Abstract
A brief survey of computer algorithms that have been developed to generate predictions of the secondary structures of RNA molecules is presented. Two particular methods are described in some detail. The first utilizes a thermodynamic energy minimization algorithm that takes into account the likelihood that short-range folding tends to be favored over long-range interactions. The second utilizes an interactive computer graphic modelling algorithm that enables the user to consider thermodynamic criteria as well as structural data obtained by nuclease susceptibility, chemical reactivity and phylogenetic studies. Examples of structures for prokaryotic 16S and 23S ribosomal RNAs, several eukaryotic 5S ribosomal RNAs and rabbit beta-globin messenger RNA are presented as case studies in order to describe the two techniques. Anm argument is made for integrating the two approaches presented in this paper, enabling the user to generate proposed structures using thermodynamic criteria, allowing interactive refinement of these structures through the application of experimentally derived data.
Full text
PDF


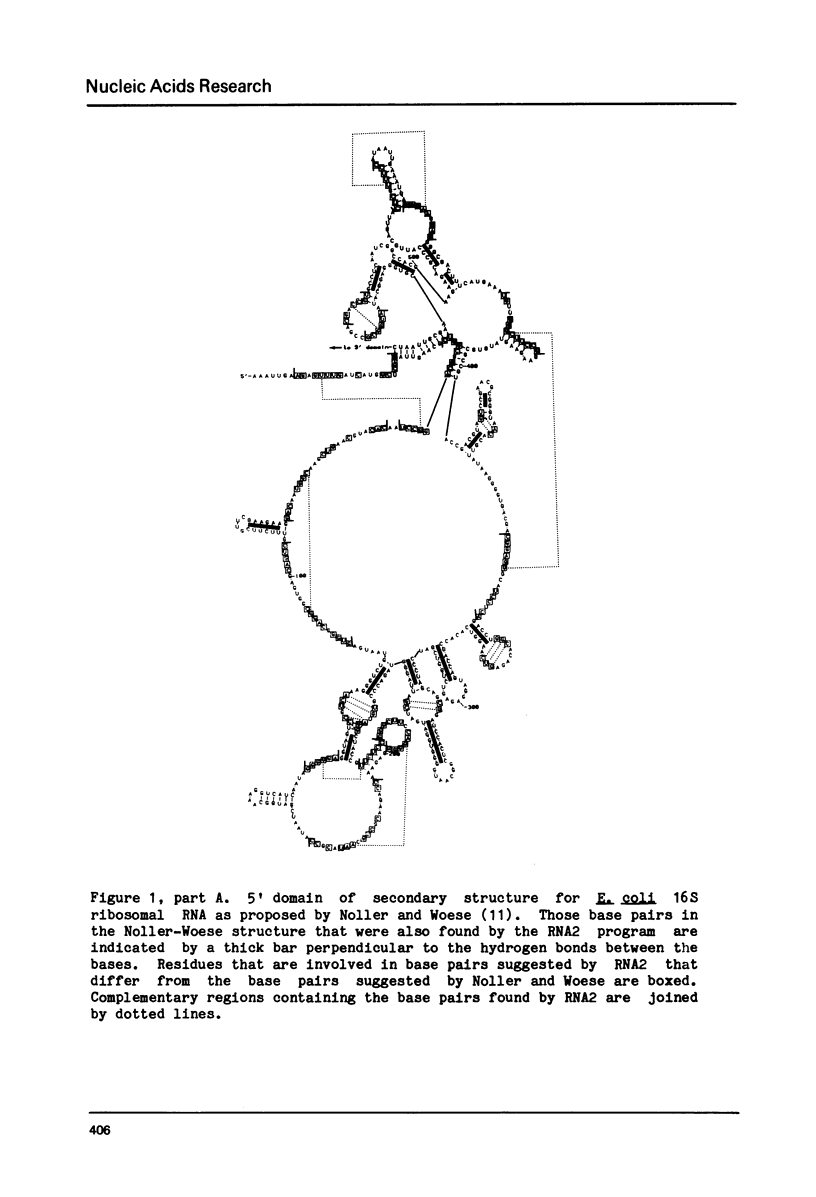
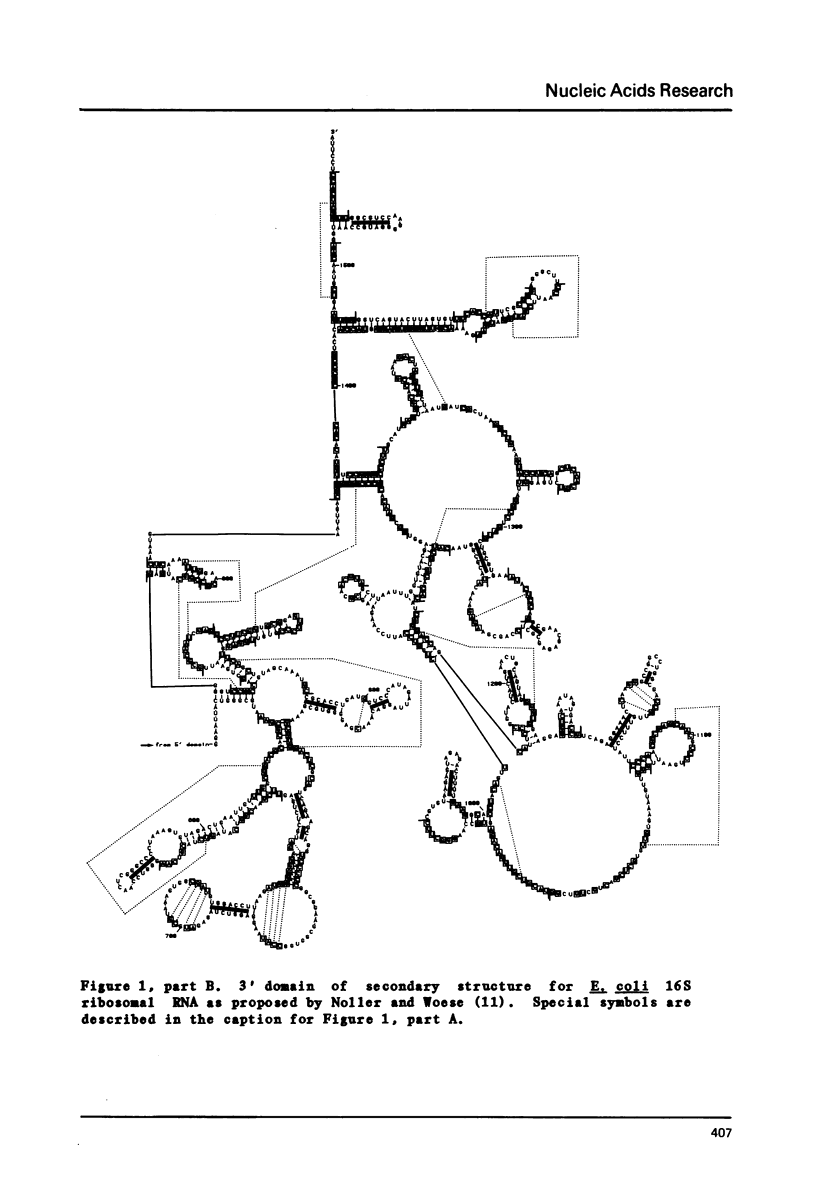




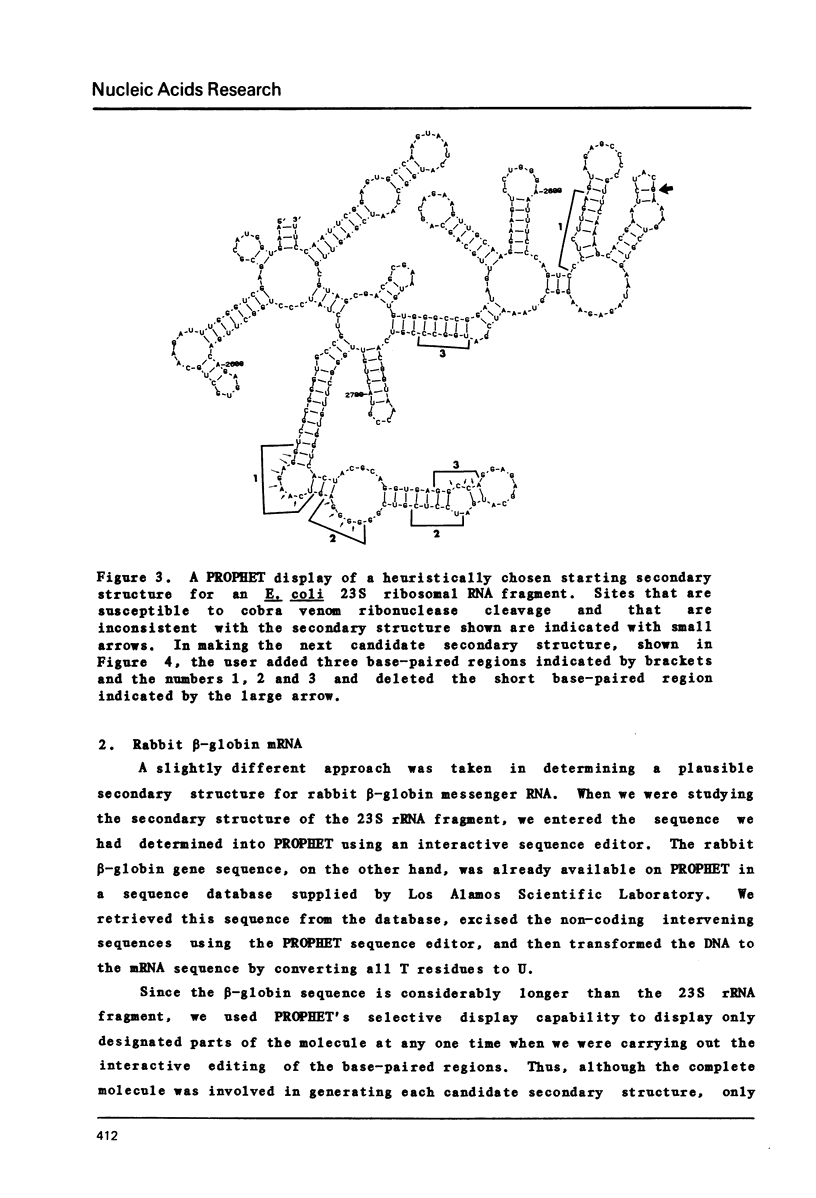
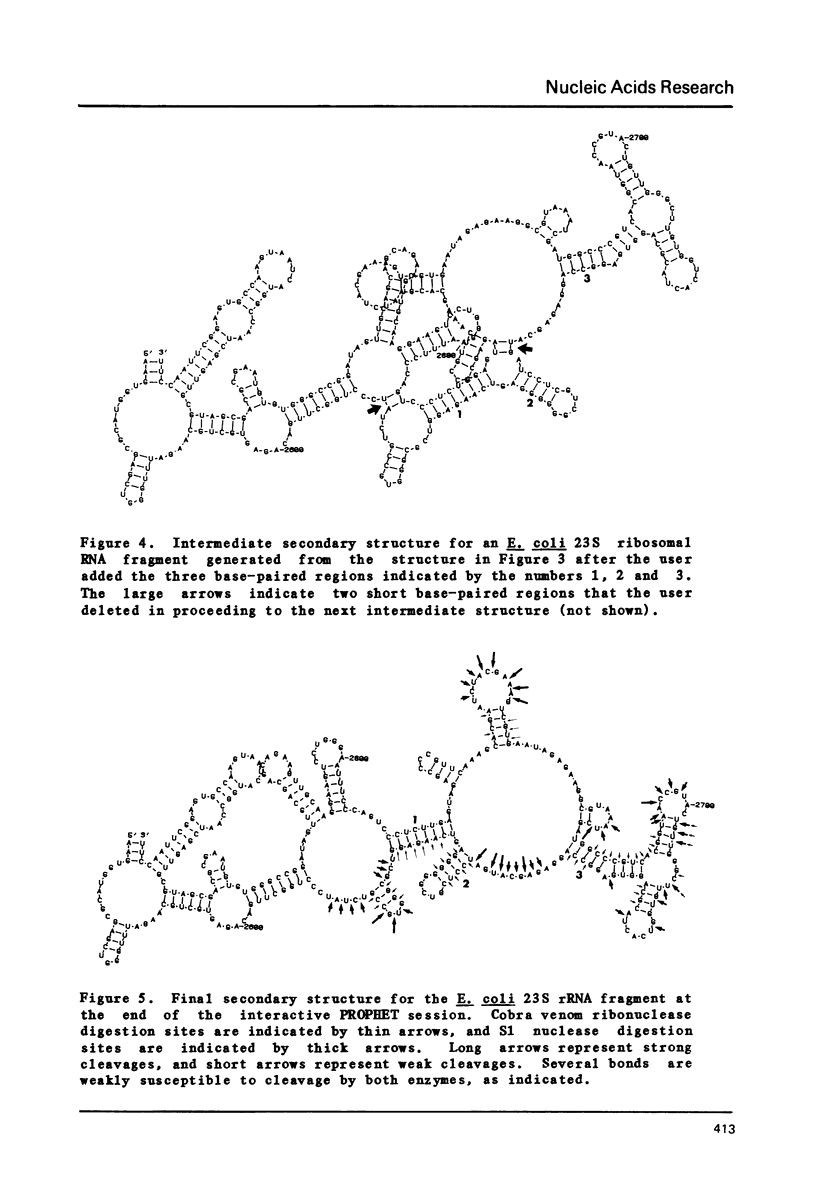



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1981 Jan 10;9(1):r25–r42. doi: 10.1093/nar/9.1.213-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRESCO J. R., ALBERTS B. M., DOTY P. Some molecular details of the secondary structure of ribonucleic acid. Nature. 1960 Oct 8;188:98–101. doi: 10.1038/188098a0. [DOI] [PubMed] [Google Scholar]
- Ghora B. K., Apirion D. Identification of a novel RNA molecule in a new RNA processing mutant of Escherichia coli which contains 5 S rRNA sequences. J Biol Chem. 1979 Mar 25;254(6):1951–1956. [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub W. F. The PROPHET system and resource sharing. Fed Proc. 1974 Dec;33(12):2390–2392. [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Spillmann S., Dohme F., Nierhaus K. H. Assembly in vitro of the 50 S subunit from Escherichia coli ribosomes: proteins essential for the first heat-dependent conformational change. J Mol Biol. 1977 Sep 25;115(3):513–523. doi: 10.1016/0022-2836(77)90168-1. [DOI] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Zuker M., Ebel J. P., Ehresmann C. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res. 1981 May 11;9(9):2153–2172. doi: 10.1093/nar/9.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Vasilenko S. K., Ryte V. C. [Isolation of highly purified ribonuclease from cobra (Naja oxiana) venom]. Biokhimiia. 1975 May-Jun;40(3):578–583. [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


