Abstract
Assessing relevant molecular differences between human-induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) is important, given that such differences may impact their potential therapeutic use. Controversy surrounds recent gene expression studies comparing hiPSCs and hESCs. Here, we present an in-depth quantitative mass spectrometry-based analysis of hESCs, two different hiPSCs and their precursor fibroblast cell lines. Our comparisons confirmed the high similarity of hESCs and hiPSCS at the proteome level as 97.8% of the proteins were found unchanged. Nevertheless, a small group of 58 proteins, mainly related to metabolism, antigen processing and cell adhesion, was found significantly differentially expressed between hiPSCs and hESCs. A comparison of the regulated proteins with previously published transcriptomic studies showed a low overlap, highlighting the emerging notion that differences between both pluripotent cell lines rather reflect experimental conditions than a recurrent molecular signature.
Keywords: human embryonic stem cells, human-induced pluripotent stem cells, proteomics, quantitation
Introduction
Human embryonic stem cells (hESCs) are capable of self-renewal and multi-lineage differentiation (i.e., pluripotency; Thomson et al, 1998). Owing to these two unique properties, they are considered as one of the most promising sources for tissue replacement therapies. However, the use of hESCs entails numerous ethical issues as they are derived from human embryos. Recently, reprogramming of somatic cells to an embryonic stem cell-like state, named induced pluripotent stem cells (iPSCs), was achieved through retroviral transfection of a defined set of transcription factors (Takahashi et al, 2007; Yu et al, 2007; Park et al, 2008b). To date, multiple somatic cells from diverse adult tissues (i.e., endoderm, mesoderm and ectoderm origins) have been successfully reprogrammed to iPSCs, including fibroblasts (Takahashi et al, 2007; Yu et al, 2007; Park et al, 2008b), blood (Loh et al, 2009), neural progenitors (Eminli et al, 2008) and fully differentiated lymphocytes (Hanna et al, 2008). Furthermore, multiple strategies have been proposed as alternatives to potentially harmful retroviruses, including drug-inducible systems (Hockemeyer et al, 2008), virus-free transposon mediated (Woltjen et al, 2009), recombinant proteins (Kim et al, 2009) and miRNAs (Miyoshi et al, 2011). Finally, human-induced pluripotent stem cells (hiPSCs) represent a unique tool to develop cellular models for many human diseases (Park et al, 2008a; Soldner et al, 2009).
Although current functional assays such as in-vitro differentiation, teratoma formation, chimera formation germline contribution and tetraploid complementation (Jaenisch and Young, 2008) have confirmed the pluripotency of hiPSCs (Takahashi et al, 2007; Yu et al, 2007; Park et al, 2008b), there might still be significant differences when compared with their natural hESC counterparts. For instance, hiPSCs have been shown to differentiate in a less efficient manner than hESCs (Feng et al, 2010; Hu et al, 2010). Consequently, an extensive molecular characterization to address differences and similarities between these two pluripotent cell lines seems to be a prerequisite before any clinical application is conducted. Despite that great efforts have been made to address how similar hESCs and hiPSCs are, the definite answer to this fundamental question is still the subject of active debate (Guenther et al, 2010; Newman and Cooper, 2010; Chin et al, 2009, 2010b). Using microarray-based approaches, several studies have reported residual levels of transcriptional memory of the parental somatic cell line in the reprogrammed hiPSCs (Chin et al, 2009; Marchetto et al, 2009; Ghosh et al, 2010; Ohi et al, 2011). However, it has also been shown that these gene expression profiles could represent lab-specific signatures due to in-vitro microenvironmental conditions rather than a recurrent molecular signature across different hiPS cell lines (Guenther et al, 2010; Newman and Cooper, 2010). In addition, epigenetic analyses have documented significant differences in the DNA methylation patterns between hiPSCs and hESCs (Deng et al, 2009; Doi et al, 2009; Lister et al, 2009; Kim et al, 2010; Polo et al, 2010; Bock et al, 2011). In fact, the transcriptional memory of hiPSCs could be partially explained by the incomplete DNA methylation at the promotor regions of somatic genes (Ohi et al, 2011). Non-coding miRNAs have an important role in the underlying mechanisms of reprogramming (Samavarchi-Tehrani et al, 2010; Subramanyam et al, 2011) and they can replace the ectopic expression of transcription factors to generate iPSCs with even higher efficiency (Anokye-Danso et al, 2011). Thus, miRNA profiles between hESCs and hiPSCs were compared and a signature in the expression of the miR-371/372/373 cluster was found (Wilson et al, 2009). Finally, genetic integrity was also studied and it was found that the reprogramming process could induce several genomic abnormalities (Mayshar et al, 2010; Hussein et al, 2011; Laurent et al, 2011).
