Abstract
Within solid tumor microenvironments, lactic acidosis and hypoxia each have powerful effects on cancer pathophysiology. However, the influence that these processes exert on each other is unknown. Here we report that a significant portion of the transcriptional response to hypoxia elicited in cancer cells is abolished by simultaneous exposure to lactic acidosis. In particular, lactic acidosis abolished stabilization of HIF-1α protein which occurs normally under hypoxic conditions. In contrast, lactic acidosis strongly synergized with hypoxia to activate the unfolded protein response (UPR) and an inflammatory response, displaying a strong similarity to ATF4-driven amino acid deprivation responses (AAR). In certain breast tumors and breast tumor cells examined, an integrative analysis of gene expression and array CGH data revealed DNA copy number alterations at the ATF4 locus, an important activator of the UPR/AAR pathway. In this setting, varying ATF4 levels influenced the survival of cells after exposure to hypoxia and lactic acidosis. Our findings reveal that the condition of lactic acidosis present in solid tumors inhibits canonical hypoxia responses and activates UPR and inflammation responses. Further, they suggest that ATF4 status may be a critical determinant of the ability of cancer cells to adapt to oxygen and acidity fluctuations in the tumor microenvironment, perhaps linking short-term transcriptional responses to long-term selection for copy number alterations in cancer cells.
Introduction
Lactic acidosis and hypoxia are two prominent microenvironmental stresses in solid tumors. Hypoxia is usually caused by an imbalance between the supply and demand of oxygen due to vascular insufficiency or increased oxygen consumption by rapidly proliferating tumors. As oxygen becomes limiting, cells adapt by up-regulation of the HIF-1α protein and activation of the hypoxia response, which shifts the energy source to glycolysis, leading to higher levels of lactic acid (lactic acidosis) with medium tumor lactate from 7–10 mM/g and up to 25.9 mM/g (1–3). Since the presence of hypoxia and lactic acidosis are mechanistically and topologically linked, many tumor cells are concurrently exposed to lactic acidosis and hypoxia (4, 5). Although our understanding of hypoxia and lactic acidosis as individual stress has advanced significantly in recent years, the interaction between these two stresses is not well defined.
Oxygen is essential for aerobic metabolism in all mammalian cells. When oxygen is limited, cells adapt through a well-coordinated gene expression program termed the “hypoxia response” (5). The hypoxia response program plays an important role in tumor initiation, progression, invasion and in many steps of the oncogenic process (5). The hypoxia response is triggered by a family of transcription factors called hypoxia-inducible factors (HIFs), which are heterodimeric protein complexes consisting of a constitutively expressed subunit, HIF-1β or ARNT, and an oxygen-sensitive inducible subunit, HIF-1α or HIF-2α (5). Although most regulation occurs at proteosome-degradation level, several other reports have also indicated that HIF-1α can be regulated at the levels of translation initiation and mRNA abundance (6–8).
Although lactic acidosis has been recognized as an important feature of the tumor microenvironment (9), relatively little is known about its influences cancer cells. Many studies of acidosis have focused on the ability of acidosis to select tumor cells with altered phenotypes and metabolism (9–11). By using microarray gene expression analysis to interrogate the link between in vitro perturbations and in vivo human tumors, we have found that lactic acidosis inhibits tumor glycolysis through the inhibition of glycolysis gene expression, Akt pathways (12) and induction of TXNIP (13). In addition, we have also shown that the hypoxia and lactic acidosis gene signatures can be associated with DNA copy number alterations (CNAs) and various oncogenic events in human cancers (14, 15).
Given the mechanistic link between hypoxia and lactic acidosis, it is not surprising that many studies have pointed to the extensive cross-talk between these two stresses. However, the nature of their interaction is not fully understood. For example, some of the hypoxia-inducible genes, including VEGF and IL8, can be also induced by acidosis (16–18), consistent with the relocalization of the VHL protein into nucleoli and thus stabilization of the HIF-1α protein (19). These studies suggest that the acidosis and hypoxia gene expression responses share certain features. On the other hand, other reports point out that several hypoxia-induced genes are repressed in the presence of acidosis (20, 21). This paradox has also been pointed out in a recent article (11), but the mechanisms for the distinct hypoxia response to lactic acidosis are unknown.
In this study, we used gene expression analysis by microarrays to examine how lactic acidosis and hypoxia interact in terms of their influence on gene expression. We found that co-existing lactic acidosis has a dramatic inhibitory effect on the expression of a subset of hypoxia response genes due to the translational inhibition of HIF-1α. On the other hand, combined lactic acidosis and hypoxia synergistically trigger gene expression of both the unfolded protein response (UPR) and the inflammatory responses. We utilize an integrative genomic analysis to identify an amplification of the ATF4 locus in certain breast tumors and cell lines. ATF4 amplification promotes increased cell survival under hypoxia and lactic acidosis and may provide a mechanistic link between the short term transcriptional response and long term selection by microenvironmental stresses.
Materials and Methods
Cell culture
MCF7 and SUM52PE breast cancer cells were cultured in DMEM (GIBCO-11995) supplemented with 10% fetal bovine serum and 1× antibiotics (penicillin, 10000 UI/ml and streptomycin, 10000 UI/ml). HT1080 stable ATF4 knockdown fibrosarcoma cells (shATF4) and the control cells (shNT) were cultured as previously reported with 0.5 μg/ml puromycin and supplemented with NEAA and 55 μM β-ME (22). Lactic acidosis condition was created with the addition of 10 mM lactic acid (Sigma) to the respective media and adjusted to the desired pH 6.7 with a final concentration of 25 mM HEPES buffer (GIBCO) and 1 N HCL. All the cells were maintained in a humidified incubator at 37°C and 5% CO2. Hypoxic conditions were generated by lowering the oxygen level to 1% oxygen in hypoxia chamber (HERA cell 150, Heraeus).
RNA isolation and microarray analysis
RNA from MCF7 cells exposed to the control, lactic acidosis, hypoxia or the combined hypoxia and lactic acidosis conditions for 24 hrs were extracted by miRVana kits (Ambion) and hybridized to Affymetrix Human genome 133A 2.0 arrays with standard protocol. CEL files (GEO (GSE29406)) were normalized by RMA, filtered by indicated criteria, hierarchically clustered with cluster 3.0, and displayed with Treeview.
