities of rat heart during ageing. Mech Ageing Dev. 1994;76(2–3):73–88. doi: 10.1016/0047-6374(94)91583-0. [DOI] [PubMed] [Google Scholar]
- Choksi KB, Roberts LJ, 2nd, DeFord JH, Rabek JP, Papaconstantinou J. Lower levels of F2-isoprostanes in serum and livers of long-lived Ames dwarf mice. Biochemical Biophys Res Comm. 2007;364:761–764. doi: 10.1016/j.bbrc.2007.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Cleeter MW, Ragan CI, Riley M, Doolittle RF, Attardi G. URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit. Science. 1986;234:614–618. doi: 10.1126/science.3764430. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Conley KE, Jubrias SA, Esselman PC (2000) Oxidative capacity and ageing in human muscle. J Physiol 526(1):203–210 [DOI] [PMC free article] [PubMed]
- Cooper JM, Mann VM, Schapira AH. Analyses of mitochondrial respiratory chain function and mitochondrial DNA deletion in human skeletal muscle: effect of ageing. J Neurol Sci. 1992;113(1):91–98. doi: 10.1016/0022-510X(92)90270-U. [DOI] [PubMed] [Google Scholar]
- Cooperstein SJ, Lazorow A. A microspectrophotometric method for the determination of cytochrome oxidase. J Biol Chem. 1951;189:665–670. [PubMed] [Google Scholar]
- Corton JC, Brown-Borg HM. Peroxisome proliferator-activated receptor gamma coactivator 1 in caloric restriction and other models of longevity. J Geron A Biol Sci Med Sci. 2005;60(12):1494–1509. doi: 10.1093/gerona/60.12.1494. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/en.141.7.2608. [DOI] [PubMed] [Google Scholar]
- Davey GP, Peuchen S, Clark JB. Energy thresholds in brain mitochondria. Potential involvement in neurodegeneration. J Biol Chem. 1998;273(21):12753–12757. doi: 10.1074/jbc.273.21.12753. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Fearnley IM, Carroll J, Shannon RJ, Runswick MJ, Walker JE, Hirst J. GRIM-19, a cell death regulatory gene product, is a subunit of bovine mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2001;276(42):38345–38348. doi: 10.1074/jbc.C100444200. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhu M, Schaub MC, Gehrig P, Roschitzki B, Lucchinetti E, Zaugg M. Phosphoproteome analysis of isoflurane-protected heart mitochondria: phosphorylation of adenine nucleotide translocator-1 on Tyr194 regulates mitochondrial function. Cardio Res. 2008;80(1):20–29. doi: 10.1093/cvr/cvn161. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Papconstantinou J, Miller RA, Harrison DA. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP. What is the mitochondrial permeability transition pore? J Mol Cell Cardiol. 2009;46(6):821–831. doi: 10.1016/j.yjmcc.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Hauck S, Bartke A. Effects of growth hormone on hypothalamic catalase and CuZn superoxide dismutase. Free Rad Biol Med. 2000;28:970–978. doi: 10.1016/S0891-5849(00)00186-6. [DOI] [PubMed] [Google Scholar]
- Hauck S, Bartke A. Free radical defenses in the liver and kidney of human growth hormone transgenic mice: possible mechanisms of early mortality. J Geron Bio Sci. 2001;56A:B153–B162. doi: 10.1093/gerona/56.4.B153. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kuznetsov AV, Clark JF, Winkler K, Kunz WS. Increase in flux control of cytochrome c oxidase in copper-deficient mottled brindled mice. J Biol Chem. 1996;271:283–288. doi: 10.1074/jbc.271.1.283. [DOI] [PubMed] [Google Scholar]
- Kwong LK, Sohal RS. Age-related changes in activities of mitochondrial electron transport complexes in various tissues of the mouse. Arch Biochem Biophys. 2000;373(1):16–22. doi: 10.1006/abbi.1999.1495. [DOI] [PubMed] [Google Scholar]
- López-Torres M, Gredilla R, Sanz A, Barja G. Influence of aging and long-term caloric restriction on oxygen radical generation and oxidative DNA damage in rat liver mitochondria. Free Rad Biol Med. 2002;32(9):882–889. doi: 10.1016/S0891-5849(02)00773-6. [DOI] [PubMed] [Google Scholar]
- McCarter RJM. Handbook of physiology. Oxford, UK: Oxford University Press; 1995. Aging. [Google Scholar]
- Muoio DM, Koves TR. Skeletal muscle adaptation to fatty acid depends on coordinated actions of the PPARs and PGC1 alpha: implications for metabolic disease. Appl Physiol Nutr Metab. 2007;32(5):874–883. doi: 10.1139/H07-083. [DOI] [PubMed] [Google Scholar]
- Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Phys Cell Phys. 2007;292(2):C670–C686. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77(3):731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Phys Endo Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Salvioli S, Bonafè M, Capri M, Monti D, Franceschi C. Mitochondria, aging and longevity—a new perspective. FEBS Lett. 2001;492(1–2):9–13. doi: 10.1016/S0014-5793(01)02199-8. [DOI] [PubMed] [Google Scholar]
- Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakalmal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci USA. 2005;102(15):5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber AJ, Brown-Borg H, Liu J, Waalkes MP, Laughter A, Staben RA, Coley JC, Swanson C, Voss KA, Kopchick JJ, Corton JC. Constitutive expression of peroxisome proliferator-activated receptor alpha-regulated genes in dwarf mice. Mol Pharmacol. 2005;67(3):681–694. doi: 10.1124/mol.104.007278. [DOI] [PubMed] [Google Scholar]
- Sugiyama S, Takasawa M, Hayakawa M, Ozawa T. Changes in skeletal muscle, heart and liver mitochondrial electron transport activities in rats and dogs of various ages. Biochem Mol Biol Int. 1993;30(5):937–944. [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/S0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- Trounce I (2000) Genetic control of oxidative phosphorylation and experimental models of defects. Human Reprod 15 Suppl 2:18-27 [DOI] [PubMed]
- Uthus EO, Brown-Borg HM. Altered methionine metabolism in long-living Ames dwarf mice. Exp Geron. 2003;38:491–498. doi: 10.1016/S0531-5565(03)00008-1. [DOI] [PubMed] [Google Scholar]
- Ventura B, Genova ML, Bovina C, Formiggini G, Lenaz G. Control of oxidative phosphorylation by complex I in rat liver mitochondria: implications for aging. Biochim Biophys Acta. 2002;1553(3):249–260. doi: 10.1016/S0005-2728(01)00246-8. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10(1):12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook R, Bonkowski MS, Strader AD, Bartke A. Alterations in oxygen consumption, respiratory quotient, and heat production in long-lived GHRKO and Ames dwarf mice, and short-lived bGH transgenic mice. J Geron A Biol Sci Med Sci. 2009;64(4):443–451. doi: 10.1093/gerona/gln075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Cortopassi G. Reproducible quantitative PCR of mitochondrial and nuclear DNA copy number using the LightCycler. Methods Mol Biol. 2002;197:129–137. doi: 10.1385/1-59259-284-8:129. [DOI] [PubMed] [Google Scholar]
- Yan LJ, Sohal RS. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci USA. 1998;95(22):12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]