Abstract
The Dcp1:Dcp2 decapping complex catalyses the removal of the mRNA 5′ cap structure. Activator proteins, including Edc3 (enhancer of decapping 3), modulate its activity. Here, we solved the structure of the yeast Edc3 LSm domain in complex with a short helical leucine-rich motif (HLM) from Dcp2. The motif interacts with the monomeric Edc3 LSm domain in an unprecedented manner and recognizes a noncanonical binding surface. Based on the structure, we identified additional HLMs in the disordered C-terminal extension of Dcp2 that can interact with Edc3. Moreover, the LSm domain of the Edc3-related protein Scd6 competes with Edc3 for the interaction with these HLMs. We show that both Edc3 and Scd6 stimulate decapping in vitro, presumably by preventing the Dcp1:Dcp2 complex from adopting an inactive conformation. In addition, we show that the C-terminal HLMs in Dcp2 are necessary for the localization of the Dcp1:Dcp2 decapping complex to P-bodies in vivo. Unexpectedly, in contrast to yeast, in metazoans the HLM is found in Dcp1, suggesting that details underlying the regulation of mRNA decapping changed throughout evolution.
Keywords: mRNA decapping, proteins, RNA, structural biology
Introduction
The cleavage of the mRNA 5′ cap structure (decapping) is an important step in gene expression, as it removes a transcript from the translational pool. Therefore, it is essential to tightly regulate the activity of the enzymes that perform the decapping reaction as premature or incomplete decapping could interfere with cellular homeostasis. The mRNA decapping reaction is catalysed by the Dcp2 enzyme (van Dijk et al, 2002; Wang et al, 2002), which works in concert with its prime activator Dcp1. Dcp1:Dcp2 activity is regulated in vivo and in vitro by several proteins, including the enhancers of decapping 1–4 (Edc1–4), Pat1 and the LSm1–7 complex (Boeck et al, 1998; Bonnerot et al, 2000; Bouveret et al, 2000; Kshirsagar and Parker, 2004; Fenger-Gron et al, 2005; Haas et al, 2010; Borja et al, 2011).
Initial insights into the mechanism and regulation of the decapping complex have been provided by a number of high-resolution crystal structures. Dcp1 (She et al, 2004) folds into an EVH1 domain (Callebaut, 2002) and interacts tightly with Dcp2 (She et al, 2006, 2008) via an N-terminal helical extension that is unique to Dcp1. The structure of Dcp2 displays two folded domains, an N-terminal α-helical regulatory domain followed by a catalytic or Nudix domain (She et al, 2006) (Figure 1A). Interestingly, the three different crystal structures of Dcp2 (She et al, 2006, 2008) (Supplementary Figure S1) show that the Dcp2 catalytic domain can adopt multiple, significantly different orientations with respect to the Dcp2 regulatory domain. In general, the available Dcp1:Dcp2 structures can be divided into open and closed forms. In solution, it has been shown that different structural states are sampled during the enzymatic cycle, and the compactness of the decapping complex increases when nucleotides are added (She et al, 2008). These observations suggest that the active state is more closed, whereas the inactive state is in a more open conformation. More recently, however, it was noted that the residues involved in substrate binding are not clustered in any of the known structures (Floor et al, 2010). This suggests that the closed structure, as observed in the high-resolution crystal structure, does not represent the catalytically active form of the decapping complex (Floor et al, 2010) and that additional structural states of the enzyme must exist.
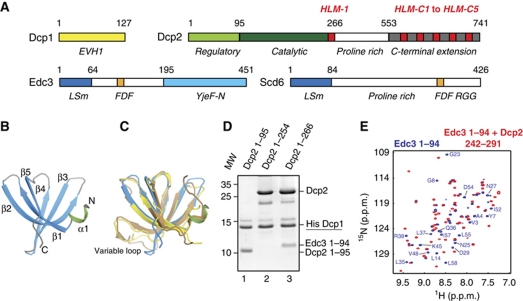
Figure 1.
Interaction between Dcp2 and Edc3. (A) Domain organization of the S. pombe Dcp1, Dcp2, Edc3, and Scd6 (Tral/Sum2) proteins. Folded protein domains are coloured in yellow (Dcp1), light green (Dcp2 regulatory domain), dark green (Dcp2 catalytic domain), cyan (YjeF-N domain), and blue (Lsm domain). The FDF motif is coloured orange, the Dcp2 linear sequence motifs that interact with the Edc3 and Scd6 LSm domains (HLM-1, HLM-C1 to HLM-C5; see below) are coloured red. (B) The solution structure of the S. pombe Edc3 LSm domain. Secondary structure elements are indicated. The short N-terminal helical turn is coloured green. All figures displaying protein structures are made using Pymol (http://www.pymol.org). (C) An overlay of the structures of the S. pombe LSm domain (blue/green) with the Edc3 LSm domains from D. melanogaster (brown, PDB: 2RM4) and H. sapiens (yellow, PDB: 2VC8). The variable region that is absent in the yeast Edc3 LSm domain is indicated. (D) Identification of the Dcp2 residues required for interaction with Edc3. His6–Dcp1, was co-expressed with different versions of Dcp2 (see Supplementary data) and supplemented with separately expressed Edc3 LSm domain. The mixed cell lysates were purified using Ni-affinity chromatography. Dcp1 interacts with Dcp2 in all cases, indicating that all Dcp2 constructs were properly folded. Only the Dcp1:Dcp2 complex that contains Dcp2 residues 255–266 binds the Edc3 LSm domain (lane 3). (E) 1H-15N NMR spectra of the 15N-labelled S. pombe Edc3 LSm domain in the absence (blue) and presence (red) of an unlabelled peptide corresponding to residues 242–291 in Dcp2. Some assignments in the isolated Edc3 LSm domain (blue) are indicated.
The Dcp2 residues C-terminal to the structured catalytic domain are rich in proline residues (Gaudon et al, 1999), harbour low overall sequence conservation, and are predicted to be largely intrinsically disordered (Figure 1A). No functional data are available for this Dcp2 region, apart from a 50 amino acid long sequence close to the C-terminal part of the catalytic domain that has been reported to interact with Edc3 in yeast (Harigaya et al, 2010).
In a cellular context, the Dcp1:Dcp2 decapping enzyme is part of an RNP (ribonucleo protein) complex that can vary in composition (Franks and Lykke-Andersen, 2008). Under certain conditions, it has been observed that these decapping complexes can cluster into large assemblies called mRNA processing bodies or P-bodies, which can be visualized using fluorescence microscopy (Sheth and Parker, 2003; Franks and Lykke-Andersen, 2008). How the clustering of the mRNA degradation machinery influences cellular function is unclear, but most of the proteins found in P-bodies are involved in translational repression and mRNA degradation (Eulalio et al, 2007). However, the molecular details of the intermolecular interactions within these large protein–RNA complexes are poorly understood. One of the proteins that influences P-body formation is the enhancer of decapping 3 (Edc3), and Edc3 deletion results in a reduction in the number and size of these cellular foci (Decker et al, 2007; Ling et al, 2008). In yeast, both the LSm domain and the YjeF-N dimerization domain of Edc3 have been shown to be functionally important for P-body formation (Decker et al, 2007).
Proteins of the Scd6 family (Lsm13–15; Trailer hitch/Tral in Drosophila melanogaster; CAR-1 in Caenorhabditis elegans; Sum2 in Schizosaccharomyces pombe) share a number of features with Edc3 (Figure 1A). First, both proteins localize to P-bodies (Tritschler et al, 2007, 2008). Second, both Edc3 and Scd6 contain an FDF motif that directly interacts with the DEAD box helicase Dhh1 in a competitive fashion (Tritschler et al, 2008, 2009a). Third, both proteins contain an N-terminal LSm domain (Albrecht and Lengauer, 2004). Finally, both proteins are part of the same RNP and interact with Dcp2 in yeast (Decourty et al, 2008; Tarassov et al, 2008; Nissan et al, 2010). The cellular function of Scd6 is not well established, although Scd6 seems to play a role in inhibiting translation initiation in yeast (Nissan et al, 2010), and Scd6 orthologues have been shown to associate with translationally repressed mRNAs (Decker and Parker, 2006). Interestingly, a functional redundancy between Edc3 and Scd6 has been proposed (Decourty et al, 2008). However, in contrast to Edc3, Scd6 has not been shown to modulate the catalytic activity of the Dcp1:Dcp2 decapping complex in vitro.