Statistical Analyses
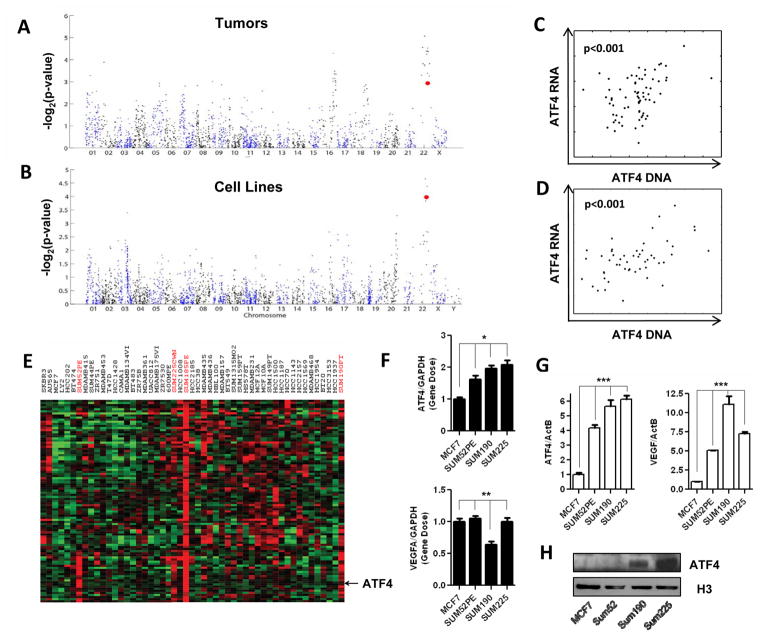
The effects of each stress on gene expression were normalized to the controlby subtracting the average expression level of the control samples. Those probe sets that varied from the baseline by at least 2-fold in at least two samples were selected for hierarchically clustering. Data was analyzed using SAM (23) and GSEA (24) as described using indicated selection criteria. The 99 probesets co-clustered with ATF4 in breast cancer cell lines were identified by examining the gene expression of the 50 breast cancer cell lines (25). The chromosomal and GO enrichment was performed using Gather (26). Association between ATF4 gene expression (Affymetrix U133 probe id 200779_at) and copy number variation was computed independently for each clone using Pearson correlation on those samples which both gene expression and CGH data (27, 28). The CGH clone that is plotted in figures 4C and 4D is the one that was closest to the ATF4 start site at position 38,248,081 on chromosome 22.
Figure 4. DNA copy number alterations of the ATF4 locus among the breast tumors and cancer cell lines.
(A, B) Strength of the correlation (−Log2(p-value), Pearson correlation) between DNA (array CGH) and ATF4 mRNA (microarrays) along all chromosomes among breast tumors (A) and cancer cell lines (B). Each spot represents the strength of correlation for one CGH clone. The clone closest to ATF4 is highlighted in red. (C, D) Scatterplot of ATF4 RNA versus DNA from the CGH clone closest to ATF4 in the genome. Figure (C) shows breast tumors and (D) shows cancer cell lines. Each spot is a single sample. (E) The clustered expression of ATF4 and adjacent chromosomal genes in breast cancer cell lines. (F, G) The DNA (F) and mRNA level (G) of ATF4 and VEGFA genes were determined in the indicated breast cancer cell lines by qPCR or RT-qPCR. Error bars are mean ± SD, significant p-values are indicated as (* p<0.001, n=3; ** p>0. 1, n=3; *** p<0.001, n=3). (H) Immunoblot detection of ATF4 and Histone-3 protein in nuclear extracts of indicated cell lines.
Realtime RT–PCR
RNA was reverse-transcribed to cDNA with the SuperScript II reverse transcription kit using random hexamers as the substrate for gene expression level measured by qPCR with Power SYBR Green PCR Mix (Applied Biosystems) following the manufacturer’s protocol. All primers used in this study are listed in Supplemental Table 1.
Western blot analysis and VEGFA ELISA
Cells under the indicated conditions were washed with cold PBS and lysed with NP40 buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40) supplemented with protease inhibitor and phosphatase inhibitor cocktail (Roche) for the extraction of total protein. The concentrations were determined with Bradford assay. Equal amounts of protein were loaded for the immunoblot analyses. Primary antibodies of HIF-1α (Novus), EGLN3 (Novus), S6K1 (whole and pho-T398, cell signaling), eIF2a (pho-Ser51, cell signaling), anti-ATF4 (Santa Cruz), and β-tubulin (Sigma) were applied following the manufacturers’ protocols. The signal was detected by the ECL plus western blotting detection system (Amersham). VEGFA concentration in culturing media was quantitated by human VEGF ELISA kit (RayBiotech, Inc).
Plasmids, RNAi transfection and retroviral infection
Cells were plated in 6-well plates allowing the cells to reach 60–70% confluence, 1 μg plasmid or 50 nM siRNAs were transfected by using lipofectamine 2000. After two days, cells were subjected to RNA, protein analysis or stress treatments. Retroviral particles were generated using the Amphotropic Phoenix packaging cell system transfected with pSM2-shATF4 and the control pSM2 plasmid. Cells were infected with retroviral supernatants twice with 4 μg/ml polybrene (Sigma). Cells were selected with 1 μg/ml puromycin for 7 days supplemented with NEAA and 55 μM β-ME and maintained in 0.5 μg/ml puromycin afterward.
FACS cell cycle and apoptosis analysis
Cells treated with the indicated stresses were trypsinized and collected, fixed in ice cold 70% ethanol overnight. Cells were washed twice with PBS buffer and resuspended in PBS containing 50 μg/ml PI and, 10 μg/ml RNase A. Flow cytometry was performed using BD FACSCanto II flow cytometer. At least 10000 cells were analyzed per sample. Statistical analyses of flow cytometry data were carried out by unpaired Student’s t-test.
Colony formation assays
A total of 104 cells were seeded in 6-well plates for two days prior to the stress treatment. The cells were treated with the indicated stress for two days, and the medium was thereafter replaced with regular culture DMEM and maintained under normoxia for additional 12 days with fresh medium replacement every 4 days. The number of colonies formed per plate was determined quantitatively following staining with crystal violet.
Results
The genomic analysis of the interaction between lactic acidosis and hypoxia
To systematically analyze the cross-talk between the responses to lactic acidosis and hypoxia, we exposed MCF7 cells to either control condition (ambient air ~21% O2, no lactate and neutral pH), lactic acidosis (ambient air, 10 mM Lactate and pH 6.7), hypoxia (1% pO2, no lactate and neutral pH) or combined lactic acidosis and hypoxia conditions (1% pO2, 10 mM Lactate and pH 6.7) for 24 hours. The level of lactate (10 mM) was in line with the median lactate levels reported in solid tumors (2, 3). The gene expression pattern of these RNA samples were interrogated with Affymetrix U133A genechips and normalized by RMA (Robust Multi-Array) algorithm.
