Abstract
Nucleotide-sequence-specific interactions mediated by double-stranded RNA (dsRNA) can induce gene silencing. Gene silencing through transcriptional repression can be induced by dsRNA targeted to a gene promoter. However, until recently, no plant has been produced that harbors an endogenous gene that remains silenced in the absence of promoter-targeting dsRNA. We have reported for the first time that transcriptional gene silencing can be induced by targeting dsRNA to the endogenous gene promoters in petunia and tomato plants, using a Cucumber mosaic virus (CMV)-based vector and that the induced gene silencing is heritable. Efficient silencing depended on the function of the 2b protein encoded in the vector, which facilitates epigenetic modifications through the transport of short interfering RNA (siRNA) to the nucleus. Here we show that gene silencing that is mediated by targeting dsRNA to a gene promoter via the CMV vector can be as strong as co-suppression in terms of both the extent of mRNA decrease and phenotypic changes. We also show that the expression of genes involved in RNA-directed DNA methylation and in demethylation are upregulated and downregulated, respectively, in Arabidopsis plants infected with CMV. Thus, along with the function of the 2b protein, that transports siRNA to the nucleus, the promoter-targeted silencing system using the CMV vector has some property that facilitates heritable epigenetic changes on endogenous genes, enabling the production of a novel class of modified plants that do not have a transgene.
Key words: Cucumber mosaic virus, epigenetic changes, histone modification, RNA-directed DNA methylation, RNA silencing, short interfering RNA, transcriptional gene silencing
Nucleotide-sequence-specific inhibition of gene expression can be induced by diverse processes that are mediated by RNA.1 Gene silencing through transcriptional repression can be induced by dsRNA targeted to a gene promoter,2 a process known as RNA-mediated transcriptional gene silencing (TGS). This phenomenon was first discovered in plants using a transgene that transcribes an inverted repeat of a promoter sequence and was later reported in cultured human cells and in Schizosaccharomyces pombe.3–7 Plant RNA viruses such as the Potato virus X (PVX), Tobacco rattle virus (TRV) and Cucumber mosaic virus (CMV) have also been used as a tool to induce TGS.8–11
There is a marked difference between transgenes and endogenous genes in plants in the feasibility of the induction of silencing by targeting RNA to a promoter.12 Transgenes can be easily silenced and the silenced state is heritable in the presence or absence of the silencing inducer.9 On the other hand, endogenous genes can be silenced only in the presence of the silencing inducer,13–15 and no plant has been produced that harbors a silenced endogenous gene after the promoter-targeting dsRNA has been removed.
We previously developed a CMV-based vector, CMV-A1, which is suitable for inducing RNA silencing.10,16,17 We found that the CMV-A1 vector, which carries the endogenous gene promoter, efficiently induced heritable gene silencing that was accompanied by epigenetic changes and that the 2b protein of CMV was involved in this efficient RNA-mediated TGS.11 Here we describe the potency and other features of this silencing system.
The Effect of RNA-Mediated TGS Is as Strong as that of Co-suppression
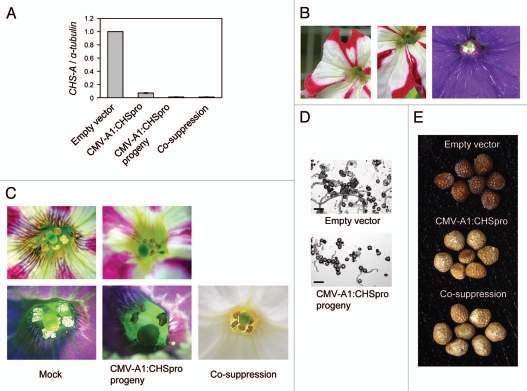
We previously targeted the promoter of the CHS-A gene for chalcone synthase in petunia (Petunia hybrida) because silencing of this gene is manifested as an altered visible phenotype on flower organs. A prominent, visible change is a reduction in pigmented area in the petal tissues of Red Star,11 a petunia variety that produces flowers with a red and white bicolor pattern as a result of naturally occurring sequence-specific degradation of CHS-A transcripts in the white sector of the petal.18 To evaluate the effect of RNA-mediated TGS, we compared the mRNA level of the CHS-A gene between plants infected with the virus carrying the CHS-A promoter (referred to as CMV-A1:CHSpro) and a transgenic line C001 that exhibits CHS-A co-suppression (Fig. 1A). We previously demonstrated that CHS-A silencing in this line is induced by degradation of RNA transcribed from both an endogenous gene and its homologous transgene.19 This co-suppressed line produces entirely white petals. Quantitative RT-PCR analysis indicated that the mRNA level for the CHS-A gene in the CMV-A1:CHSpro-infected plants was as low as that in the C001 line (Fig. 1A).