Here, we address how Edc3 and Scd6 interact with the Dcp1:Dcp2 decapping complex and thereby influence its enzymatic activity. To this end, we solved the structure of the S. pombe Edc3 LSm domain in complex with a sequence motif derived from Dcp2. Based on this structure, we identified additional conserved sequence motifs in Dcp2 that interact with the LSm domains of Scd6 and Edc3 in a mutually exclusive manner. We show that deletion of the C-terminal motifs of Dcp2 that interact with the Edc3 and Scd6 LSm domains results in the failure of the Dcp1:Dcp2 decapping complex to localize to P-bodies. In summary, our data reveal the molecular details of a network of competing interactions that can modulate Dcp1:Dcp2 decapping activity and cellular localization.
Results
S. pombe Dcp2 residues 255–266 interact with the Edc3 LSm domain
The Edc3 protein is composed of an N-terminal LSm domain (Like-Sm) (Albrecht and Lengauer, 2004; Tritschler et al, 2007) that is connected to a C-terminal YjeF-N domain (Ling et al, 2008) (Figure 1A). The linker region between the two folded domains is predicted to be unstructured and contains an FDF motif that interacts with the RNA helicase Dhh1/Me31b (Tritschler et al, 2007, 2009a). In yeast, the Edc3 LSm domain has been shown to interact with a 50 amino acid long region in Dcp2 that follows the catalytic domain (Harigaya et al, 2010). To obtain insight into this interaction, we solved the solution structure of the S. pombe Edc3 LSm domain.
The Edc3 LSm domain folds into a canonical five-stranded β-sheet that is preceded by a short α-helical turn (Figure 1B; Supplementary Figure S2A; Supplementary Table SIA). The yeast Edc3 LSm structure is similar to the structures of the D. melanogaster and Homo sapiens Edc3 LSm domains (Figure 1C) (Tritschler et al, 2007) although the variable loop between β-strands 3 and 4 is shorter in the yeast protein. Many (L)Sm proteins form hexa- or heptameric rings (e.g. LSm1–7 and Hfq) (Wilusz and Wilusz, 2005). However, the S. pombe Edc3 LSm domain is monomeric in solution, as previously described for the D. melanogaster and H. sapiens Edc3 LSm domains (Tritschler et al, 2007).
To identify which residues in Dcp2 are responsible for the interaction with Edc3, we performed pull-down experiments with His6–Dcp1:Dcp2 decapping complexes. We used Dcp2 constructs of increasing length (Figure 1D) and found that a Dcp2 region located between residues 255 and 266 is required for the interaction with Edc3 (Figure 1D, lane 3 versus lanes 1 and 2). Both in Saccharomyces cerevisiae (Harigaya et al, 2010) and in S. pombe, a Dcp2 region C-terminal of the catalytic domain is thus important for the interaction with Edc3.
In the reverse experiment, we performed NMR titration experiments to identify residues in the Edc3 LSm domain that interact with Dcp2. To this end, we added NMR-inactive Dcp2 to 15N-labelled Edc3 LSm domain (Figure 1E, blue). Edc3 residues that come in close spatial proximity with Dcp2 are expected to experience chemical shift perturbations upon binding. Indeed, >50% of the residues in the Edc3 LSm domain sense the presence of Dcp2 (Figure 1E, blue versus red). The large number of chemical shift changes, however, prevented the accurate identification of an Edc3 binding site for Dcp2 residues 242–291. This could reflect a small secondary conformational effect in the Edc3 LSm domain upon Dcp2 binding, but given the marked sensitivity of amide group chemical shifts to even small structural movements, any such change may be very small.
The Edc3 LSm domain interacts with Dcp2 in an unprecedented manner
Previous studies have shown that LSm domains use two distinct modes to interact with their binding partners. In one mode, they establish intermolecular interactions between β-strand 4 of one monomer and β-strand 5 of another monomer (Kambach et al, 1999), resulting in the assembly of a six- or seven-member LSm ring (Wilusz and Wilusz, 2005). The other mode of interaction is between LSm rings and RNA, in which binding sites on both the proximal and distal faces have been reported and often involve residues in the loop connecting β-strands 2 and 3 and residues in the loop that connects β-strands 4 and 5 (Toro et al, 2001; Thore et al, 2003; Link et al, 2009; Leung et al, 2011). As such, we expected that the monomeric Edc3 LSm domain would exploit one of these binding sites to interact with Dcp2.
The NMR titration experiments (Figure 1E) did not reveal which residues in Edc3 contact Dcp2 directly. To determine the molecular details of this interaction, we assigned the resonances of the complex of the Edc3 LSm domain and Dcp2 residues 242–291 and solved the solution structure (Figure 2A and B; Supplementary Figure S2B; Supplementary Table SIB). In the complex, the LSm domain fold remains unchanged (Supplementary Figure S2C–E), whereas Dcp2 residues 257–266 fold into an amphipathic α-helical structure (shown in red in Figure 2). Due to the Dcp2 interaction, the hydrophobic core of the Edc3 LSm domain increases significantly in size, which explains the large number of chemical shift changes in the NMR spectrum of the Edc3 LSm domain upon complex formation with Dcp2 (Figure 1E). The residues in the Edc3-binding site that are closest to Dcp2 are located in the Edc3 N-terminal α-helix (F6), β-strand 2 (F28), the loop between β-strands 4 and 5 (T49 and I52), and β-strand 5 (D54, L55, and I57) (Figure 2B). These residues form a hydrophobic surface on the LSm domain that contacts four residues on one side of the Dcp2 helix (S257, L260, L261, and L264) (Figure 2B). In particular, the methyl groups of the Dcp2 leucine residues make ample interactions with the aromatic and long hydrophobic residues in Edc3 and contribute significantly to the tight binding. Indeed, mutating either L260 or L264 in Dcp2 to alanine reduces the strength of the interaction, whereas a double mutation in which both of these leucines are replaced with alanines completely abolishes the Edc3:Dcp2 interaction (Supplementary Figure S3A). In summary, we identified that Dcp2 residues S257 to L264 form a linear motif that interacts tightly with Edc3 (Figures 1D, 2A and B).
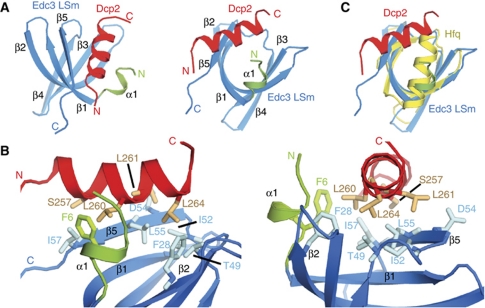
Figure 2.
The structure of the Edc3:Dcp2 complex. (A) The solution structure of the S. pombe Edc3 LSm domain in complex with Dcp2 residues 257–266. Edc3 residues are in blue and green (see Figure 1B), and Dcp2 residues are in red. (B) A view into the binding pocket of the Edc3:Dcp2 complex. Sidechains that provide the main intermolecular contacts are displayed with sticks. (C) An overlay of the Edc3:Dcp2 complex (blue, green, and red) with a structure of a canonical Hfq LSm domain (yellow, PDB: 1KQ1). The N-terminal helix is significantly shorter in the Edc3 LSm domain (green) compared with the canonical (L)Sm fold (yellow). The resultant space on the Edc3 surface that becomes available is used for interaction with the C-terminal end of the Dcp2 helix.