The influence of hypoxia, lactic acidosis, and combined hypoxia and lactic acidosis on gene expression was derived by a zero transformation process in which we compared transcript levels for each gene in cells cultured under each of the three stress conditions to the average transcript levels in control samples. Based on the filtering criteria of two fold changes in more than two arrays, 1956 probesets were selected and arranged by hierarchical clustering (Fig 1A). Such analysis indicated that lactic acidosis, hypoxia and combined hypoxia and lactic acidosis, each elicited a distinct cellular response.
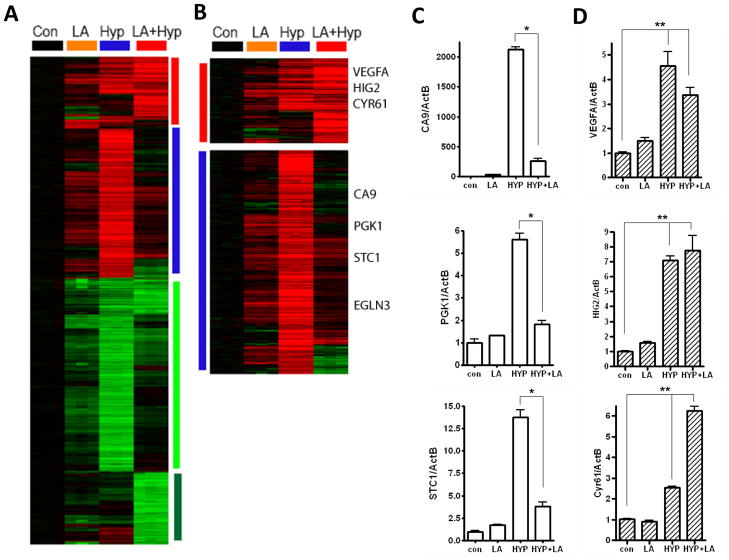
Figure 1. The transcriptional response of MCF7 to lactic acidosis (LA), hypoxia (Hyp), and the combined hypoxia and lactic acidosis (LA+Hyp) conditions.
(A) The gene expression profiles of MCF7 in response to lactic acidosis (10 mM Lactic acid, pH 6.7), hypoxia (1% O2) and the combined condition(10 mM Lactic acid, pH 6.7 in 1% O2) at 24 hrs. 1956 probe sets were selected to have at least two fold change in more than two arrays, and arranged by hierarchical clustering as shown. (B) The gene clusters of hypoxia induced genes which were sensitive (blue vertical bar) and resistant (red vertical bar) to the inhibitory effects of lactic acidosis were shown with selected gene names. (C) The levels of indicated lactic acidosis sensitive hypoxia genes (CA9, PGK1, STC1) in response to individual treatment were measured using RT-qPCR (n=3). (D) The levels of indicated lactic acidosis resistant hypoxia genes (VEGFA, HIG2, Cyr61) in response to individual treatment at 24 hrs were measured using RT-qPCR (n=3). Error bars are mean ± SD, significant p-values are indicated as (* p<0.001; ** p<0.01).
The most prominent feature for the transcriptional responses to combined hypoxia and lactic acidosis was the abolishment of most of the hypoxia response with the addition of lactic acidosis (Fig 1A, B). Both the induction and repression of respective large clusters of hypoxia-induced genes were abolished with the simultaneous presence of lactic acidosis (Fig 1A, B). For example, hypoxia activates the glycolytic pathway and stimulates glucose influx and consumption by induction of the glycolytic genes. It is evident that the glycolytic genes were dramatically repressed with the addition of lactic acid under hypoxia conditions (Fig S1A–B).
To further examine the hypoxia-induced genes in detail, we used Significant Analysis of Microarray (SAM) (23) to compare between the control and hypoxia samples to identify 717 probe sets as hypoxia-affected genes with 0% false discovery rate (FDR)(Fig S2A). Among the 448 induced probe sets (hypoxia-induced genes), there were many well-known hypoxia-inducible genes, including CA9, PGK1, ELGN3, GLUT-1. During combined hypoxia and lactic acidosis, the induction of the majority of the hypoxia-induced genes was abolished under lactic acidosis, and were thus classified as lactic acidosis-sensitive hypoxia genes (e.g., CA9, PGK1, ELGN3, BNIP3, STC1 and GLUT-1 (Fig 1B, S2). This distinct sensitivity to lactic acidosis for several hypoxia-inducible genes has also been observed in several previous studies (11, 20, 21). Intriguingly, a smaller subset of hypoxia-inducible genes still maintained their high expression levels under lactic acidosis. These genes, termed lactic acidosis-resistant hypoxia genes include VEGFA, HIG2 and CYR61 (Fig 1B, S2). Among all the hypoxia response genes, LA-resistant component was highly enriched in the DNA binding factors and epithelial cell differentiation using GOrilla (29)(Table 2).
We used real-time PCR to verify the ability of lactic acidosis to affect hypoxia-inducible genes based on the microarray analysis of MCF-7 cells. Among the lactic acidosis-sensitive hypoxia genes, we found that expression levels of CA9, PGK1 and STC1 were indeed strongly induced by hypoxia, but significantly repressed by coexisting lactic acidosis (10 mM lactate and pH 6.7) (Fig 1C). Among the lactic acidosis-resistant hypoxia genes, we verified the induction of VEGFA, HIG2 and CYR61 under hypoxia and the maintenance of persistent high levels even in the co-presence of lactic acidosis (Fig 1D). These results were also reproducible in MDA231 and HT1080 cells (Fig S2B-C) suggesting that the observation is generalizable to multiple cell types. Our global analysis of the transcriptional response has revealed that such distinctions of lactic acid sensitivity vs. resistance can be extended to a large number of hypoxia-inducible genes. Taken together, these results demonstrate that lactic acidosis had a profound and dramatic influence on the hypoxia response by abolishing most of hypoxia-inducible gene expression.
During long-term exposure to hypoxia alone, we noted significant accumulation of lactic acid in the culture media (Fig S1C–D). Lactate concentration was increased to 35mM in long term hypoxia, while only 15 mM in normoxia condition. Extracellur pH dropped to ~6.8. Similar to combined hypoxia and lactic acidosis, CA9 gene induction was attenuated during long-term hypoxia, while VEGFA induction was not affected (Fig S1E). These data suggest that combined hypoxia and lactic acidosis mimics long-term hypoxia.
