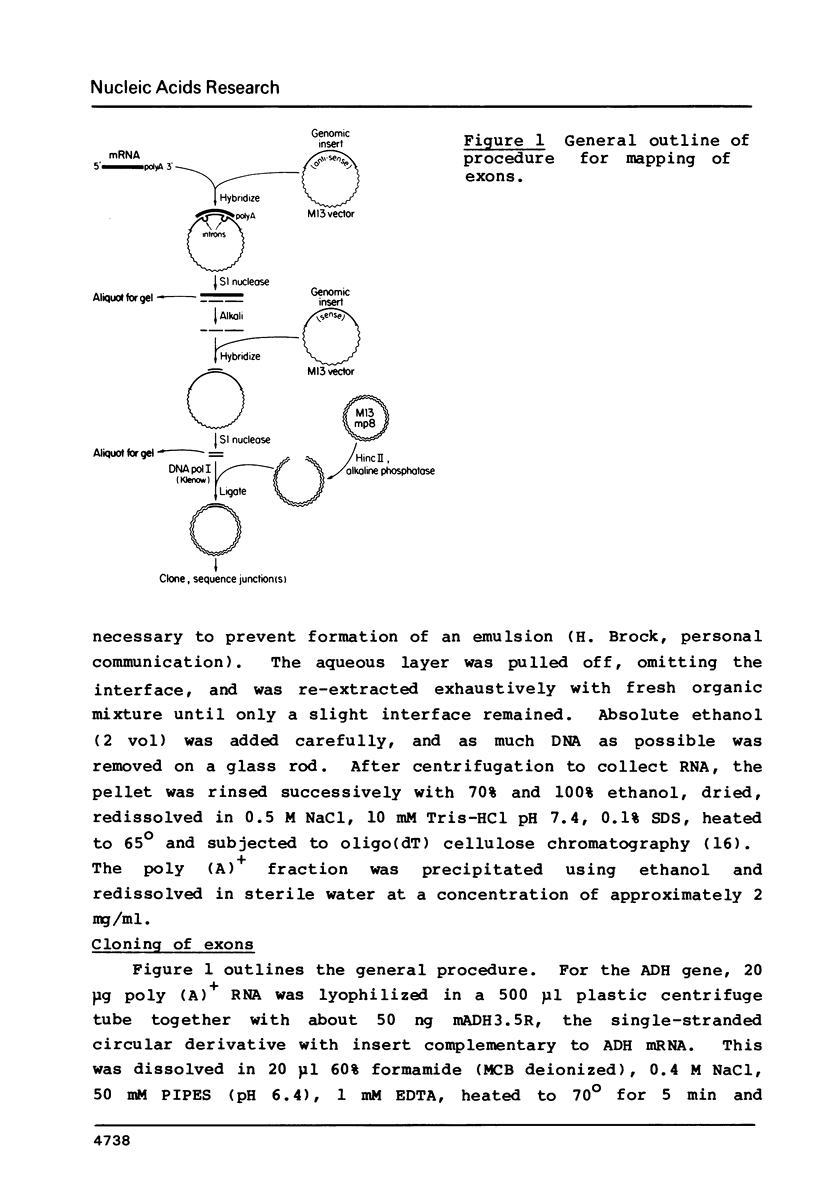
Abstract
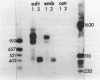
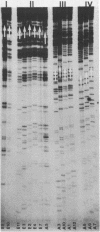
A novel method has been developed for determining the location of RNA termini and intron-exon boundaries using genomic clones. RNA is hybridized to a single-stranded M13 phage derivative of the genomic fragment of interest. S1 nuclease digestion results in an RNA-DNA hybrid corresponding to any transcript protected by the insert. Hydrolysis of the RNA with alkali, hybridization of the DNA with the opposite-strand M13 derivative of the genomic fragment and S1 nuclease digestion yields a mixture of pure exons. This mixture is analyzed by agarose gel electrophoresis and is cloned using excess blunt-ended M13 phage vector. Plaques that contain inserts are identified by transfer to nitrocellulose and hybridization to the genomic insert and are used without further purification. Cloning junctions are then determined by partial sequence analysis. These very nearly correspond to intron-exon boundaries or to either end of the transcript. When applied to the alcohol dehydrogenase gene from Drosophila, this method revealed clear differences between the 5' ends of embryo and adult transcripts both by blot hybridization and by analysis of 23 independent exon clones. In embryos, the mature transcript is apparently derived from three exons and in adults from four with the difference lying in the 5' untranslated portion of the transcript. The method should be particularly valuable for mapping and cloning transcripts that are rare or are not polyadenylated.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981 Jul 10;9(13):3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Place A. R., Powers D. A., Sofer W. Alcohol dehydrogenase gene of Drosophila melanogaster: relationship of intervening sequences to functional domains in the protein. Proc Natl Acad Sci U S A. 1981 May;78(5):2717–2721. doi: 10.1073/pnas.78.5.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Place A. R., Wang N., Pentz E., Sofer W. Deletions at intervening sequence splice sites in the alcohol dehydrogenase gene of Drosophila. Nucleic Acids Res. 1982 Nov 25;10(22):7261–7272. doi: 10.1093/nar/10.22.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Wang N., Reddy A., Weinberg E., Sofer W. Alcohol dehydrogenase in Drosophila: isolation and characterization of messenger RNA and cDNA clone. Nucleic Acids Res. 1980 Dec 11;8(23):5649–5667. doi: 10.1093/nar/8.23.