Abstract
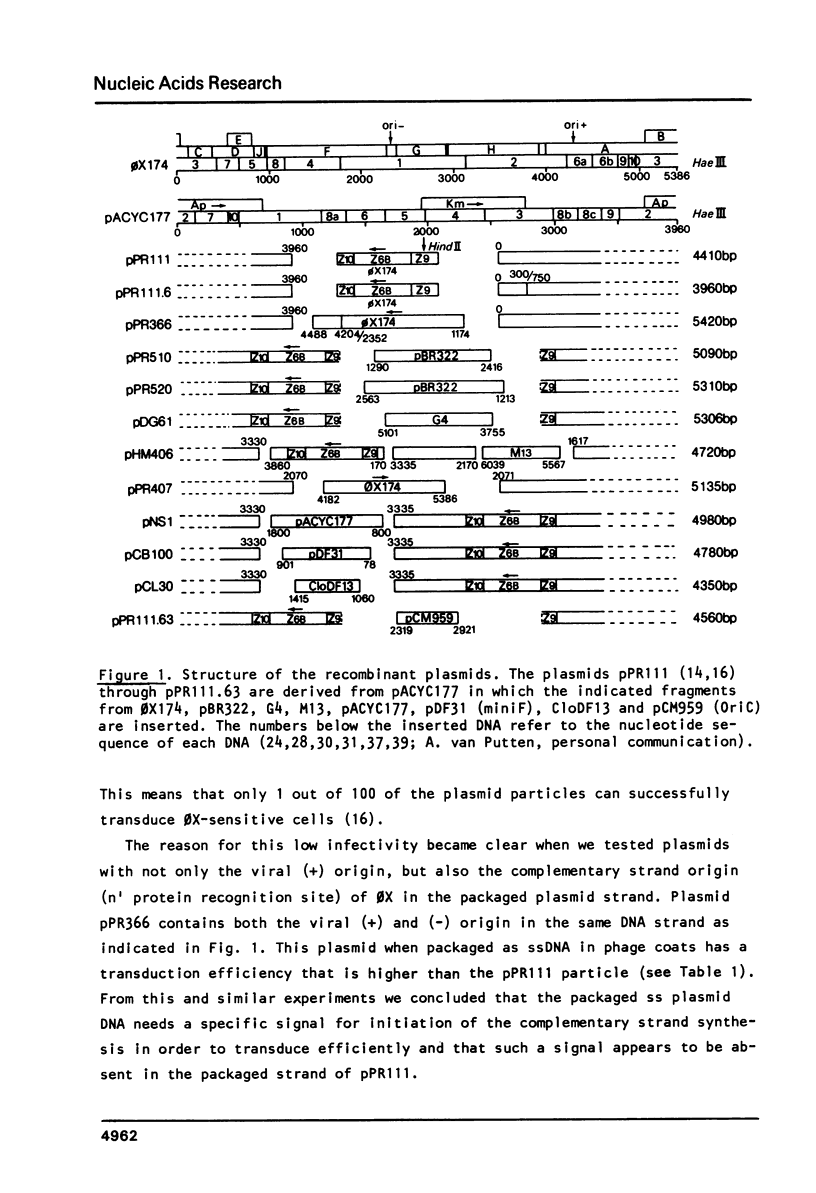
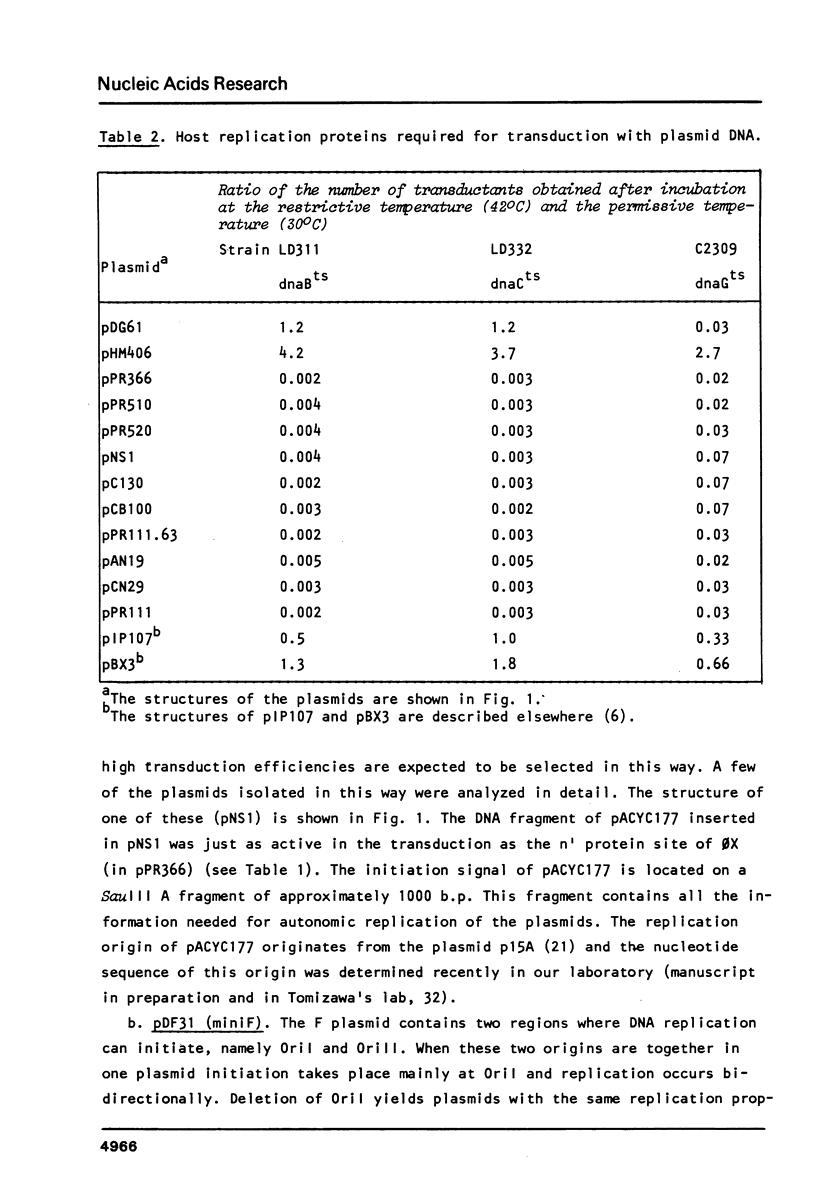
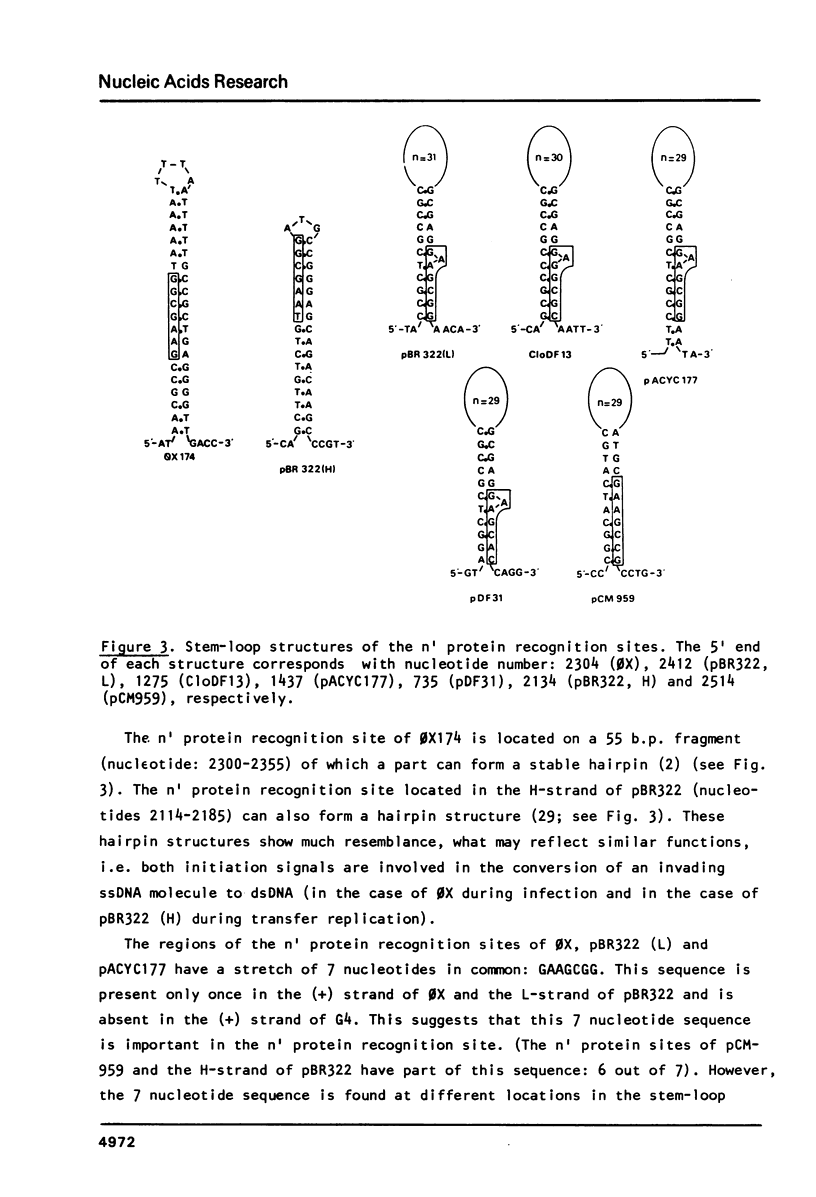
The bacteriophage 0X174 origin for (+) strand DNA synthesis, when inserted in a plasmid, is in vivo a substrate for the initiator A protein, that is produced by infecting phages. The result of this interaction is the packaging of single-stranded plasmid DNA into preformed phage coats. These plasmid particles can transduce 0X-sensitive cells; however, the transduction efficiency depends strongly on the presence in the packaged DNA strand of an initiation signal for complementary strand DNA synthesis. A plasmid with the complementary (-) strand origin of 0X inserted in the same strand as the viral (+) origin transduces 50-100 times more efficient than the same plasmid without the (-) origin of 0X. The transduction efficiency of such a particle is comparable to the infection efficiency of the phage particle. It is shown that in this system the 0X (-) origin can be replaced by the complementary strand origins of the bacteriophages G4 and M13. We have used this system to isolate sequences, from E. coli plasmids (pACYC177, CloDF13, miniF and OriC) and from the E. coli chromosome that can function as initiation signals for the conversion of single-stranded plasmid DNA to double-stranded DNA. All isolated origins were found to be dependent for their activity on the dnaB, dnaC and dnaG proteins. We conclude that these signals were all primosome-dependent origins and that primosome priming is the major mechanism for initiation of the lagging strand DNA synthesis in E. coli. The assembly of the primosome depends on the sequence-specific interaction of the n' protein with single-stranded DNA. We have used the isolated sequences to deduce a consensus recognition sequence for the n' protein. The role of a possible secondary structure in this sequence is discussed.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama A., Hayashi M. In vitro packaging of plasmid DNAs into phi X174 bacteriophage capsid. Nature. 1982 Jun 24;297(5868):704–706. doi: 10.1038/297704a0. [DOI] [PubMed] [Google Scholar]