Lactic acidosis represses the hypoxia response by the inhibition of HIF-1α synthesis
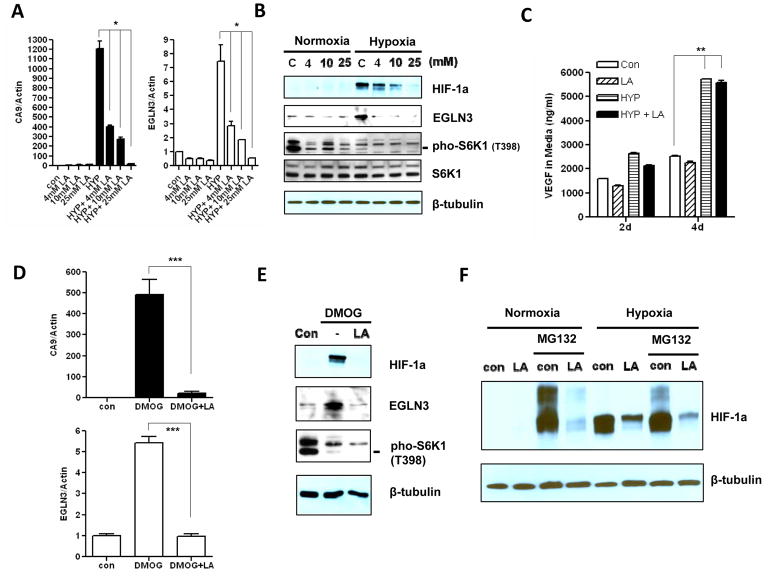
Tumor hypoxia switches the balance of cellular energy production by enhancing glycolysis-dependent lactate generation. Intratumoral lactate concentration can reach 25 mM in some extreme cases. A decrease in intracellular as well as extracellular pH is a by-product of this process. We therefore examined how different lactate levels affect the hypoxia response in the context of acidosis (pH 6.7). We found that higher levels of lactate had more pronounced inhibitory effects on the expression of the hypoxia genes CA9 and EGLN3 in the context of acidic conditions (Fig 2A). Lactate alone had little effect on hypoxic genes induction (data not shown). Consistent with the attenuated gene induction by hypoxia under combined hypoxia and lactic acidosis, we observed that lactic acidosis repressed accumulation of HIF-1α protein and its target gene EGLN3 under hypoxia (Fig 2B). Consistent with sustained VEGFA mRNA induction in combined hypoxia and lactic acidosis, VEGFA secretion was increased similar to hypoxia condition (Fig 2C). The reduction of HIF-1α protein correlated with increased lactate level under acidic conditions. Although the hypoxia response mostly relies on HIF-1α in MCF7 cells, we also observed instability of the HIF-2α protein under combined hypoxia and lactic acidosis conditions. These data suggest that lactic acidosis represses the hypoxic gene response by abolishing HIF-1α protein accumulation under hypoxia.
Figure 2. The repression of hypoxia response by lactic acidosis is mediated by inhibition of HIF-1α protein synthesis.
(A) The mRNA levels of CA9 and EGLN3 in MCF7 cells measured by RT-qPCR treated with indicated lactic acid concentration (pH 6.7) either in normoxia or in hypoxia condition for 24 hrs (n=3). (B) Immunoblot detection of HIF-1α and EGLN3 protein expressions, and S6K phosphorylation at Thr-398 in MCF7 cells treated as (A). (C) The concentration of VEGFA in media was detected by ELISA assay at indicated days and treatments (10 mM Lactic acid pH 6.7. n=3). (D) The mRNA levels of CA9 and EGLN3 in MCF7 treated with control, DMOG or DMOG plus lactic acidosis (10 mM Lactic acid pH 6.7) in normoxia condition (n=3). (E) Immunoblot detection of HIF-1α and EGLN3 protein expressions, and S6K phosphorylation at Thr-398 in MCF7 cells treated as (C). (F) The HIF-1α protein expression level was determined in MCF7 cells treated with lactic acidosis (10 mM Lactic acid, pH 6.7) in either normoxia or hypoxia condition for 18 hrs, then further treated with the protease inhibitor MG132 (10 μM) for additional 4 hrs. Error bars are mean ± SD, significant p-values are indicated as (* p<0.001; ** p<0.0001; *** p<0.001).
Under normoxia, HIF-1α protein is rapidly ubiquitylated and targeted for proteasomal degradation mediated by a pVHL-containing E3 ubiquitin ligase complex. Under hypoxic conditions, HIF-1α protein is poorly hydroxylated since the activity of prolyl hydroxylase is inhibited and is therefore resistant to VHL-directed degradation and becomes stabilized. We observed that the mRNA level of HIF-1α was not changed under different stress conditions. To understand at which step lactic acidosis deregulates the HIF-1α protein, we used Dimethyloxallyl Glycine (DMOG), a cell permeable, competitive inhibitor of prolyl hydroxylase (PHD) (30), to stabilize HIF-1α protein under normoxia. As expected, DMOG induced the hypoxia genes CA9 and EGLN3 and led to HIF-1α protein accumulation under normoxia (Fig 2D–E). However, lactic acidosis repressed the induction of the hypoxia genes CA9 and EGLN3 even in the presence of DMOG (Fig 2D). Consistent with that result, the HIF-1α protein was unable to accumulate in the DMOG co-treated with lactic acidosis condition (Fig 2E). Moreover, MG132, a proteasome inhibitor that stabilizes HIF-1α protein in cells by blocking its proteosomal degradation (31), was also unable to reverse the HIF-1α inhibition by lactic acidosis, either in normoxic or hypoxic conditions (Fig 2F). As previously reported, lactic acidosis triggers the activation of AMPK and the inhibition of mTOR (13), which control protein synthesis. Therefore, we examined the phosphorylation status of p70S6K1 protein to monitor the mTOR activity. Lactic acidosis exerted greater inhibition on p70S6K1 protein phosphorylation when combined with either hypoxia or DMOG treatment, in comparison to either hypoxia or DMOG treatment alone (Fig 2B, 2E and Fig S3A–B). Hypoxia mostly inhibits the mRNA level of HIF-1α (Fig S3C). It reflects that HIF-1α is generally under post-transcription regulation. We also examined polysome profiles for HIF-1α and ATF4 under hypoxic or combined hypoxia with lactic acidosis condition. Cytoplasmic extracts from these MCF7 cells were sedimented through sucrose gradients. The distribution of the HIF-1α, ATF4 mRNAs within each fraction was determined by RT-PCR (Fig S3D–F). With lactic acid, HIF1a mRNA shifted to low polysome fractions (Fig S3E), while ATF4 mRNA increased to high polysome fractions (Fig S3F). Taken together, these data suggested that lactic acidosis represses the hypoxia response, not through the promotion of HIF-1α degradation, but rather, through the synthesis of HIF-1α.