The mode of interaction of the Edc3 LSm domain with Dcp2 differs significantly from previously reported LSm interactions and presents a novel mode of binding between LSm domains and protein ligands. In canonical (L)Sm proteins an N-terminal α-helix covers the surface of the protein formed by β-strands 2 and 5, and the loop preceding β-strand 5. In the LSm domain of Edc3, the corresponding helix is much shorter and contains only a single turn (shown in green for Edc3 and yellow for one monomer of the hexameric Hfq protein in Figure 2C). As a consequence, an interaction surface becomes available that is used to contact the C-terminal half of the Dcp2 helix. In addition, the N-terminal half of the Dcp2 helix binds to the central residues in β-strand 5, which is used to contact the neighbouring monomer in multimeric LSm ring-like structures. Therefore, both the shorter N-terminal helix and the monomeric nature of the Edc3 LSm domain are important characteristics that allow the unprecedented mode of interaction with Dcp2. Although a Dcp2 fragment comprising residues 257–264 possess helical propensity, it is disordered in isolation in solution and only folds into a stable helical conformation in the presence of the Edc3 LSm domain (Supplementary Figure S3B).
To probe the relevance of our structure in the context of the active Dcp1:Dcp2 decapping complex, we compared NMR spectra of the Dcp1:Dcp2 (residues 1–289) decapping complex in the absence and presence of the Edc3 LSm domain. After the formation of the trimeric complex containing Dcp1:Dcp2 and Edc3, resonances that correspond to the Dcp2 helix appear at the same frequency as observed in the dimeric Edc3:Dcp2 complex (Supplementary Figure S4). This clearly indicates that the Dcp2 LSm interaction motif is structurally identical in the Dcp1:Dcp2 decapping complex and in the structure of the minimal complex that we present here.
Dcp2 contains multiple linear motifs that interact with Edc3
Close inspection of the Dcp2 residues that interact with Edc3 revealed that the primary amino acid sequence has a helical propensity. From the structure, it can be inferred that the amino acids of importance are SxxLLxxL (Figure 2B). Interestingly, the C-terminal extension of Dcp2 contains multiple similar sequence motifs that are conserved between different yeast species (Supplementary Figure S5). In the C-terminal Dcp2 extension of S. pombe (residue 553–741), there are at least five of these conserved motifs (Figures 1A and 3A; Supplementary Figure S5) located between residues 556 and 563 (SAQLLQAL), 581 and 589 (SLSLLTLL), 648 and 655 (QFDLLKVS), 693 and 700 (SPGFVKIL), and 721 and 729 (DDHFLSYL).
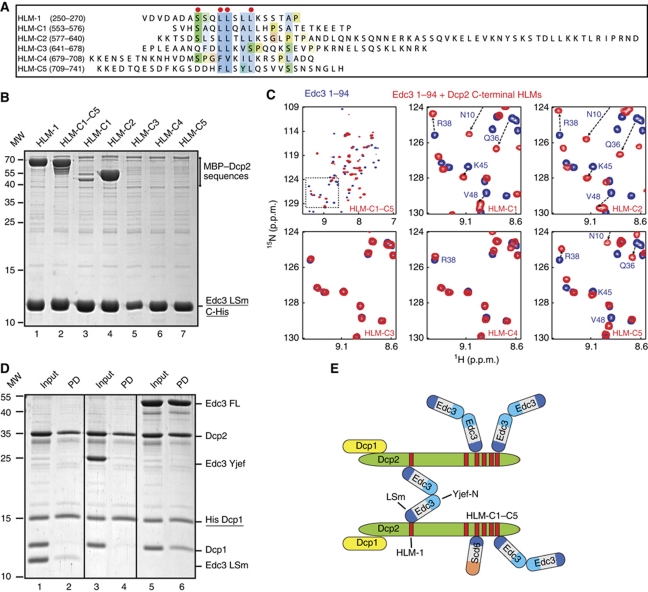
Figure 3.
The Dcp2 C-terminal region interacts with Edc3. (A) Sequence alignment of different HLMs in S. pombe Dcp2. The top sequence (HLM-1) is located close to the Dcp2 catalytic domain and is used to solve the Edc3:Dcp2 structure (Figure 2). The residues that contact the Edc3 LSm domain are marked with a red dot. The lower five sequences (HLM-C1 to HLM-C5) are located in the Dcp2 C-terminal extension (Supplementary Figure S5). The residue numbers of the Dcp2 fragments are indicated in brackets. (B) Pull-down experiments with the C-terminally His6-tagged Edc3 LSm domain and MBP-tagged LSm interaction motifs. The Edc3 LSm domain interacts strongly with the Dcp2 HLM-1 sequence (lane 1, Figures 1 and 2). A strong interaction is also observed between the last ∼200 Dcp2 residues (HLM-C1–C5, lane 2). The latter interaction is mediated through the HLM-C2 sequence (lane 4) and through the HLM-C1 sequence (lane 3). The interaction with the other HLMs in the Dcp2 C-terminus are too weak to be detected under the experimental conditions. The corresponding MBP–HLM proteins were present in the input of all pull-down experiments (Supplementary Figure S5C). (C) NMR spectra of 15N-labelled Edc3 LSm domain without (blue) and with (red) different HLMs derived from the Dcp2 C-terminus. Specific interactions are observed upon addition of the complete Dcp2 C-terminal extension (HLM-C1–C5) or upon addition of three (LSM-C1, LSM-C2, and LSM-C5) of the five fragments. Note that the same residues in the Edc3 LSm are affected upon addition of the HLM-1 sequence (Figure 1E). (D) Edc3 can interact with two decapping complexes simultaneously. Purified His6-tagged Dcp1:Dcp2 (residues 1–289) and untagged Dcp1:Dcp2 were mixed together with different Edc3 constructs (LSm domain, YjeF-N domain, or FL). The protein mixture was applied to Ni-affinity resin that selected for His6-tagged Dcp1 (underlined). The bound protein was eluted from the matrix prior to SDS–PAGE analysis. Lanes 5 and 6 show that untagged Dcp1 co-purified with His6-tagged Dcp1 in the presence of FL Edc3. (E) Summary of the Edc3:Dcp2 interactions we identified here. The Dcp2 C-terminus can interact with the Edc3 LSm domain (and the Scd6 LSm domain, see below) at multiple sites. The Edc3 dimer can bind two decapping complexes simultaneously.
Similar linear sequence motifs were previously identified in Dcp2 from S. cerevisiae (Gaudon et al, 1999). These motifs were termed helical leucine-rich motifs (HLMs) and in analogy we will refer to the linear motifs we identified here in S. pombe Dcp2 as HLM-1 for the first motif (residues 257–264) and HLM-C1 to HLM-C5 for the C-terminal motifs. Apart from these motifs, the sequence conservation of the Dcp2 C-terminus is very low. In addition, the C-terminal extension is predicted to be unstructured in isolation, which is confirmed by NMR spectra of Dcp2 residues 553–741 (Supplementary Figure S6A).
The presence of several motifs in the Dcp2 C-terminal extension that resembled the sequence that interacts with the Edc3 LSm domain prompted us to probe for interactions between this region of Dcp2 and Edc3. To this end, we performed pull-down experiments using the His6-tagged Edc3 LSm domain and the Dcp2 C-terminal extension (residues 553–741; Figure 3B; Supplementary Figure S6C). As a positive control, we used a construct of Dcp2 residues 96–291 that includes HLM-1 and that strongly interacts with Edc3 (Figures 1D and 3B, lane 1). Interestingly, the Dcp2 C-terminal extension shows a strong interaction with the Edc3 LSm domain (Figure 3B, lane 2), which corroborates our hypothesis that this region of Dcp2 contains at least one motif that strongly interacts with Edc3. To probe which of the HLM-C1 to HLM-C5 sequences is responsible for Edc3 binding, we performed co-purifications with His6-tagged Edc3 and five Dcp2 protein fragments, each of which contained a potential interaction motif (Figure 3B, lanes 3–7; Supplementary Figure S6C). We observed that the fragment containing HLM-C2 (lane 4) interacted with the Edc3 LSm domain to a similar extent as HLM-1 (lane 1). In addition, a weaker interaction between Edc3 and a fragment containing HLM-C1 was observed (lane 3). In contrast, interactions with the other motifs were not detected in this assay. Hence, we conclude that HLM-C2 is the main contributor to the binding between the Dcp2 C-terminal extension and Edc3.