Figure 1.
Characterization of the changes induced by CMV-A1:CHSpro infection in petunia plants. (A) The mRNA level of the CHS-A gene (mean ± SE; n = 3) in the petal tissues of V26 plants infected with the empty vector (negative control) or CMV-A1:CHSpro, the progeny of virus-infected plants, and plants that had CHS-A co-suppression. The CHS-A mRNA level relative to the α-tubulin mRNA level in empty vector-inoculated plants was assigned a value of 1. (B) Nonpigmented sectors generated in the pigmented portions of the petals in the self-pollinated progeny of the CMV-A1:CHSpro-infected plant. Left and center, var. Red Star; right, V26 line. (C) Changes in anther phenotype maintained in the progeny of CMV-A1:CHSpro-infected plant. Upper row, var. Red Star; lower row, V26 line. The phenotype of male-sterile transgenic line C001 that exhibits CHS-A co-suppression is also shown. (D) In vitro germination of pollen. Bars indicate 50 µm. (E) Changes in pigmentation of seed coat tissues in V26: upper, seeds produced by empty-vector infected plants; center, seeds produced by CMV-A1:CHSpro-infected plants; lower, seeds produced by male-sterile transgenic line C001.
In the progeny of CMV-A1:CHSpro-infected petunia plants, white sectors were often generated inside the pigmented portions of the petal (Fig. 1B). This phenomenon suggests that strong de novo CHS-A silencing occurred in the lineage of pigmented cells during development. This observation is consistent with the fact that CHS-A mRNA level was very low even though the petal tissues were pigmented; the mRNA level was presumably only slightly higher than the threshold mRNA level that allows pigmentation in the petal tissues. Our previous data indicate that the estimated threshold level of transcripts of a gene involved in flavonoid synthesis associated with pigmentation in a plant tissue is about 3% of the steady-state mRNA level of the gene.20
Plants having petals with a greater white area produced less pollen compared with the control plants.11 The outward phenotype of anthers in CMV-A1:CHSpro-infected plants and their progeny was very similar to that of a plant line that exhibits CHS-A co-suppression; pollen grains remained clumped within the anther case, and anthers degenerated without exposing a large amount of pollen grains on the surface of the anther at anthesis (Fig. 1C). This male-sterile phenotype is ascribed to a reduction in flavonol content brought about by CHS-A silencing.21 In vitro pollen germination assay according to the method of Jahnen et al. showed that the frequency of pollen germination was lower in the progeny of CMV-A1:CHSpro-infected plants than in the control plants (Fig. 1D).
In addition to the changes in the anther and petal tissues, the seed coat of CMV-A1:CHSpro-infected plants did not have typical brown pigmentation, and the color was very similar to that of the CHS-A co-suppressed line (Fig. 1E). Taken together, these observations suggest that the effect of promoter-targeted silencing was equivalent to that of co-suppression in terms of both the extent of mRNA decrease and phenotypic changes.