Despite intensive efforts in molecular characterization, direct assessment of protein levels has yet to be incorporated into these integrative systems-level analyses. Protein levels are tuned by intricate mechanisms of gene expression regulation and it has recently been documented that mRNA and protein levels poorly correlate in mouse ESCs (Lu et al, 2009). Proteomics is, however, more labor-intensive and often lacks the profiling depth that can be obtained at the transcript level. Mass spectrometry (MS)-based proteomics is, currently, the most powerful tool to globally profile proteomes and has also been used to study different aspects of the stem cell biology (Swaney et al, 2009; Van Hoof et al, 2009; Rigbolt et al, 2011). Here, we use in-depth quantitative proteomics to gain insights into the differences and similarities in the protein content of two hiPS cell lines (IMR90 and 4Skin), their precursor fibroblast cell lines and one hES (HES-3) cell line, all grown and maintained under the same experimental conditions, providing novel molecular signatures that may assist in filling a gap in our understanding of pluripotency.
Results
Confirmation of pluripotency and experimental design
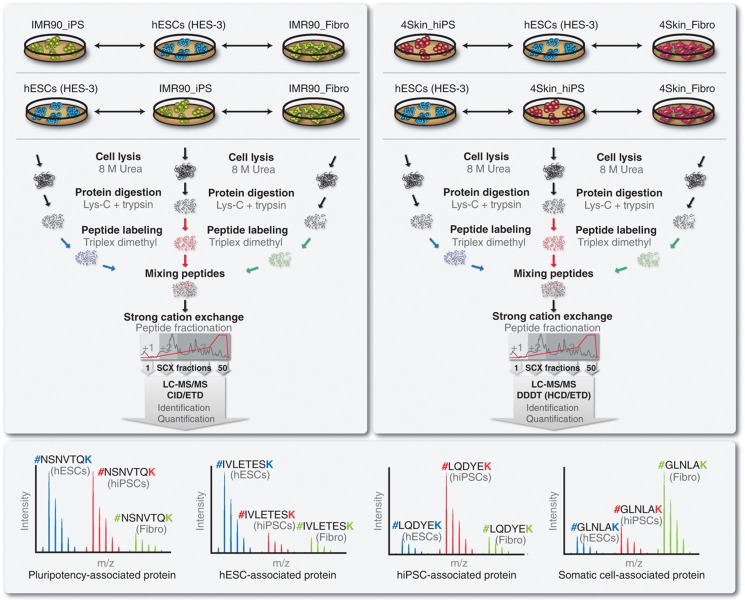
To study the degree of similarity, at the protein level, between hiPSCs and hESCs, two MS-based proteomic experiments using two different hiPS cell lines were conducted (Figure 1). In Experiment 1, IMR90_iPS were compared to hESCs (HES-3) and to the parental cell line, IMR90_Fibro. In Experiment 2, 4Skin_iPS, hESCs (HES-3) and the somatic cells, 4Skin_Fibro, were analyzed. Both hiPS cell lines were derived through the reprogramming of IMR90 fetal fibroblasts and foreskin fibroblasts, by ectopic expression using retroviruses carrying SOX2, OCT4, NANOG and LIN28 transgenes (Yu et al, 2007). Upon extended culture, hiPSCs adopt a gene expression profile which more closely resembles that of the hESCs (Chin et al, 2009). For this study, the two hiPS cell lines were analyzed at late passage. However, long-term culture conditions might induce genomic instability (Baker et al, 2007), which might compromise the pluripotency of these cell lines. Therefore, we confirmed the pluripotency of both hiPS cell lines by checking the expression of known hESCs markers (e.g., OCT4, podocalyxin and tra-1-60), karyotypic stability and in-vivo differentiation capabilities (Supplementary Figure S1). Characterization of hESCs (HES-3 cell line) was described elsewhere (Chin et al, 2010a).