To investigate whether the HLMs that did not interact with Edc3 in pull-down assays exhibited weak affinity for Edc3, we used NMR titration experiments. We followed changes in the spectrum of the 15N-labelled Edc3 LSm domain upon addition of the different Dcp2 C-terminal motifs (Figure 3C). In agreement with the pull-down experiments, both the complete C-terminal region (HLM-C1–C5) and the two isolated motifs (HLM-C1 and HLM-C2) induce large chemical shift changes in the Edc3 spectrum (Figure 3C, top panels). In addition, we observed chemical shift changes upon the addition of the most C-terminal motif (HLM-C5), indicating that the last motif also interacts with Edc3. The other two motifs in the Dcp2 C-terminal tail do not interact significantly with Edc3, which can be judged from the very slight (HLM-C4) or nonexistent chemical shift changes (HLM-C3) in the NMR spectra of the Edc3 LSm domain. The absence of binding for these two motifs (HLM-C3 and HLM-C4) correlates well with the lack of helical propensity in these Dcp2 sequences (Figure 3A).
All motifs that interact with Edc3 induce similar changes in the NMR spectrum, indicating that all HLMs interact with the Edc3 LSm domain in a structurally similar way, as displayed in the structure of the Edc3:HLM-1 complex (Figure 2A and B). In the reverse experiment, we probed the effect of the Edc3 LSm domain addition on the spectrum of the disordered 15N-labelled Dcp2 C-terminal residues. As expected, we clearly observed binding and resonances indicative of α-helical structure appeared (Supplementary Figure S6B). This shows that the leucine-rich motifs in the disordered Dcp2 C-terminus fold into an α-helical conformation when interacting with the Edc3 LSm domains, as we observed for the HLM-1 sequence.
The Edc3–Dcp2 interaction can promote clustering of decapping complexes
Our findings show that Dcp2 contains multiple HLMs that can interact with the Edc3 LSm domain (Figure 3B and C). Interestingly, Edc3 forms dimers in solution that are mediated by the C-terminal YjeF-N domain (Ling et al, 2008). This observation prompted us to probe if one Edc3 dimer could also interact with two Dcp1:Dcp2 decapping complexes simultaneously.
To test this, we incubated equal amounts of purified His6-tagged Dcp1:Dcp2 and untagged Dcp1:Dcp2 in the presence of different versions of the Edc3 protein. We then purified the protein mixture over Ni-NTA resin to select for protein complexes that contained at least one copy of His6-tagged Dcp1. When the decapping complex was supplemented with the Edc3 LSm domain (Figure 3D, lane 1), a complex consisting of His6-tagged Dcp1, Dcp2, and the Edc3 LSm domain was purified (Figure 3D, lane 2). This result is fully consistent with the experiment shown in Figure 1D (lane 3), in which the Edc3 LSm domain is shown to co-purify with His6–Dcp1:Dcp2. When a mixture of decapping complexes consisting of His6–Dcp1:Dcp2 and untagged Dcp1:Dcp2 was supplemented with the Edc3 YjeF-N dimerization domain (Figure 3D, lane 3), the Ni-NTA purified complex only contained the His6–Dcp1:Dcp2 complex (Figure 3D, lane 4). This indicates that the Edc3 YjeF-N dimerization domain does not directly interact with the Dcp1:Dcp2 decapping complex. However, when full-length (FL) Edc3 was added to the mixture of His6–Dcp1:Dcp2 and Dcp1:Dcp2 (Figure 3D, lane 5), the Ni-NTA purified complex did contain untagged Dcp1 (Figure 3D, lane 6). This clearly shows that untagged Dcp1 co-purifies with His6-tagged Dcp1 in the presence of Dcp2 and FL Edc3. This result can be explained by the formation of a complex that contains His6–Dcp1:Dcp2 coupled to an untagged Dcp1:Dcp2 complex through a FL Edc3 dimer.
These in vitro experiments indicate that one Edc3 dimer is able to act as a scaffold that can bind two decapping complexes simultaneously. This finding, taken together with the fact that Dcp2 can interact with multiple Edc3 proteins, suggests a redundant network of interactions between Dcp2 and Edc3 (Figure 3E).
The Scd6 LSm domain interacts with Dcp2
In addition to Edc3, other proteins that interact with yeast Dcp2 have been identified (Fromont-Racine et al, 2000; Decourty et al, 2008; Tarassov et al, 2008; Nissan et al, 2010). These include the DEAD box helicase Dhh1, Pat1, and the LSm domain containing protein Scd6 (Figure 1A). Given the similarities between the LSm domains of Edc3 and Scd6, we tested whether the Scd6:Dcp2 interaction is direct and mediated through the Scd6 LSm domain and the Dcp2 HLMs. To this end, we performed pull-down assays (Figure 4A) and found that, in contrast to the Edc3 LSm domain (Figure 3B), the interaction of the Scd6 LSm domain with HLM-1 in Dcp2 was too weak to be detected in this assay. However, using NMR spectroscopy as a more sensitive technique, we observed chemical shift changes in the NMR spectrum of the Scd6 LSm domain upon addition of HLM-1, HLM-C1, HLM-C2, and HLM-C5. This indicates that the Scd6 LSm domain specifically interacts with Dcp2 (Figure 4B), albeit with a lower affinity than the Edc3 LSm domain.
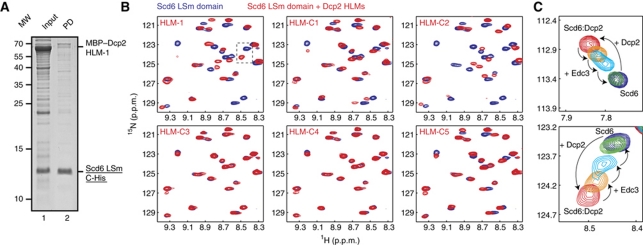
Figure 4.
Interaction between the Scd6 LSm domain and the Dcp2 HLMs. (A) Pull-down experiment of the Scd6 LSm domain with MBP–Dcp2 96–291. Both proteins are present in the E. coli cell lysate (Input, lane 1); however, the interaction is too weak to allow for co-purification (PD, pull-down, lane 2). As the Edc3 LSm domain:Dcp2 complex co-purified under the same conditions (Figure 3B), we conclude that Edc3 interacts stronger with Dcp2 than Scd6. (B) NMR spectra of the 15N-labelled Scd6 LSm domain in the absence (blue) and presence (red) of Dcp2 HLM-1 and HLM-C1 to HLM-C5. Specific binding between the Scd6 LSm domains is observed for the same motifs that interact with the Edc3 LSm domain (Figure 3C). (C) The Edc3 LSm domain competes with the Scd6 LSm domain for Dcp2 residues 242–291 (HLM-1). The NMR spectra of the 15N-labelled Scd6 LSm domain in isolation (blue) and in complex with the HLM-1 sequence (red) are shown. Unlabelled Edc3 LSm domain was added to the sample (orange, cyan, and green), which resulted in the dissociation of the Scd6:Dcp2 complex due to the formation of the NMR-invisible Edc3:Dcp2 complex. This indicates a much higher affinity for the Edc3:Dcp2 complex than for the Scd6:Dcp2 complex, as observed in the pull-down assays (Figures 3B and 4A) (see also Supplementary Figure S7).