- Arai K., Arai N., Shlomai J., Kobori J., Polder L., Low R., Hübscher U., Bertsch L., Kornberg A. Enzyme studies of phi X174 DNA replication. Prog Nucleic Acid Res Mol Biol. 1981;26:9–32. [PubMed] [Google Scholar]
- Arai N., Kornberg A. Rep protein as a helicase in an active, isolatable replication fork of duplex phi X174 DNA. J Biol Chem. 1981 May 25;256(10):5294–5298. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Borrias W. E., Hagenaar M., Van Den Brekel R., Kühlemeijer C., Weisbeek P. J. Functional relationship between bacteriophages G4 and phi X174. J Virol. 1979 Aug;31(2):288–298. doi: 10.1128/jvi.31.2.288-298.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden D. W., Twersky R. S., Calendar R. Escherichia coli deoxyribonucleic acid synthesis mutants: their effect upon bacteriophage P2 and satellite bacteriophage P4 deoxyribonucleic acid synthesis. J Bacteriol. 1975 Oct;124(1):167–175. doi: 10.1128/jb.124.1.167-175.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. D., Bennett G. N., Lee F., Schweingruber M. E., Yanofsky C. RNA polymerase interaction at the promoter--operator region of the tryptophan operon of Escherichia coli and Salmonella typhimurium. J Mol Biol. 1978 May 15;121(2):153–177. doi: 10.1016/s0022-2836(78)80003-5. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derstine P. L., Dumas L. B., Miller C. A. Bacteriophage G4 DNA synthesis in temperature-sensitive dna mutants of Escherichia coli. J Virol. 1976 Sep;19(3):915–924. doi: 10.1128/jvi.19.3.915-924.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub R., Figurski D., Helinski D. R. Bidirection replication from a unique origin in a mini-F plasmid. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1138–1141. doi: 10.1073/pnas.74.3.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub R., Sparks R. B., Helinski D. R. Bidirectional replication of the mini-ColE1 plasmid pVH51. J Bacteriol. 1979 Apr;138(1):257–260. doi: 10.1128/jb.138.1.257-260.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godson G. N., Barrell B. G., Staden R., Fiddes J. C. Nucleotide sequence of bacteriophage G4 DNA. Nature. 1978 Nov 16;276(5685):236–247. doi: 10.1038/276236a0. [DOI] [PubMed] [Google Scholar]
- Heidekamp F., Baas P. D., Jansz H. S. Nucleotide sequences at the phi X gene A protein cleavage site in replicative form I DNAs of bacteriophages U3, G14, and alpha 3. J Virol. 1982 Apr;42(1):91–99. doi: 10.1128/jvi.42.1.91-99.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., Baas P. D., Jansz H. S., van Arkel G. A., Weisbeek P. J. Nucleotide sequence of the origin of replication in bacteriophage phiX174 RF DNA. Nature. 1978 Feb 2;271(5644):417–420. doi: 10.1038/271417a0. [DOI] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murotsu T., Matsubara K., Sugisaki H., Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981 Nov;15(2-3):257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Nomura N., Low R. L., Ray D. S. Identification of ColE1 DNA sequences that direct single strand-to-double strand conversion by a phi X174 type mechanism. Proc Natl Acad Sci U S A. 1982 May;79(10):3153–3157. doi: 10.1073/pnas.79.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugimoto K., Takanami M., Hirota Y. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol Gen Genet. 1980 Apr;178(1):9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Polder L., Arai K., Kornberg A. Replication of phi X174 dna with purified enzymes. I. Conversion of viral DNA to a supercoiled, biologically active duplex. J Biol Chem. 1981 May 25;256(10):5233–5238. [PubMed] [Google Scholar]