Promotive Function of Viral Infection in the Induction of Epigenetic Changes
Previously, we found that the phenotypic changes were accompanied by epigenetic changes including cytosine methylation and histone modification.11 CMV encodes the 2b protein, which binds to siRNA and is localized in the nucleus.23 We found that these features are associated with efficient siRNA transport to the nucleus and that they facilitate RNA-directed DNA methylation (RdDM) and histone modifications, which eventually lead to RNA-mediated TGS.11
To identify additional feature(s) that may contribute to the efficient induction of epigenetic changes, here we analyzed changes in gene expression of the host plant after viral infection. Arabidopsis plants that were infected with CMV were examined for changes in the levels of genes known to be involved in RNA-mediated TGS. The mRNA levels of NRPD1, NRPE1, NRPD2 and AGO4 all increased after CMV infection. On the other hand, the mRNA level of ROS1, which is known to be involved in cytosine demethylation, decreased (Fig. 2). These results suggest that CMV infection upregulates the genes that drive RNA-mediated TGS and down-regulates the gene that antagonizes the pathway. We thus believe that these changes presumably facilitate RNA-mediated TGS.
Figure 2.
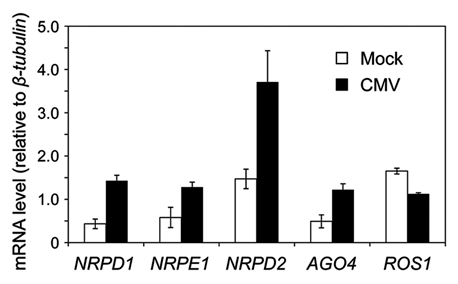
Changes in the mRNA level of the NRPD1, NRPE1, NRPD2, AGO4 and ROS1 genes in the Arabidopsis thaliana (Columbia ecotype) plants infected with CMV. Total RNA was isolated from mock-inoculated plants or plants infected with CMV-Y at 14 d postinoculation. The expression levels of these genes were analyzed with an eXpress profiling multiplex RT-PCR assay (Beckman, USA). The mRNA level of the β-tubulin gene was used as a control. The data are means and standard errors obtained from three replicates. Gene-specific sequences of the primers were as follows: for the NRPD1 gene, 5′-TAC GCT GCT TAT GAT GGC AC-3′ and 5′- GCT GCC TCA GAT AAT GCA CA-3′; for the NRPE1 gene, 5′-ACA GGA ACA ACC AAG ATG CC-3′ and 5′-CCT GAG CCT GAG ATG GAG AC-3′; for the NRPD2 gene, 5′-TCA AGA TGG GGA AAC GAA AG-3′ and 5′-TGT GGA CAA GTC GCT GGT AG-3′; for the AGO4 gene, 5′-TGA GGC ATT ACC ACC TCC TC-3′ and 5′-CCC CTT GTT CCA AAT CCT TT-3′; for the ROS1 gene, 5′-CTA ATT GCA ATG CAT GTC CG-3′ and 5′-CCT TGC TCT CTC TGG AAT GG-3′; for the β-tubulin gene, 5′-GTC AAT ACG TCG GCG ATT CT-3′ and 5′-CAT GGT ACC AGG CTC CAG AT-3′.
A previous study indicated that the mRNA levels of numerous genes of the host plant are either upregulated or downregulated after CMV infection,24 whereas the regulatory mechanisms of many of these genes are still unknown. Similarly, why the expression of genes involved in RdDM changed after viral infection remains unsolved. In these circumstances, it is conceivable that CMV has acquired a mechanism to fine-tune the expression of host genes to levels suitable for infection via RdDM during the arms race between plants and virus.
Another feature that may promote heritable epigenetic changes is the distribution of CMV within meristematic tissues, where heritable changes can be made on the genome; the accumulation of CMV-derived siRNAs has actually been detected in shoot meristems of tobacco.25 A combinatory effect of these features may account for the efficient induction of RNA-mediated, heritable epigenetic changes in the CMV vector system.