Interestingly, motifs that interact with the Edc3 LSm domain also bind to the Scd6 LSm domain, whereas the motifs that are not recognized by the Edc3 LSm domain also fail to interact with the Scd6 LSm domain. In addition, the number of residues in Scd6 that experience shift changes during the NMR titration experiments are comparable to the results obtained for Edc3. These results strongly suggest a structurally similar mode of interaction. In conclusion, the LSm domains of Edc3 and Scd6 can interact with the same HLMs in Dcp2.
The Edc3 and Scd6 LSm domains compete for binding to Dcp2 HLM sequences
To confirm the finding that Edc3 and the Scd6 LSm domains recognize the exact same binding sites in Dcp2, we performed competition assays (Figure 4C). To this end, we prepared a complex of the 15N-labelled Scd6 LSm domain and the NMR-inactive HLM-1 (Dcp2 residues 241–291). NMR spectra clearly show that the Scd6 LSm domain (Figure 4C, blue spectrum) formed a complex with the Dcp2 HLM-1 sequence (Figure 4C, red spectrum). A stepwise addition of the tighter binding Edc3 LSm domain resulted in a release of HLM-1 from the Scd6 LSm domain (Figure 4C, orange, blue, and green). In a control experiment, where we added the Edc3 Yjef-N domain, the Scd6:Dcp2 complex does not dissociate (Supplementary Figure S7). In summary, our data show that the Edc3 and Scd6 LSm domains compete for the same Dcp2-binding motifs and that both interactions are mutually exclusive.
Both Edc3 and Scd6 stimulate mRNA decapping in vitro
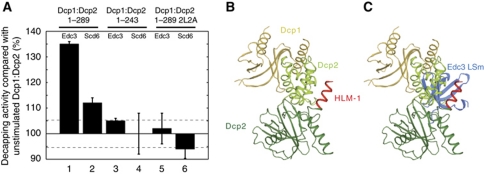
Previous studies have shown that the interaction of Edc3 with the Dcp1:Dcp2 decapping complex results in increased catalytic activity (Harigaya et al, 2010; Nissan et al, 2010). Given that the Scd6 LSm domain interacts with Dcp2 in a structurally similar way, we hypothesized that the Scd6 LSm domain should also stimulate Dcp1:Dcp2 decapping activity. To test this, we performed in vitro decapping assays using purified proteins.
As expected, the decapping activity of a Dcp1:Dcp2 complex containing FL Dcp1 and residues 1–289 of Dcp2 (which include the regulatory and catalytic domains and the HLM-1 sequence) was enhanced by addition of the Edc3 LSm domain (Figure 5A, lane 1). Interestingly, the Scd6 Lsm domain also stimulates Dcp1:Dcp2 decapping activity (lane 2). Nevertheless, the activation mediated by Scd6 was weaker, reflecting its lower affinity for Dcp2 (Figures 3 and 4). The stimulatory effect of the LSm domains of Edc3 or Scd6 required the interaction with HLM-1 in Dcp2, as Dcp2 that lacks HLM-1 (Dcp2 1–243) and Dcp2 that carried mutations in HLM-1 that abolished Edc3 and Scd6 binding (L260A L264A) were not stimulated (Figure 5A, lanes 3–6).
Figure 5.
mRNA decapping activity is stimulated by the interaction of the Edc3 and Scd6 LSm domains with Dcp2. (A) Different versions of the Dcp1:Dcp2 decapping complex were incubated with or without a 10-fold molar excess of the Edc3 or Scd6 LSm domains. The increase in activity with respect to the corresponding isolated decapping complexes (set to 100% activity) is reported. The activity of the Dcp1:Dcp2 decapping complex that contains the LSm interaction motif (HLM-1) is increased after addition of the Edc3 (lane 1) or Scd6 (lane 2) LSm domains. If the HLM-1 sequence is deleted (lanes 3 and 4) or mutated such that the LSm domain interaction is abolished (lanes 5 and 6), decapping is no longer stimulated. This clearly shows that the stimulation of mRNA decapping is a direct consequence of Dcp2:LSm domain interaction. (B) The structure of the Dcp1:Dcp2 complex in the closed and inactive conformation (Supplementary Figure S1, 2QKM) (She et al, 2008). The HLM-1 sequence is disordered in the open conformations of the decapping complex and not visible in the structure (Supplementary Figure S1). (C) The Edc3:Dcp2 structure (blue and red, same orientation as shown in Figure 2A) is docked onto the closed Dcp1:Dcp2 structure. This results in extensive steric clashing between the Edc3 LSm domain and the Dcp2 regulatory domain. The Dcp1:Dcp2 complex can thus not adopt the closed conformation displayed in the crystal structure when bound to Edc3/Scd6.
It should be noted that the decapping activation capability of Scd6 was not previously detected when the decapping complex was supplemented with an equimolar amount of Scd6 (Nissan et al, 2010). This finding reflects the low affinity of the Scd6 LSm domain for Dcp2. For that reason, we used a 10-fold excess of Scd6 in order to occupy the HLM-1 to a larger extend. To be consistent, we also used a 10-fold molar excess of Edc3, although this was not required and a similar stimulation was observed with equimolar amounts.
Dcp2 can adopt distinct conformations, termed open and closed, in which the relative domain orientation of the Dcp2 regulatory and catalytic domains varies significantly (Supplementary Figure S1). Interestingly, we noticed that superimposing the Edc3 LSm domain onto the closed crystal form of Dcp1:Dcp2 (Figure 5B) results in extensive steric clashes between Edc3 and the Dcp2 regulatory domain (Figure 5C). In order to interact with Edc3 or Scd6, the Dcp2 HLM-1 must thus dissociate from the Dcp2 regulatory domain. The loss of this Dcp2 intramolecular interaction most likely results in the opening of the decapping complex (Supplementary Figure S1). In the open conformation the HLM-1 region is freely available for the interaction with the Edc3/Scd6 LSm domain (Supplementary Figure S3B). Interestingly, it has been suggested that the closed conformation is not the catalytically active conformation (Floor et al, 2010). The binding of activators to the decapping complex could thus destabilize an inactive conformation, which would lead to a higher enzymatic activity.
The Dcp2 C-terminal HLMs are important for P-body localization in vivo
In vivo, components of the mRNA degradation machinery localize to P-bodies (Eulalio et al, 2007). Edc3 has been shown to play an important role in the formation of P-bodies, in addition to its role in activating the Dcp1:Dcp2 decapping complex. Budding yeast strains that lack Edc3 show a strong reduction in the size and number of P-bodies (Decker et al, 2007), suggesting that the Edc3 protein functions as a scaffold on which components of the mRNA degradation machinery assemble.
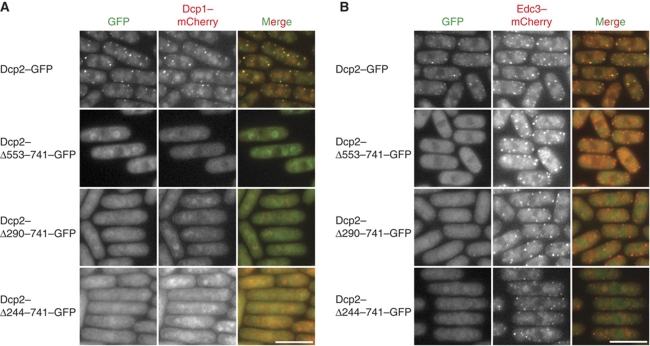
To probe the importance of the interaction between the Dcp2 HLMs and Edc3 for the proper localization of the Dcp1:Dcp2 complex to P-bodies, we performed fluorescence microscopy studies in the fission yeast S. pombe. We replaced the endogenous dcp2+ gene either by a C-terminally GFP-tagged version (dcp2+–GFP) or by truncated constructs, where HLM-C1–C5 (dcp2–Δ553–741–GFP), the entire C-terminus except the first HLM (dcp2–Δ290–741–GFP), or the C-terminus including all HLMs (dcp2–Δ244–741–GFP) was deleted. These Dcp2 versions were integrated in a strain expressing dcp1+–mCherry.