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Santangelo G. M. Analysis in Cos-1 cells of processing and polyadenylation signals by using derivatives of the herpes simplex virus type 1 thymidine kinase gene. Mol Cell Biol. 1983 Feb;3(2):267–279. doi: 10.1128/mcb.3.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Goldberg D. A. Isolation and partial characterization of the Drosophila alcohol dehydrogenase gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5794–5798. doi: 10.1073/pnas.77.10.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Tatchell K., Hall B. D., Nasmyth K. A. Isolation of a gene from Drosophila by complementation in yeast. Nature. 1981 Jan 1;289(5793):33–37. doi: 10.1038/289033a0. [DOI] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Schibler U. Comparison of mRNA precursors in plasmacytomas producing closely related kappa chains. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3678–3682. doi: 10.1073/pnas.76.8.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozek C. E., Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983 Jan;32(1):23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalenghe F., Turco E., Edström J. E., Pirrotta V., Melli M. Microdissection and cloning of DNA from a specific region of Drosophila melanogaster polytene chromosomes. Chromosoma. 1981;82(2):205–216. doi: 10.1007/BF00286105. [DOI] [PubMed] [Google Scholar]
- Shore D., Langowski J., Baldwin R. L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Mahowald A. P. Identification and genetic localization of mRNAs from ovarian follicle cells of Drosophila melanogaster. Cell. 1979 Mar;16(3):589–598. doi: 10.1016/0092-8674(79)90032-1. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher D. R., Sawyer L. Secondary-structure prediction from the sequence of Drosophila melanogaster (fruitfly) alcohol dehydrogenase. Biochem J. 1980 Jun 1;187(3):884–886. doi: 10.1042/bj1870884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Hagenbüchle O., Schibler U. A single mouse alpha-amylase gene specifies two different tissue-specific mRNAs. Cell. 1981 Feb;23(2):451–458. doi: 10.1016/0092-8674(81)90140-9. [DOI] [PubMed] [Google Scholar]
- Zachar Z., Bingham P. M. Regulation of white locus expression: the structure of mutant alleles at the white locus of Drosophila melanogaster. Cell. 1982 Sep;30(2):529–541. doi: 10.1016/0092-8674(82)90250-1. [DOI] [PubMed] [Google Scholar]
- Ziff E. B. Transcription and RNA processing by the DNA tumour viruses. Nature. 1980 Oct 9;287(5782):491–499. doi: 10.1038/287491a0. [DOI] [PubMed] [Google Scholar]