Lactic acidosis synergizes with hypoxia to activate the Unfolded Protein Response (UPR) and the inflammation response
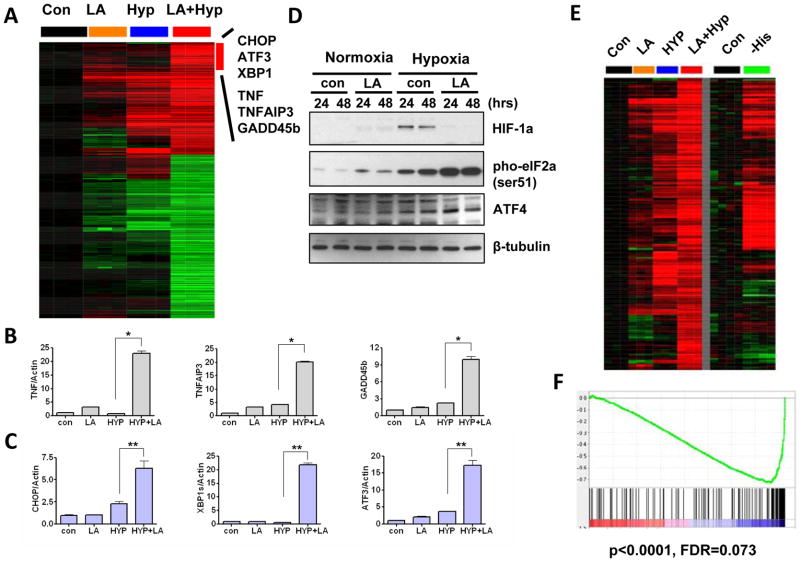
Combined hypoxia and lactic acidosis treatments caused dramatic gene expression changes with many genes either induced or repressed (Fig 1A). We used SAM to systematically identify genes that were affected by combined lactic acidosis and hypoxia. We identified 499 probesets with 0% FDR (Fig S2A), with 205 induced and 294 repressed probesets (Fig 3A). Gather (26) was used to analyze the shared features of these genes to identify three prominent features. As indicated above, the first group of genes represented lactic acidosis-resistant hypoxia genes including VEGFA, HIG2 and CYP61. The second group of genes is enriched in MAPK signaling pathway and Toll-like receptor signaling pathway (KEGG). Among those, there were several inflammatory response genes, including TNFα, TNFAIP3 and GADD45B (Fig 3A). The induction of these genes under combined hypoxia and lactic acidosis was further verified using real-time PCR (Fig 3B). The induction of these genes is known to be mediated by the NF-κB pathway. Interestingly, the regulators of NF-κB, such as BCL3, ZFP36, and NFKBIA, were also induced by these conditions. These data suggest that combined hypoxia and lactic acidosis conditions leads to the activation of the NF-κB pathway.
Figure 3. The transcriptional response triggered by combined hypoxia and lactic acidosis.
(A) Genes synergistically induced by combined hypoxia and lactic acidosis with the names of selected genes shown. (B, C) The mRNA levels of the inflammation response genes TNF, TNFAIP3 and GADD45B (B) and unfolded protein response genes CHOP, XBP1s and ATF3 (C) in MCF7 cells treated with 10 mM lactic acid (pH 6.7) either in normoxia or in hypoxia condition for 24 hrs. Error bars are mean ± SD, significant p-values are indicated as (* p<0.001, n=3; ** p<0.001, n=3). (D) Immunoblot detection of HIF-1α and ATF4 protein expressions, and eIF2α phosphorylation at Ser-51 in MCF7 treated10 mM lactic acid (pH 6.7) either in normoxia or in hypoxia condition for 24 or 48 hrs. (E) A comparison of gene expression profiles induced by combined hypoxia and lactic acidosis in MCF7 and induced by amino acid deprivation in HepG2. (F) GSEA reveals a significant enrichment of genes induced by combined hypoxia and lactic acidosis among the genes induced by histidine deprivation in HepG2.
Interestingly, the third group of genes induced by combined lactic acidosis and hypoxia is linked to the Unfolded Protein Response (UPR) pathway or ER stress pathway, as represented by CHOP, XBP-1 and ATF3 genes (Fig 3A). We verified the dramatic induction of ATF3, CHOP and XBP-1 splicing form (XBP-1s) mRNA under combined hypoxia and lactic acidosis conditions (Fig 3C) by real-time PCR. Importantly, UPR pathway is also activated in long-term hypoxia alone as indicated by XBP1s induction (Fig S1E). The UPR shares features with the amino acid response (AAR) pathway, as both stresses lead to phosphorylation of the eukaryotic translation initiation factor 2α (eIF2α) on serine 51. The phosphorylated eIF2α suppresses general protein synthesis but promotes the translation of select mRNA species with unique features in their 5′ untranslated regions, including ATF4 (32). Under the combined lactic acidosis and hypoxia conditions, we found that eIF2α protein was highly phosphorylated on the Ser51 residue followed by increased ATF4 protein accumulation (Fig 3D). Hypoxia (1% O2) alone induced a more modest eIF2α phosphorylation. Consistent with that, ATF4 protein remained at moderate-low levels and the UPR genes were weakly induced by hypoxia alone. We also found that combined lactic acidosis and hypoxia induced UPR genes in HT1080 and MDA231 cells (Fig S2B–C).
To further compare the gene expression changes under the combined lactic acidosis and hypoxia to the AAR, we compared our stress gene expression dataset in MCF7 to an independent gene expression dataset of histidine deprivation by histidinyl tRNA synthetase inhibitor histidinoh (HisOH) in HepG2 hepatoma cells (33). We found that most of the genes induced by the combined hypoxia and lactic acidosis were also induced by amino acid depletion in the HepG2 cells (Fig 3E). The Gene Set Enrichment Analysis (GSEA) (24) also revealed a very strong enrichment of the stress genes in the AAR datasets (p<0.0001, FDR= 0.073) (Fig 3F). These data indicated a high degree of similarity between hypoxia and lactic acidosis and AAR. In contrast, the genes repressed by combined hypoxia and lactic acidosis were not enriched in the amino acid depletion dataset either visually in the heatmap or by statistical analysis using GSEA (p=0.84, FDR=0.712) (Fig S2A). All of our evidence suggests that the combined lactic acidosis and hypoxia condition in the tumor microenvironment, while repressing many canonical hypoxia responses, elicited a strong UPR response and inflammation response that exhibited significant similarity to the ATF4-driven AAR response.