Dcp1–mCherry and Dcp2–GFP localized to the same cytoplasmic foci, but this localization was abolished in all three Dcp2 truncations and both Dcp1 as well as the truncated Dcp2 proteins were homogenously distributed throughout the cytoplasm (Figure 6A). Additionally, we observed that Dcp2–Δ290–741–GFP and Dcp2–Δ244–741–GFP were no longer excluded from the nucleus (Figure 6A and B). These changes in localization were not caused by differences in Dcp2 abundance, as FL Dcp2 and all truncation mutants were expressed to similar levels (Supplementary Figure S8C).
Figure 6.
The C-terminal HLMs of Dcp2 are required for P-body localization of Dcp1 and Dcp2. Fluorescence micrographs of S. pombe cells expressing dcp2+–GFP or truncated versions thereof in combination with dcp1+–mCherry (A) or edc3+mCherry (B). Dcp2–GFP co-localizes with Dcp1–mCherry (A) and with Edc3–mCherry (B) in distinct cytoplasmic foci (P-bodies). (A) Both Dcp1 and Dcp2 fail to localize to P-bodies, if the C-terminal HLMs of Dcp2 are deleted. (B) Edc3–mCherry still forms P-bodies in all dcp2 mutant strains, indicating that the redundant process of P-body formation is not significantly disrupted by the Dcp2 truncations. All images were scaled and processed in the same way. The length of the scale bars corresponds to 10 μm.
In order to address whether the absence of the HLM motifs from Dcp2 generally influenced the formation of P-bodies, we asked whether other known components of the mRNA degradation machinery were still able to assemble in such foci. However, neither for Edc3–mCherry (Figure 6B) nor for Lsm7–mCherry (Supplementary Figure S8A) we were able to observe an obvious difference in the number or size of cytoplasmic foci when the dcp2 truncation mutants were expressed in place of endogenous dcp2+. Furthermore, the absence of Dcp1 and Dcp2 from P-bodies did not impair cell growth (Supplementary Figure S8B). Only the truncation of Dcp2 that also removed HLM-1 (dcp2–Δ244–741–GFP) caused a slight but reproducible temperature-dependent growth defect (Supplementary Figure S8B).
In summary, we conclude that the C-terminal HLMs (HLM-C1–C5) of Dcp2, some of which interact strongly with Edc3 (Figure 3B), are essential for the recruitment of the Dcp1:Dcp2 decapping complex to P-bodies, but localization of the decapping complex to P-bodies is dispensable for P-body formation. The deletion of all HLMs, including HLM-1, shows an effect on cellular growth, possibly because in this mutant Dcp2 decapping activity cannot be modulated by Edc3 and/or Scd6.
An LSm-interacting motif that mediates Edc3 binding is present in metazoan Dcp1
The LSm interaction motifs that are found in yeast Dcp2 sequences are not readily identifiable in metazoan Dcp2 sequences. Indeed, the LSm interaction motif that is C-terminal to the Dcp2 catalytic domain (HLM-1) is absent in metazoan Dcp2, and the C-terminal extension contains only a few less conserved regions that not clearly resemble HLMs. On the other hand, it has been reported that the D. melanogaster Dcp1 protein interacts with the LSm domains of Edc3 and Tral (Tritschler et al, 2007, 2008), where the Edc3:Dcp1 interaction is mediated through a conserved Dcp1 sequence motif that was termed motif-1 (Tritschler et al, 2009b).
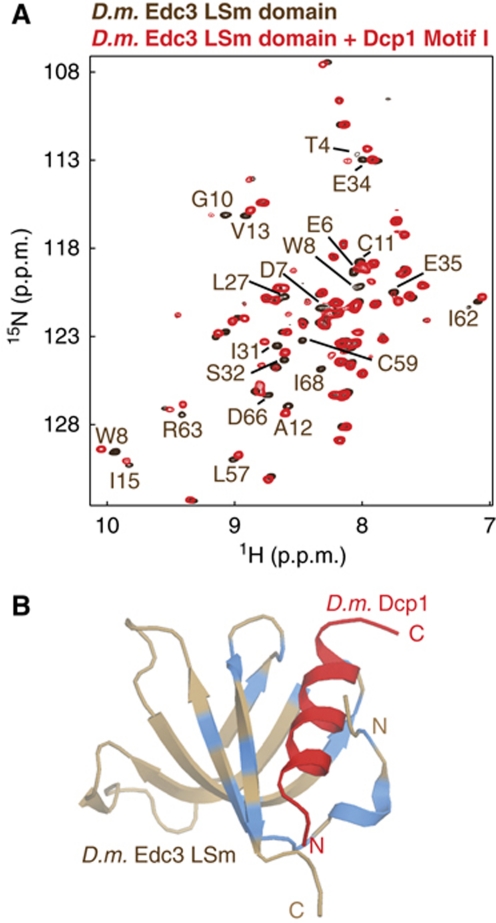
This finding might seem to contradict the results we obtained here for the yeast decapping complex, in which Dcp2 interacts with Edc3 and Scd6. However, close inspection of metazoan Dcp1 sequences (Supplementary Figure S9) revealed that motif-1 (SIFNMLT) has the propensity to fold into an amphipatic helical structure, as observed for the HLMs. To investigate whether the D. melanogaster Dcp1 motif-1 interacts with the D. melanogaster Edc3 LSm domain, we performed NMR titration experiments. Interestingly, we observed significant chemical shift perturbations in the Edc3 LSm domain upon addition of a peptide containing the Dcp1 motif-1, which demonstrates a direct interaction (Figure 7A). Mapping these changes onto the D. melanogaster Edc3 LSm structure (Tritschler et al, 2007) clearly shows that the conserved Dcp1 motif-1 binds to the same surface as the Dcp2 LSm interaction motif in the S. pombe Edc3:Dcp2 complex (Figure 7B). Taken together, these findings show that motif-1 functions like a HLM in metazoan Dcp1.
Figure 7.
D. melanogaster motif-1 in Dcp1 resembles a metazoan HLM. (A) NMR spectra of the Edc3 LSm domain from D. melanogaster in the absence (brown) and presence (red) of the D. melanogaster Dcp1 motif-1. Residues that experience significant chemical shift changes are indicated. (B) Model of the D. melanogaster Edc3 LSm domain in complex with Dcp1 motif-1. The model was constructed by superimposing the D. melanogaster Edc3 LSm domain (brown, PDB:2RM4; see Figure 1C) onto the yeast Edc3 LSm domain Dcp2 complex structure. Residues of the D. melanogaster Edc3 LSm domain that are affected by the interaction with Dcp1 motif-1 (Figure 7A) are coloured blue. The location of the Dcp2 helix in the S. pombe Dcp2:Edc3 structure is indicated in red (Figure 2A). The chemical shift changes observed for the D. melanogaster Dcp1 Motif-1:Edc3 complex agree well with the S. pombe Dcp2:Edc3 structure and suggest a similar mode of binding.
Discussion
The decapping enzyme Dcp2 requires the interaction with its protein partners for full decapping activity. These partners, or decapping activators, include Dcp1, which interacts with Dcp2 directly to form the conserved core of the decapping complex and Edc1–4, Scd6/Tral, Pat1, and the Lsm1–7 ring (Parker and Song, 2004; Franks and Lykke-Andersen, 2008). The detailed mechanism by which these activators stimulate decapping has remained largely unknown.