Figure 1.
Experimental workflow and overview of the proteomic experiments performed. To characterize the proteomes of human-induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs), two MS-based experiments, using two independent hiPS cell lines, were conducted. Experiment 1 (top-left panel) focused on hiPS_IMR90, hESCs and IMR90 fetal fibroblasts (cell line used for reprogramming). Experiment 2 (top-right panel) focused on hiPS_4Skin, hESCs and parental 4Skin fetal fibroblasts. Proteins were extracted and digested with Lys-C and trypsin. Peptides were labeled using triplex dimethyl chemistry, equally mixed and prefractionated by using strong cation exchange (SCX). Two biological replicas were performed for each experiment, where labels were swapped between hiPSCs and hESCs. SCX fractions were analyzed by high-resolution LC-MS/MS. Experiment 1 was analyzed with an LTQ Orbitrap XL using both CID and ETD fragmentations, whereas Experiment 2 was analyzed with an LTQ Orbitrap Velos using a data-dependent decision tree (DDDT) using HCD and ETD. The peak intensities of the identified peptides reflect their relative abundance in the samples (bottom panel).
All the six samples were subjected for proteomic analysis (Figure 1). Basically, proteins were extracted in a buffer containing 8 M urea and subsequently cleaved into peptides using a double digestion with Lys-C and trypsin (Figure 1). Metabolic labeling presents some caveats in hESCs (Van Hoof et al, 2007) and, so far, has not been applied to hiPSCs; whereas label-free approaches are less suitable for large multidimensional separation-based strategies. Therefore, we applied our in-house developed peptide labeling that uses solid-phase extraction and triplex dimethyl labeling chemistry (Boersema et al, 2009). Two biological replicas were conducted for each experiment, where labels were swapped between the hESCs and hiPSCs (parental fibroblasts were kept constant; Figure 1). In order to ensure maximal protein identification, we reduced sample complexity by a strong cation exchange (SCX) chromatography. Subsequently, Experiment 1 was analyzed by high-resolution LC-MS/MS with electron transfer dissociation (ETD) as well as collision-induced dissociation (CID) for peptide sequencing. Experiment 2 was analyzed with a data-dependent decision tree using higher-energy collision dissociation (HCD) and ETD with either Orbitrap or linear ion trap readout (Frese et al, 2011). MS intensities of the ‘light’, ‘intermediate’ and ‘heavy’ peaks accurately reflect the relative abundance of peptides in the three cell types (Figure 1).