The amplification of the ATF4 locus in breast tumors and cell lines
Previously, we dissected the lactic acidosis and hypoxia gene signatures into distinct sub-signatures in a breast tumor dataset (14). Some of the sub-signatures were also highly enriched in particular chromosomal locations, and the expression patterns of those sub-signatures were highly correlated with DNA copy number alterations (CNAs) and gene dosage. One such sub-signature showed strong chromosomal enrichment in the ATF4 locus at 22q13 in both tumors and breast cancer cell lines. Examining the expression of ATF4 alone, we find that there is strong evidence of correlation between ATF4 gene expression and CNA in the 22q13 chromosomal region in both tumors (Fig 4A) and cancer cell lines (Fig 4B). There was a strong positive correlation between the array CGH (DNA dosage) and corresponding gene expression of ATF4 (RNA expression) in both breast tumors (Fig 4C) and cancer cell lines (Fig 4D). When gene expression from 50 breast cancer cell lines was arranged by hierarchical clustering, ATF4 expression was co-clustered with 98 other probesets in four breast cancer cell lines (Fig 4E). When these ATF4-coclustered genes (Table S3) were analyzed by
Gather, there is a significant enrichment at chromosome 22q13, where ATF4 locus resides (Table S4). These genes are also enriched in gene ontology of amino acid metabolisms known to be ATF4 target genes (Table S5). Experimentally, we verified ATF4 gene dosage in several cell lines noted to have higher expression of ATF4 and genes in the adjacent chromosomal regions. We found that there are higher levels of ATF4 gene dosage in SUM52PE, SUM190 and SUM225 cells in agreement with the statistical analysis data, while there is no increase of VEGF DNA dosage (Fig 4F). Consistent with the level of ATF4 gene dosage in cells, ATF4 mRNA and protein increases significantly (Fig 4G and 4H). VEGF, as an ATF4 transcriptional target, also has higher expression level in those cells (Fig 4G).
ATF4 modulates cellular survival under hypoxia and lactic acidosis
Due to fluctuations in the hemodynamics of distant blood vessels, both hypoxia and lactic acidosis exhibit significant intermittent nature (34). We assessed cell growth and death in hypoxia and the combined hypoxia and lactic acidosis stresses. While cell growth was inhibited under these stresses, there was no apparent cell death in MCF7, HT1080, and SUM52PE (Fig 5A, and Fig S4A). However, when the cells were recovered from the stress and maintained in normoxia for 3 days, we noted significant cell death in cells treated with combined hypoxia and lactic acidosis (Fig 5A). The colony formation assay further showed that lactic acidosis synergizes with hypoxia to induce severe cell death, while transient exposure of hypoxia alone has no noticeable effect on survival (Fig 5B).
Figure 5. Varying levels of ATF4 impact the survival of breast cancer cells under hypoxia and lactic acidosis.
(A) The amount of cell death in MCF7 after 3 days treatment with either hypoxia or combined hypoxia and lactic acidosis condition (empty), and 3 days recovery from stresses by re-plating regular culture medium and maintaining in normoxia condition (hatched). The cell death was assessed by the Propidium Iodide (PI) stained sub-G1 population using FACS. (B) Clonogenic survival of MCF7 treated with indicated stress for 2 days then replated with regular medium every four days for 12 days. Cell colonies were stained with crystal violet. (C) MCF7 transfected with 50 nM either control siRNA or siATF4 were treated with hypoxia or combined hypoxia and lactic acidosis. The levels of cell death were determined at 3 days stress treatment and 2 or 3 days post-stresses. Cell death was measured by the PI stained sub-G1 population (n=4). (D) The levels of cell death of MCF7 cells which have been transfected with either control vector or ATF4 expression constructs were treated, collected and assessed as (C) (n=3). (E) Cell death was measure in HT1080 shNT/shATF4 cells treated as (C) (n=3). (F) The mRNA levels of VEGFA, ASNS and HIG2 in the early passage of MCF7 cells stably infected with empty vector pSM2 and shATF4. (G) The mRNA level of ATF4 gene and cell death were measured at early and late passage of MCF7 cells stably infected with empty vector pSM2 and shATF4 (n=3). Error bars are mean ± SD, significant p-values are indicated as (* p<0.01; ** p<0.05; *** p<0.01; # p< 0.001; ## p<0.01).
Given the significant induction of UPR and inflammation response under hypoxia and lactic acidosis, cell death under combined stresses is probably modulated by these two biological processes. In addition, the expression level of ATF4 varies significantly among different breast tumors, partially due to DNA amplification. Therefore, we evaluated the role of varying levels of ATF4 in the ability of cancer cells to sustain the presence of stresses. We manipulated ATF4 gene expression in cells by both siRNA-mediated gene silencing and overexpression of ATF4 expression constructs. We observed that knockdown of ATF4 significantly sensitized cells to cell death when the cells recovered from the combined hypoxia and lactic acidosis stress (Fig 5C). Consistent with this observation, the overexpression of ATF4 significantly protected cells from death under the same conditions (Fig 5D). Similarly, ATF4 exhibited protection effects in HT1080 stable shATF4 cells (Fig 5E) and SUM52PE siATF4 cells (Fig S4A) when cells recovered from the combined hypoxia and lactic acidosis stress. Manipulation of ATF4 expression levels had less protective effects on hypoxia post-treated cells, but this is likely because hypoxia led to less cell apoptosis (Fig 5C–E and Fig S4A) than the combined conditions. These results suggested that ATF4 is crucial for cell survival during the post-stress recovery phase of the combined hypoxia and lactate acidosis. Therefore, the gain of ATF4 gene dosage due to DNA amplification in some tumors and cell lines might be an adaptive event in fluctuating hypoxia and lactic acidosis in the tumor microenvironment.
It has been reported that ATF4 knockout MEFs and shATF4 established HT1080 cells require the presence of NEAAs and antioxidant such as β-mercaptoethanol (β-ME) to survive (22, 32). However, we observed that cell death is visible in shATF4 established MCF7 and SUM52 breast cancer cells even in the presence of NEAAs and β-mercaptoethanol, and this affect is different than that seen in HT1080 shATF4 and ATF4−/− MEF cells (Fig 5F and Fig S4C). Due to selective pressures, shATF4 established cells in late passages gradually lost the inhibitory effect of shATF4 on ATF4 gene expression since the mRNA level of ATF4 returned back to near normal levels. Cell death was induced by knockdown ATF4 in the early passages and became barely visible in the late passages (Fig 5F and Fig S4C).