Here, we identify the molecular basis of a network of interactions between the Dcp1:Dcp2 decapping complex and the decapping components Edc3 and Scd6. In this interaction network, both Edc3 and Scd6 use an N-terminal LSm domain to interact with helical leucine-rich sequence motifs in Dcp2 (HLMs) (Gaudon et al, 1999) (Figures 2, 3, 4). Multiple HLM sequences are present in Dcp2, close to the catalytic domain and in the disordered C-terminal extension (Figures 1A and 3). The presence of multiple HLMs suggests that multiple activators can bind at various positions in the decapping complex, resulting in the assembly of decapping complexes that differ in Edc3/Scd6 content and that potentially differ in substrate specificity. In addition, we show that multiple decapping complexes can be clustered through Edc3. Our results thus reveal a plastic network of intermolecular interactions that can result in mRNA decapping clusters of varying composition.
We unravelled the structural basis of the interactions between the decapping complex and the activator proteins by solving the structure of the Edc3 LSm domain in complex with HLM-1 from Dcp2 (Figure 2). The structure of the complex displays a novel mode of LSm domain interaction with ligands. Upon complex formation, the disordered HLM sequence adopts a helical conformation that binds close to the N-terminal ends of β-strands 2 and 5. This novel mode of interaction is possible only as a result of two specific features of the Edc3 and Scd6 LSm domains: (1) the monomeric nature of the LSm domain that makes β-strand 5, that is normally used for intermolecular contacts, available for the interaction with the N-terminal end of the HLM helix and (2) the very short N-terminal helix in the Edc3 and Scd6 LSm domain compared with canonical (L)Sm folds, which makes space available for the interaction with the C-terminal end of the HLM helix.
Based on the finding that two different LSm domains are able to interact with four different HLM sequences in yeast, it is clear that a certain degree of sequence variation is tolerated in both the interaction domains and the recognition sequences (Figure 3A). This plasticity is especially true for the HLM sequence that, according to the structure of the complex, must possess a helical propensity. Based on the interacting motifs, the HLMs preferably contain an SxxLLxLL motif; however, more than a single leucine-to-alanine mutation is required to abolish binding, indicating the lack of a very stringent requirement for these amino acids (Figure 3; Supplementary Figure S3A). As such, it is tempting to speculate that additional LSm-interacting HLMs are present in other components of the mRNA degradation machinery. One such protein is Rps28B that interacts with Edc3 (Ito et al, 2001; Decourty et al, 2008) and contains a sequence that is reminiscent of an HLM (EDDILVLM between residues 51 and 58 in S. pombe). Interestingly, in S. cerevisiae, the deadenylation-independent degradation of the Rps28B mRNA depends on one hand on the Edc3 protein and on the other hand on an interaction between the Rps28B mRNA 3′ UTR and the Rps28B protein (Badis et al, 2004). It is thus tempting to speculate that the Dcp1:Dcp2 decapping complex can recruit the Rps28B mRNA through Edc3, where the dimeric Edc3 protein uses one LSm domain to interact with Dcp2 and uses the other LSm domain to interact with the HLM in the Rps28B protein–mRNA complex.
We show that there are two main biological consequences of the interaction of Edc3/Scd6 with the decapping complex. First, the interaction between the Edc3 or Scd6 LSm domains and the first HLM sequence in Dcp2 (HLM-1, residues 257–266) results in an increase in the catalytic activity of the decapping complex (Figure 5). We argue that the molecular basis of this modulation of decapping activity results from the destabilization of the inactive closed conformation of the enzyme. Indeed, the interaction of the decapping complex with the Edc3 LSm domain is incompatible with the closed conformation of the decapping complex that has been suggested to be catalytically inactive (Floor et al, 2010). The interaction of Edc3 or Scd6 with HLM-1 in Dcp2 thus induces a shift of the structural equilibrium away from catalytically inactive conformations (Figure 5C). In general, the level of Edc3-induced activation of the Dcp1:Dcp2 decapping complex is less than twofold (Figure 5) (Harigaya et al, 2010; Nissan et al, 2010). From a structural point of view, this is not surprising, as the population of the closed inactive state that is prevented by the interaction of the decapping complex with the Edc3/Scd6 LSm domain is only one of the possible conformations that the decapping complex can adopt. Interestingly, removal of all HLMs from fission yeast Dcp2 (Figure 6) results in a growth defect at higher temperatures (Supplementary Figure S8B). As the truncated decapping complex still possesses catalytic activity (Floor et al, 2010) (Figure 5A), this suggests that cellular function is impaired due to a loss of the regulation of the decapping complex activity. To what extend individual mRNA degradation rates are influenced remains to be determined. It is, however, interesting to note that removal of all C-terminal residues from S. cerevisiae Dcp2 results in increased Rps28B and MFA2pG mRNA levels (Harigaya et al, 2010), suggesting that HLM-1 not only has an important role in modulating the activity of the decapping complex in vitro, but also in vivo.
The second biological function of the HLMs results from the LSm interaction sites located in the C-terminal region of Dcp2. Using fluorescence microscopy of fission yeast strains that carry different versions of the Dcp2 protein, we show that HLM-C1 to HLM-C5 are required for the localization of the decapping complex to processing bodies (Figure 6A). Interestingly, a single HLM (HLM-1) is not sufficient to target the decapping complex to P-bodies (Figure 6A). Upon deletion of all HLMs from Dcp2, Edc3, and Lsm7 still localized to cytoplasmic foci. This shows that Dcp2 is not essential for P-body formation and is in agreement with the fact that no single protein has been identified that is absolutely essential for the formation of these cytoplasmic foci in yeast (Decker et al, 2007; Teixeira and Parker, 2007). Nevertheless, it has been shown that Dcp2 contributes to the assembly or maintenance of P-bodies (Teixeira and Parker, 2007). This correlates well with our in vitro findings that show that Edc3 is able to promote a clustering of decapping complexes through HLMs in Dcp2 (Figure 3D and E).
The basic set of proteins that regulate mRNA degradation is conserved from yeast to humans. However, in higher eukaryotes, additional proteins (e.g. Edc4/Ge-1) are required for Dcp1:Dcp2 catalytic activity (Fenger-Gron et al, 2005), indicating that differences in the regulation of mRNA decapping may have evolved. The HLMs in yeast Dcp2 are not present in metazoan Dcp2 sequences. However, we showed that the conserved motif-1 in the metazoan Dcp1 resembles an HLM-like motif that directly interacts with metazoan Edc3 LSm domain. The observation that both yeast and metazoan Edc3 are able to interact with Dcp1:Dcp2 indicates that the basic interactions between decapping complex components are conserved but that the details have changed during evolution. These changes can be explained by the low evolutionary pressure on the location of small linear motifs in the protein complexes, as only a small number of point mutations in a disordered region of a protein are required to relocate these motifs (Neduva and Russell, 2005). Currently, there is no high-resolution structure of the Dcp1:Ge-1:Dcp2 decapping complex available. Thus, it is not possible to speculate whether the interaction of the Edc3/Scd6 Lsm domain with Dcp1 will be able to enhance the catalytic activity by inducing conformational changes in Dcp2, similar to the observation presented here for the yeast decapping machinery. It is worth noting, however, that it was previously shown that metazoan Dcp1 trimerizes through a unique domain at the C-terminus (Tritschler et al, 2009b). As a result, the metazoan Dcp1:Dcp2 complex contains multiple binding motifs for Edc3, similar to the multiple binding motifs in the monomeric yeast Dcp2 protein. Interestingly, the removal of the trimerization domain from metazoan Dcp1 results in the failure of this decapping complex to localize to P-bodies (Tritschler et al, 2007, 2009b), indicating that a single Edc3:Dcp1 binding event is not sufficient for the P-body localization. This correlates with our findings here that show that a single HLM is not sufficient for the P-body localization of the fission yeast decapping complex (Figure 6A).