In-depth quantitative proteomic analysis of hESCs, hiPSCs and fibroblasts
An overview of the proteomic results is presented in Supplementary Table S1. Briefly, a total of 348 LC-MS/MS analyses (including technical and biological replicates) were performed leading to 4 551 920 MS/MS sequencing events (cumulative value of CID, HCD and ETD spectra). We confidently identified 1 593 446 peptide spectrum matches at a peptide false discovery rate (FDR) below 1% (Mascot Ion Score>20). In Experiment 1 (IMR90), a total of 6873 unique protein groups were identified (3994 in common between both biological experiments; Supplementary Table S2). On the other hand, Experiment 2 (4Skin) consists of 8548 unique protein groups (5516 identified in both biological replicas; Supplementary Table S3). Combining all the data sets, we identified 10 628 unique protein groups (3001 proteins were identified at the intersection of all four data sets). Most importantly, the vast majority of the proteins in our data set (80–90%) were identified on the basis of at least two unique peptides with an average of 9±15 peptides per protein (Supplementary Figures S2 and S3). To the best of our knowledge, the coverage obtained in this study represents the largest achieved by any proteomics screen on pluripotent cells. Typically, proteomic studies are biased toward the detection of highly expressed genes; nevertheless, our data set includes numerous proteins known to be of low abundance in mammalian cells. We classified, by protein class, all the 10 628 identified proteins (7631 contained official gene symbols with functional annotation) and found 649 transcription factors, 247 kinases (48% of the putative human kinome (Manning et al, 2002)), and proteins that are difficult to detect by MS, such as membrane proteins (1494 proteins were predicted by TMHMM to contain transmembrane helices). Interestingly, we also confirmed the existence, at the protein level, of genes where only transcript evidence was available (1876 proteins were annotated as ‘hypothetical’ or ‘putative uncharacterized’). Furthermore, we compared our data set with two of the largest proteomic analyses carried out to date in hESCs (Van Hoof et al, 2009; Rigbolt et al, 2011). Remarkably, we found a high overlap as we identified ∼90% of the reported proteins by these studies (∼2200 were unique to our current analysis). The transcriptional circuitry involved in pluripotency is controlled by a core of three transcription factors: SOX2 (Yuan et al, 1995; Avilion et al, 2003), NANOG (Mitsui et al, 2003) and OCT4 (Niwa et al, 2000). We confidently identified the protein product of these genes and several other well-known hESC markers, such as DNMT3B, UTF1, PODXL, GRB7 and BRIX (Adewumi et al, 2007). Taken together, these results indicate the comprehensiveness of our data (mammalian cells express 10 000–15 000 transcripts (Jongeneel et al, 2003)) and thus it can serve as a reliable resource for those interested in the pluripotent stem cell proteome.