Final Remarks
Recent reports indicate that induction of epigenetic changes is potentially useful for plant breeding.26–29 However, just as mutagenesis causes mutations randomly in a genome, treatment of plants with an agent that induces epigenetic changes (e.g., a demethylating agent) also causes random changes. In contrast, our method allows induction of epigenetic changes on a target gene. Another advantage of the RNA-mediated TGS using the CMV vector is that the progeny plants do not have any transgene because the virus is eliminated during meiosis. Plants that are produced by this system have altered traits but do not carry a transgene, thus constituting a novel class of modified plants. The scheme of our silencing system is shown in Figure 3. The changes in the expression of host genes involved in RdDM or demethylation as a consequence of CMV infection suggest that CMV may have sophisticated epigenetic control of host genes and that it uses this control to infect the host. Thus, aside from using CMV for practical applications, we can also use CMV to provide new insights into the interactions between the virus and its host plant.
Figure 3.
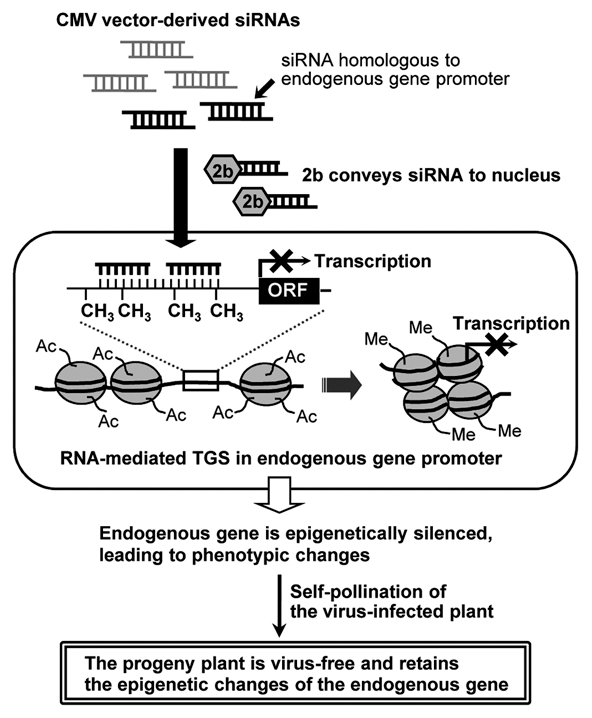
Scheme of induction of promoter-targeted silencing and epigenetic changes by the CMV vector. The CMV vector containing an endogenous gene promoter sequence produces siRNAs in the infected plants via the RNA silencing machinery. The 2b protein (2b) of CMV functions as an RNA silencing suppressor that inhibits posttranscriptional gene silencing (PTGS), but meanwhile it enhances nuclear transport of siRNAs. The siRNAs homologous to the endogenous gene promoter are conveyed by 2b and possibly by other mechanisms such as diffusion. The siRNAs are targeted to the promoter sequence of the endogenous gene in the nucleus, resulting in RNA-directed DNA methylation and histone modification. These epigenetic modifications eventually induce TGS of the endogenous gene and consequent phenotypic changes. In addition, the epigenetic modifications are maintained in the self-pollinated progeny, and thus the progeny plants have an altered phenotype but do not carry promoter-targeting dsRNAs. Ac and Me indicate acetylation and methylation of histone tails, respectively. ORF, open reading frame.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 3.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 5.Schramke V, Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301:1069–1074. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- 6.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 7.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol. 2001;11:747–757. doi: 10.1016/s0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 10.Otagaki S, Arai M, Takahashi A, Goto K, Hong JS, Masuta C, Kazazawa A. Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant Biotechnol. 2006;23:259–265. [Google Scholar]
- 11.Kanazawa A, Inaba J, Shimura H, Otagaki S, Tsukahara S, Matsuzawa A, et al. Virus-mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J. 2011;65:156–168. doi: 10.1111/j.1365-313X.2010.04401.x. [DOI] [PubMed] [Google Scholar]
- 12.Okano Y, Miki D, Shimamoto K. Small interfering RNA (siRNA) targeting of endogenous promoters induces DNA methylation, but not necessarily gene silencing, in rice. Plant J. 2008;53:65–77. doi: 10.1111/j.1365-313X.2007.03313.x. [DOI] [PubMed] [Google Scholar]