Taken together, our data suggest that the induction of ATF4-driven gene expression program is essential for cell survival during recovery from combined hypoxia and lactic acidosis. The CNAs in the ATF4 locus in a subset of tumors and cell lines may promote cancer cell survival and a growth advantage in hypoxia and lactic acidosis tumor microenvironment as well as in culture.
Discussion
In this report, we have found that lactic acidosis significantly modulates the canonical hypoxia response mediated by HIF-1α inhibition to the UPR/AAR gene expression program mediated by eIF2α-ATF4. Such a transition is essential for cellular survival during recovery from the combined stresses of hypoxia and lactic acidosis. Here, we show that lactic acidosis negatively regulates the hypoxia pathway by inhibiting HIF-1α synthesis. Previous studies have found that hypoxia alone can induce translation inhibition either by inhibiting mTOR and S6K1 kinase activity and subsequently inducing phosphorylation of EIF4BP, or by inducing the phosphorylation of eIF2α (35). However, a careful examination of polysome profiles (36) under hypoxia show that there is no reduced ribosome occupancy and translation efficiency of HIF-1α mRNA. This is consistent with induced HIF-1α protein accumulation under hypoxia. Extreme acidity (pH=5.5) can lead to eIF2α phosphorylation and translation inhibition in astrocytes (37). eIF2α phosphorylation has also been shown to regulate the protein levels of HIF-1α (38). But the effects of either of these stresses had modest effects on the translation efficiency of HIF-1α. In contrast, the simultaneous presence of both hypoxia and lactic acidosis dramatically induced S6K1 dephosphorylation and eIF2α phosphorylation, which may account for the inhibition of HIF-1α protein during stress. Interestingly, glucose deprivation was shown to inhibit the translation of HIF-1α under hypoxia (39). Given the high similarity of the gene expression response between lactic acidosis and glucose deprivation (13), it is likely that both lactic acidosis and glucose deprivation share similar mechanisms on the translation inhibition of HIF-1α.
Our results have shown that activation of ATF4 and UPR occur in 1% pO2 with the presence of lactic acidosis. A previous study showed that ATF4 is only activated in MCF-7 until anoxia with extreme low O2 (< 0.01%)(40) thought to rarely occur in human tumors. Since solid tumors are frequently associated with regions of poor perfusion, tumor cells are often exposed to both lactic acidosis and hypoxia spatially. Our findings indicate that ATF4 activation and UPR gene induction may be more prevalent in tumors than previously thought. Indeed, recent reports point to UPR genes (e.g., ATF3, Ero1L, etc) being induced in solid tumors (41). In addition, these observations suggest a hierarchy among stress responses to cope with different levels of metabolic stresses. During moderate hypoxia, hypoxia gene expression prevails to deal with the metabolic stress, whereas in response to combined hypoxia and lactic acidosis or extreme anoxia (<0.01% pO2), the hypoxia response becomes inadequate and inhibited to switch to UPR and ATF4-driven responses. One possible reason for the extreme metabolic stresses under hypoxia and lactic acidosis may be due to the inhibition of hypoxia-driven glycolysis by co-existing lactic acidosis to create a significant ATP reduction (noted in (12)). During the transition from HIF-1α to ATF4, the significant overlap between their target genes (e.g. VEGF) may offer a mechanism for orderly maintenance of certain conserved responses while switching among stress-specific responses to cope with the distinct challenges of different degrees of metabolic stresses.
Many histopathology studies have shown areas of necrosis surrounded by regions with expression of HIF target genes. Since these regions are also expected to be exposed to lactic acidosis, how do we reconcile these observations with our findings? It is important to point out that while lactic acidosis attenuates the hypoxia response, it usually does not completely shut off hypoxia response when compared with non-hypoxic area. Furthermore, in the regions of necrosis, there may be many additional metabolic stresses, such as deprivation of glucose and other nutrients, in addition to hypoxia and lactic acidosis. The simultaneous presence of these multiple stresses may contribute to necrosis in the solid tumors not entirely observed under combined hypoxia and lactic acidosis used in this study.
While our current studies reveal the ability of lactic acidosis to inhibit hypoxia responses during short term exposure, these results may also provide a mechanism for long-term selection. One important aspect of tumor progression is genomic instability and high frequency of somatic mutations (42). Long term exposure to these stresses may select for tumors with certain mutations which modulate the hypoxia pathway and become resistant to the inhibitory effects of lactic acidosis. Activation of the HER2, PI3K and EGFR pathways or inactivation of pVHL-dependent degradation pathways caused by various somatic mutations and CNAs, may trigger and further enhance the hypoxia pathway by a mechanism that is resistant to inhibition by lactic acidosis. These pathways are often associated with the hypoxia pathway in human cancers (15). In addition, such stresses select for tumors bearing certain mutations which confer survival advantages. Over time, tumor cells with gain-of-function mutations will clonally expand and become the dominant components of tumors, a process sometimes termed somatic evolution (9, 43–45). For example, hypoxia enriches for tumor cells that lack p53 (46) and glucose deprivation selects for tumor cells that bear K-ras mutations (47). Similarly, CNAs of the ATF4 locus in a subset of breast cancer cells lead to high expression levels of ATF4 and ATF4-driven VEGF and provide a survival advantage under hypoxia and lactic acidosis. While observed in a small subset of breast tumor and cell lines, the gain at the ATF4 locus is likely to be relevant for survival under different stresses and may be targeted therapeutically using agents targeting UPR. In addition, our results also suggest a mechanism that bridges the gap between the short-term transcriptional responses and long-term adaptive somatic mutations resulting in CNAs of human tumors and cell lines. A similar approach may be helpful to identify additional CNAs which offer survival advantages under distinct sets of microenvironmental stresses.
These findings advance our understanding of the link between survival mechanisms under stresses and “hard-wired” CNAs to circumvent the barriers of stresses presented by hypoxia and lactic acidosis. ATF4 overexpression alone has been shown to be sufficient to trigger a partial transcription program and anti-stress response (48). Therefore, the hard-wired CNAs of ATF4 may lead to a constitutive and/or high induction levels of ATF4 protein to provide survival advantages under various stresses and may lead to drug resistance and treatment failure (49). The tumors harboring ATF4 CNAs may be especially addicted to high levels of ATF4 and may be more likely to respond to the agents that specifically target ATF4 and/or the UPR pathway.