Overall, 5835 proteins were quantified in Experiment 1, 3537 of which were found in common between the two biological replicas (Supplementary Table S2). In the same way, quantitative measurements for 7154 proteins were obtained in Experiment 2, where 4718 proteins were measured in the two biological replicas (Supplementary Table S3). We further focused on the 2683 proteins confidently quantified in all our experiments and data sets. The analysis of variability in our technical (Supplementary Figure S4) and biological (Supplementary Figure S5) replicas demonstrated high quantification accuracy and reproducible proteomic measurements for both experiments with Pearson correlation factors between 0.84 and 0.96. Remarkably, ∼85% of our protein ratios showed <35% variability (Supplementary Figures S6D, S7D, S8D and S9D). Of note, we obtained accurate measurements for proteins changing in abundance more than 100-fold. Furthermore, using the extracted ion chromatograms of the three most abundant peptides per protein (Grossmann et al, 2010), we estimated the absolute abundance of the identified proteins within the samples spanning six orders of magnitude (Supplementary Tables S2 and S3).
High similarity in the proteomes of hESCs and hiPSCs
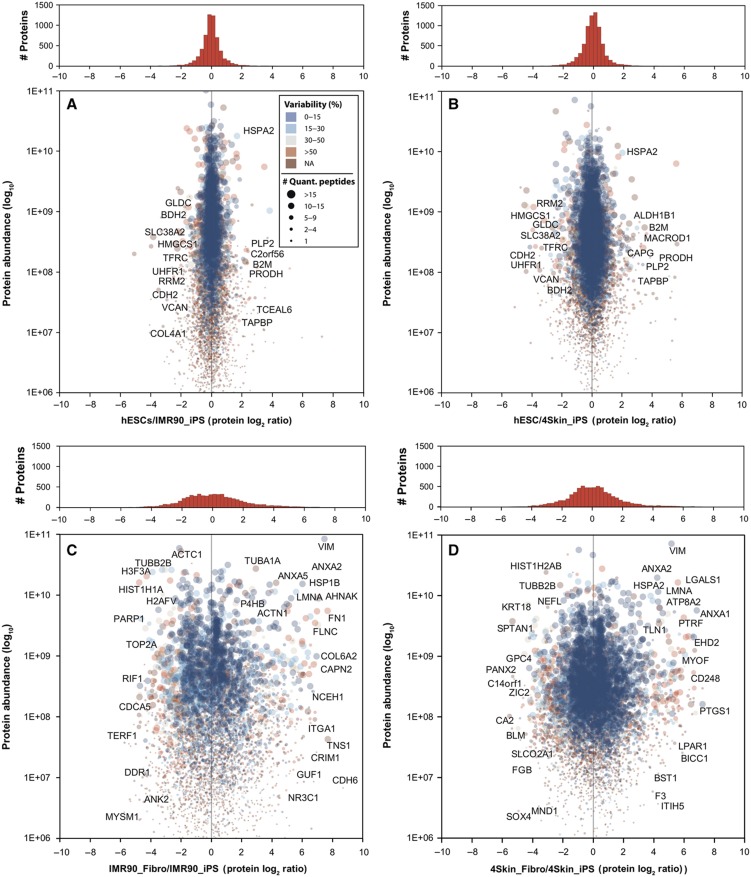
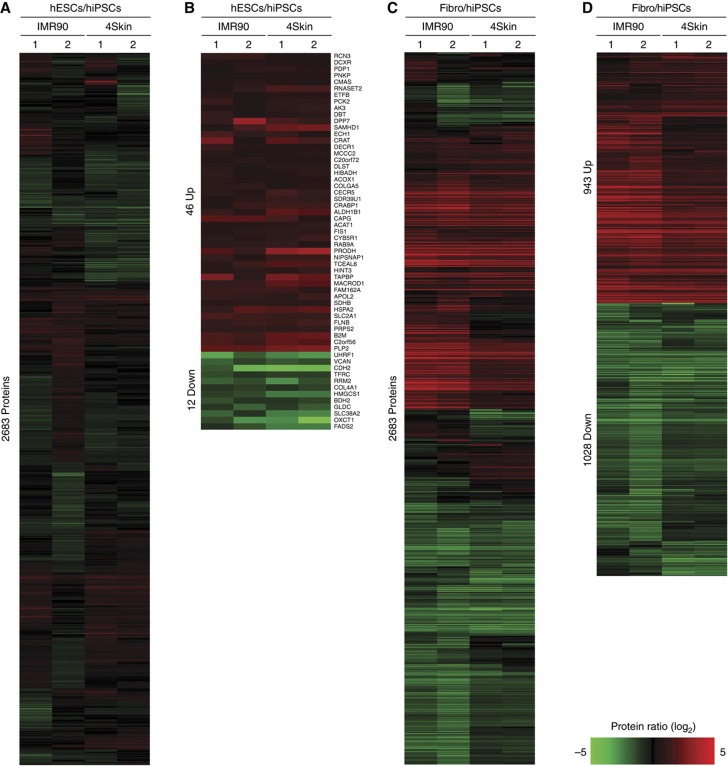
Besides the fact that both hiPSCs and hESCs are pluripotent, it is still not clear how similar both cell lines are at the proteome level. Thus, we compared the protein levels of hESCs and hiPSCs and found a very high degree of similarity. In Figure 2, the absolute protein abundance (log10 scale) is plotted against the relative protein ratios (log2 scale) for the hESC/IMR90_iPS (Figure 2A) and hESC/4Skin_iPS (Figure 2B) comparisons. The vast majority of the proteins showed minor or no changes between hiPSCs and hESCs (as seen in the histograms of frequencies). As expected, pluripotency markers including SOX2, NANOG, OCT4, LIN28 and SALL4 were found in almost identical levels between hESCs and hiPSCs in both experiments. We then sought to define those proteins that differentially expressed between hESCs and hiPSCs. For this purpose, we used the significance analysis of microarrays (SAMs) test (Tusher et al, 2001): a commonly used statistical test in transcriptomic studies (see Materials and methods section). SAM has been recently shown to be applicable for quantitative proteomic data sets as well (Roxas and Li, 2008) and is particularly useful because it provides an estimation of FDRs for a defined set of significant changes. Only those proteins quantified in both experiments (i.e., IMR90 and 4Skin) and in both biological replicas were subjected to statistical analysis using SAM (i.e., 2683 proteins). Figure 3A shows the log2 ratios for the hESCs/hiPSCs comparisons of the 2683 ‘confidently’ quantified proteins represented as a heatmap plot. After SAM analysis, we found 58 proteins significantly regulated (FDR=1.27%, Supplementary Figure S10A) between the two hiPS cell lines and the hESCs: 46 proteins hESCs>hiPSCs and 12 proteins hESCs<hiPSCs (Figure 3B and Supplementary Table S4).