- Soeller W. C., Marians K. J. Deletion mutants defining the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7253–7257. doi: 10.1073/pnas.79.23.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuitje A. R., Spelt C. E., Veltkamp E., Nijkamp H. J. Identification of mutations affecting replication control of plasmid Clo DF13. Nature. 1981 Mar 19;290(5803):264–267. doi: 10.1038/290264a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Griffith J., Geider K., Schaller H., Kornberg A. Initiation of deoxyribonucleic acid synthesis. VII. A unique location of the gap in the M13 replicative duplex synthesized in vitro. J Biol Chem. 1974 May 25;249(10):3049–3054. [PubMed] [Google Scholar]
- Zechel K., Bouché J. P., Kornberg A. Replication of phage G4. A novel and simple system for the initiation of deoxyribonucleic acid synthesis. J Biol Chem. 1975 Jun 25;250(12):4684–4689. [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Identification of two Escherichia coli factor Y effector sites near the origins of replication of the plasmids (ColE1 and pBR322. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6521–6525. doi: 10.1073/pnas.77.11.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]
- van der Ende A., Langeveld S. A., Teertstra R., van Arkel G. A., Weisbeek P. J. Enzymatic properties of the bacteriophage phi X174 A protein on superhelical phi X174 DNA: a model for the termination of the rolling circle DNA replication. Nucleic Acids Res. 1981 May 11;9(9):2037–2053. doi: 10.1093/nar/9.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A., Teertstra R., Weisbeek P. J. Initiation and termination of the bacteriophage phi X174 rolling circle DNA replication in vivo: packaging of plasmid single-stranded DNA into bacteriophage phi X174 coats. Nucleic Acids Res. 1982 Nov 11;10(21):6849–6863. doi: 10.1093/nar/10.21.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]