Supplementary Material
Acknowledgments
We recognize research support from the NIH (NCI R01CA125618 to J.T.C, R01CA94214 to C.K.) and Komen Foundation grant KG090869 to J.T.C. We also appreciate the technical assistance and helpful discussions with members of Chi lab. The funding sources have no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; or in the decision to submit the manuscript for publication.
References
- 1.Quennet V, Yaromina A, Zips D, Rosner A, Walenta S, Baumann M, et al. Tumor lactate content predicts for response to fractionated irradiation of human squamous cell carcinomas in nude mice. Radiother Oncol. 2006;81:130–5. doi: 10.1016/j.radonc.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Brizel DM, Schroeder T, Scher RL, Walenta S, Clough RW, Dewhirst MW, et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2001;51:349–53. doi: 10.1016/s0360-3016(01)01630-3. [DOI] [PubMed] [Google Scholar]
- 3.Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfor K, Rofstad EK, et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–21. [PubMed] [Google Scholar]
- 4.Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Semin Radiat Oncol. 2004;14:198–206. doi: 10.1016/j.semradonc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 6.Jung HJ, Park JW, Lee JS, Lee SR, Jang BC, Suh SI, et al. Silibinin inhibits expression of HIF-1alpha through suppression of protein translation in prostate cancer cells. Biochem Biophys Res Commun. 2009;390:71–6. doi: 10.1016/j.bbrc.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi H, Pino MS, Zeng M, Shirasawa S, Chung DC. Oncogenic KRAS and BRAF differentially regulate hypoxia-inducible factor-1alpha and -2alpha in colon cancer. Cancer Res. 2009;69:8499–506. doi: 10.1158/0008-5472.CAN-09-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, et al. Gene Expression Programs in Response to Hypoxia: Cell Type Specificity and Prognostic Significance in Human Cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang JS, Gillies RD, Gatenby RA. Adaptation to hypoxia and acidosis in carcinogenesis and tumor progression. Semin Cancer Biol. 2008;18:330–7. doi: 10.1016/j.semcancer.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moellering RE, Black KC, Krishnamurty C, Baggett BK, Stafford P, Rain M, et al. Acid treatment of melanoma cells selects for invasive phenotypes. Clin Exp Metastasis. 2008;25:411–25. doi: 10.1007/s10585-008-9145-7. [DOI] [PubMed] [Google Scholar]
- 11.Willam C, Warnecke C, Schefold JC, Kugler J, Koehne P, Frei U, et al. Inconsistent effects of acidosis on HIF-alpha protein and its target genes. Pflugers Arch. 2006;451:534–43. doi: 10.1007/s00424-005-1486-3. [DOI] [PubMed] [Google Scholar]
- 12.Chen JL, Lucas JE, Schroeder T, Mori S, Wu J, Nevins J, et al. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet. 2008;4:e1000293. doi: 10.1371/journal.pgen.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JL, Merl D, Peterson CW, Wu J, Liu PY, Yin H, et al. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas JE, Kung HN, Chi JT. Latent factor analysis to discover pathway-associated putative segmental aneuploidies in human cancers. PLoS computational biology. 2010;6:e1000920. doi: 10.1371/journal.pcbi.1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatza ML, Kung HN, Blackwell KL, Dewhirst MW, Marks JR, Chi JT. Analysis of tumor environmental response and oncogenic pathway activation identifies distinct basal and luminal features in HER2-related breast tumor subtypes. Breast Cancer Res. 2011;13:R62. doi: 10.1186/bcr2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukumura D, Xu L, Chen Y, Gohongi T, Seed B, Jain RK. Hypoxia and acidosis independently up-regulate vascular endothelial growth factor transcription in brain tumors in vivo. Cancer Res. 2001;61:6020–4. [PubMed] [Google Scholar]
- 17.Chiche J, Brahimi-Horn MC, Pouyssegur J. Tumor hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Duan B, Wang DG, Deng XH, Zhang GY, Xu L, et al. Coupling between NMDA receptor and acid-sensing ion channel contributes to ischemic neuronal death. Neuron. 2005;48:635–46. doi: 10.1016/j.neuron.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. 2004;6:642–7. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- 20.Sorensen BS, Hao J, Overgaard J, Vorum H, Honore B, Alsner J, et al. Influence of oxygen concentration and pH on expression of hypoxia induced genes. Radiother Oncol. 2005;76:187–93. doi: 10.1016/j.radonc.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 21.Sorensen BS, Alsner J, Overgaard J, Horsman MR. Hypoxia induced expression of endogenous markers in vitro is highly influenced by pH. Radiother Oncol. 2007;83:362–6. doi: 10.1016/j.radonc.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 22.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–96. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107:6994–9. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–33. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 27.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 31.Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–7. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 32.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 33.Shan J, Lopez MC, Baker HV, Kilberg MS. Expression profiling after activation of the amino acid deprivation response in HepG2 human hepatoma cells. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00217.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 35.Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–16. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116–31. doi: 10.1261/rna.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vantelon N, Rioux-Bilan A, Ingrand S, Pain S, Page G, Guillard O, et al. Regulation of initiation factors controlling protein synthesis on cultured astrocytes in lactic acid-induced stress. Eur J Neurosci. 2007;26:689–700. doi: 10.1111/j.1460-9568.2007.05698.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang MJ, Lin S. A region within the 5′-untranslated region of hypoxia-inducible factor-1alpha mRNA mediates its turnover in lung adenocarcinoma cells. J Biol Chem. 2009;284:36500–10. doi: 10.1074/jbc.M109.008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas JD, Dias LM, Johannes GJ. Translational repression during chronic hypoxia is dependent on glucose levels. RNA. 2008;14:771–81. doi: 10.1261/rna.857308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29:4424–35. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 41.Marotta D, Karar J, Jenkins WT, Kumanova M, Jenkins KW, Tobias JW, et al. In Vivo Profiling of Hypoxic Gene Expression in Gliomas Using the Hypoxia Marker EF5 and Laser-capture Microdissection. Cancer Research. 2011;71:779–89. doi: 10.1158/0008-5472.CAN-10-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nature reviews Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 43.Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 45.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 46.Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 47.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan J, Ord D, Ord T, Kilberg MS. Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J Biol Chem. 2009;284:21241–8. doi: 10.1074/jbc.M109.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rzymski T, Milani M, Singleton DC, Harris AL. Role of ATF4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle. 2009;8:3838–47. doi: 10.4161/cc.8.23.10086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.