In summary, we provide the first structural insights into the interactions that regulate the activity and cellular localization of the mRNA decapping complex. Based on these and previous results, it appears that proteins involved in mRNA degradation have multiple functions. Edc3 not only enhances the catalytic activity of the Dcp1:Dcp2 mRNA decapping complex but also is responsible for the localization of the complex to P-bodies. Scd6 was previously reported to repress translation (Nissan et al, 2010), but we have shown here that it also has a direct stimulatory effect on the decapping complex. The same dual activity, translational repression and enhancement of mRNA decapping, has been shown for Pat1 and the helicase Dhh1 (Fischer and Weis, 2002; Coller and Parker, 2005; She et al, 2008). Taken together, these findings suggest a high level of crosstalk between different steps of the mRNA degradation pathway. Unravelling the molecular basis of the interactions between all decapping factors and their biological consequences is important, and these aspects will need to be addressed in order to understand how cells determine the fate of specific mRNAs.
Materials and methods
Molecular cloning and protein purification
The DNA sequences of Edc3, Scd6 (Sum2), Dcp1, and Dcp2 were amplified from S. pombe genomic DNA or from cDNA and ligated into modified pET vectors (Supplementary Table SII). Proteins were overexpressed at 20°C overnight in the Escherichia coli strain BL21 (DE3) Codon Plus RIL (Stratagene). To incorporate NMR-active nuclei, cells were grown in M9 minimal medium containing 15NH4Cl and/or 13C6-labelled glucose as the sole nitrogen and carbon sources. Proteins were purified from the cell lysate in buffer A (50 mM sodium phosphate, pH 7.5, 10 mM imidazole, 150 mM NaCl). The soluble fraction was applied to an affinity matrix (Ni-NTA; Qiagen), washed and eluted with 350 mM imidazole, followed by cleavage of the affinity tag using TEV protease. The resultant protein was purified to homogeneity using size exclusion chromatography (Superdex 200 or Superdex 75, GE Healthcare) in 25 mM HEPES buffer pH 7.3, 125 mM NaCl, and 1 mM DTT.
NMR spectroscopy
All NMR spectra were recorded at 30°C on Bruker AVIII-600 and AVIII-800 spectrometers. Backbone sequential assignments on a 15N/13C-labelled samples were completed using HNCA, HNCACB, CC(CO)NH-TOCSY, HNCO, and HN(CA)CO experiments. Sidechain assignments were obtained from 3D-CC(CO)NH-TOCSY and 3D-CCH-TOCSY spectra. For the isolated Edc3–LSm domain, 93.5% (backbone)/87.5% (sidechains) of the 58 structured residues were assigned. For the Edc3–LSm domain, Dcp2 complex 98% (backbone)/92% (sidechains) of the 71 structured residues were assigned. Distance restraints were derived from a set of six NOESY spectra (see Supplementary data) using an 80-ms mixing time. χ1 and χ2 diherdal angle restraints were derived from HNHB spectra and intra-residual NOE contacts (see Supplementary data), backbone conformational restraints were generated based on secondary chemical shift information derived from TALOS (Cornilescu et al, 1999). Structures were calculated with XPLOR (NIH version 2.9.4) (Schwieters et al, 2003) based on structural distance and angle restraints (Supplementary Tables SI–SIII).
Decapping assays
In vitro decapping assays were performed under single turnover conditions as previously described (Borja et al, 2011) using 0.05 μM Dcp1:Dcp2 with or without a 10-fold molar excess of the LSm domain of the decapping activator Edc3 or Scd6. It should be noted that an equimolar amount of Edc3 is able to significantly stimulate decapping. Nevertheless, we added the same excess activator protein in all experiments to be able to compare both experiments directly. The decapping reaction was started by addition of the 32P-labelled capped RNA substrate. After incubation at 30°C for 5 min, the reaction was stopped by the addition of 0.1 M EDTA. The fraction of released m7GDP at the end point of the reaction was determined using TLC analysis. The activity of the decapping complex without the addition of an activator was set to 100%. The stimulatory effect of the decapping activators was calculated compared with the unstimulated decapping complex. The mean value and s.d. were obtained from three independent experiments.
S. pombe strains
S. pombe strains that express dcp2+–GFP, dcp2–Δ553–741–GFP, dcp2–Δ290–741, dcp2–Δ244–741, edc3+–mCherry, dcp1+–mCherry, or lsm7+–mCherry from the endogenous genomic locus were created using a PCR-based gene targeting method (Bahler et al, 1998) (see Supplementary data and Supplementary Table SIV).
Data deposition
Chemical shift assignments have been deposited in the BMRB (Edc3 LSm domain: 18041; Edc3 LSm domain: Dcp2 complex: 18042), and coordinates have been deposited in the PDB (Edc3 LSm domain: 4A53; Edc3 LSm domain: Dcp2 complex: 4A54).
Supplementary Material
Acknowledgments
We thank Silke Hauf for support with the in vivo experiments and for critical reading of the manuscript. We are grateful to J Ullmann for excellent technical assistance and thank Andrei Lupas, Silke Wiesner, Murray Coles, Ancilla Neu, and Markus Mund for discussions and Tonja Wolff for help with the yeast strains. We thank the Yeast Genetic Resource Center (YGRC) for the Sum2 cDNA clone. JK was supported by a stipend of the Boehringer Ingelheim Fonds. This study was supported by the Max Planck Society and a Marie Curie reintegration Grant to RS (FP7/2007–2013, Grant agreement no. 239164).
Author contributions: SAF, VT, JK, JEB, EI, and RS designed the research. SAF, VT, JK, JEB, NAH, and RS performed the experiments. SAF, VT, JK, JEB, and RS analysed the data. EI and RS wrote the paper. SAF, VT, JK, JEB, NAH, EI, and RS commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albrecht M, Lengauer T (2004) Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett 569: 18–26 [DOI] [PubMed] [Google Scholar]
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A (2004) Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell 15: 5–15 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB (1998) Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol 18: 5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B (2000) The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol 20: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borja MS, Piotukh K, Freund C, Gross JD (2011) Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA 17: 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B (2000) A Sm-like protein complex that participates in mRNA degradation. EMBO J 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I (2002) An EVH1/WH1 domain as a key actor in TGFbeta signalling. FEBS Lett 519: 178–180 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R (2005) General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R (2006) CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol 173: 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R (2007) Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, Jacquier A (2008) Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci USA 105: 5821–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J (2005) Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Fischer N, Weis K (2002) The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J 21: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor SN, Jones BN, Hernandez GA, Gross JD (2010) A split active site couples cap recognition by Dcp2 to activation. Nat Struct Mol Biol 17: 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J (2008) The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Mayes AE, Brunet-Simon A, Rain JC, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs JD, Legrain P (2000) Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17: 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudon C, Chambon P, Losson R (1999) Role of the essential yeast protein PSU1 in p6anscriptional enhancement by the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J 18: 2229–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E (2010) HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 189: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R (2010) Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol 30: 1446–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375–387 [DOI] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R (2004) Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J (2011) Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SH, Decker CJ, Walsh MA, She M, Parker R, Song H (2008) Crystal structure of human Edc3 and its functional implications. Mol Cell Biol 28: 5965–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennan RG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA 106: 19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neduva V, Russell RB (2005) Linear motifs: evolutionary interaction switches. FEBS Lett 579: 3342–3345 [DOI] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R (2010) Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 39: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- She M, Decker CJ, Chen N, Tumati S, Parker R, Song H (2006) Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol 13: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H (2004) Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol 11: 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H (2008) Structural basis of dcp2 recognition and activation by dcp1. Mol Cell 29: 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW (2008) An in vivo map of the yeast protein interactome. Science 320: 1465–1470 [DOI] [PubMed] [Google Scholar]
- Teixeira D, Parker R (2007) Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18: 2274–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Mayer C, Sauter C, Weeks S, Suck D (2003) Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J Biol Chem 278: 1239–1247 [DOI] [PubMed] [Google Scholar]
- Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J 20: 2293–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O (2009a) Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell 33: 661–668 [DOI] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Motz C, Igreja C, Haas G, Truffault V, Izaurralde E, Weichenrieder O (2009b) DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc Natl Acad Sci USA 106: 21591–21596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V (2008) Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol 28: 6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O (2007) A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol 27: 8600–8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M (2002) The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J (2005) Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol 12: 1031–1036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.