- 13.Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JN, Kooter JM. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol. 2001;11:436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- 14.Cigan AM, Unger-Wallace E, Haug-Collet K. Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J. 2005;43:929–940. doi: 10.1111/j.1365-313X.2005.02492.x. [DOI] [PubMed] [Google Scholar]
- 15.Heilersig BH, Loonen AE, Janssen EM, Wolters AM, Visser RG. Efficiency of transcriptional gene silencing of GBSSI in potato depends on the promoter region that is used in an inverted repeat. Mol Genet Genomics. 2006;275:437–449. doi: 10.1007/s00438-006-0101-4. [DOI] [PubMed] [Google Scholar]
- 16.Nagamatsu A, Masuta C, Senda M, Matsuura H, Kasai A, Hong JS, et al. Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol J. 2007;5:778–790. doi: 10.1111/j.1467-7652.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- 17.Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, et al. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathogens. 2011;7:e1002021. doi: 10.1371/journal.ppat.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koseki M, Goto K, Masuta C, Kanazawa A. The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by the sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 2005;46:1879–1883. doi: 10.1093/pcp/pci192. [DOI] [PubMed] [Google Scholar]
- 19.Kanazawa A, O'Dell M, Hellens RP. Epigenetic inactivation of chalcone synthase-A transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol. 2007;48:638–647. doi: 10.1093/pcp/pcm028. [DOI] [PubMed] [Google Scholar]
- 20.Nagamatsu A, Masuta C, Matsuura H, Kitamura K, Abe J, Kanazawa A. Downregulation of flavonoid 3′-hydroxylase gene expression by virus-induced gene silencing in soybean reveals the presence of a threshold mRNA level associated with pigmentation in pubescence. J Plant Physiol. 2009;166:32–39. doi: 10.1016/j.jplph.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Mo Y, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahnen W, Lush WM, Clarke AE. Inhibition of in vitro pollen tube growth by isolated S-glycoproteins of Nicotiana alata. Plant Cell. 1989;1:501–510. doi: 10.1105/tpc.1.5.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. Characterization of silencing suppressor 2b of Cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007;48:1050–1060. doi: 10.1093/pcp/pcm074. [DOI] [PubMed] [Google Scholar]
- 24.Marathe R, Guan Z, Anandalakshmi R, Zhao H, Dinesh-Kumar SP. Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol Biol. 2004;55:501–520. doi: 10.1007/s11103-004-0439-0. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki T, Ohki ST. Shoot meristem tissue of tobacco inoculated with Cucumber mosaic virus is infected with the virus and subsequently recovers from infection by RNA silencing. J Gen Plant Pathol. 2004;70:363–366. [Google Scholar]
- 26.Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y, Sano H. Epigenetic inheritance in rice plants. Ann Bot. 2007;100:205–217. doi: 10.1093/aob/mcm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauben M, Haesendonckx B, Standaert E, Van Der Kelen K, Azmi A, Akpo H, et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc Natl Acad Sci USA. 2009;106:20109–20114. doi: 10.1073/pnas.0908755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinders J, Wulff BB, Mirouze M, Mari-Ordonez A, Dapp M, Rozhon W, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyko A, Blevins T, Yao Y, Golubov A, Bilichak A, Ilnytskyy Y, et al. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS One. 2010;5:9514. doi: 10.1371/journal.pone.0009514. [DOI] [PMC free article] [PubMed] [Google Scholar]