Figure 2.
Quantitative proteomic comparisons of hESCs, two hiPSCs and their precursor fibroblast cell lines. Protein abundances (Grossmann et al, 2010) are plotted against protein ratios for the hESCs/IMR90_iPS (A), hESCs/4Skin_iPS (B), IMR90_Fibro/IMR90_iPS (C) and 4Skin_Fibro/4Skin_iPS (D) comparisons. The size of the spot reflects the number of unique peptides used to calculate the protein ratio. The color code reflects the variability (i.e., relative standard deviation) of the peptide ratios for each protein. On top, the histograms of frequencies show the density of proteins in each analysis using a bin size of 0.25 (log2). Some of the proteins that are discussed in the text are shown in the plots. NA, not applicable.
Figure 3.
Proteome differences between hESCs, hiPSCs and their precursor fibroblasts. 2683 proteins were quantified in the four data sets: IMR90 and 4Skin experiments (Biological replicas 1 and 2). Relative protein abundances are represented as heatmaps for hESCs/hiPSCs (A) and fibroblasts/hiPSCs (C) comparisons. Using significance analysis of microarrays (SAM; Tusher et al, 2001; Roxas and Li, 2008), 58 proteins (2.2%) were found significantly regulated between hESCs/hiPSCs (B) and 1927 (73.4%) between fibroblasts/hiPSCs (D). The figure was created using the MultiExperiment Viewer software (Saeed et al, 2006). Red and green colors indicate upregulated and downregulated events, respectively. Genes were further grouped using hierarchical clustering (distance metric was Cosine correlation and linkage method was average).
Next, we tested whether the proteins differentially expressed between hiPSCs and hESCs were functionally linked. To this end, we used GO enrichment analyses using all the 10 658 identified proteins in this study as the background data set (see Materials and methods section). The 46 proteins upregulated in hESCs were enriched (P<0.05, binomial test) in GO terms related to antigen processing (e.g., β-2 microglobulin (B2M), TABP) and metabolism of amino acids (e.g., SDHB, ACOX1) and lipids (e.g., APOL2, SOAT1) among others (Supplementary Figure S11A). On the other hand, the 12 proteins that we found highly expressed in hiPSCs were mainly related to cell-adhesion and ectoderm and mesoderm development (e.g., VCAN, COL4A1, CDH2; P<0.05, binomial test; Supplementary Figure S11B). Taken together, our results indicate that the reprogramming process remodeled the proteome of both fibroblast cell lines to a profile that closely resembles the pluripotent hESCs proteome: 97.8% of the ‘confidently’ quantified proteins (i.e., 2683 proteins) showed nonsignificant changes. Nevertheless, a small fraction of their proteomes, 58 proteins (2.2%), was found significantly changing between hiPSCs and hESCs. Functional analyses on this subset of proteins revealed enrichment in certain biological processes, including cell communication and immune system.
Profound differences in the proteomes of hiPSCs and their parental fibroblast cell lines
The inclusion of the two parental fibroblast cell lines in our analysis allowed us to study changes in the proteome at both the starting and end points of the reprogramming process. As expected, this comparison revealed completely different proteomes: the vast majority of the proteins showed differential expression between the parental fibroblasts and the reprogrammed pluripotent cells (Figure 2). We observed a remarkable number of highly abundant proteins displaying very extreme ratios in the fibroblasts (more than 100-fold in many cases). Some of these proteins corresponded to fibroblast markers, such as VIM, COL1A1, COL1A2 and THBS1, which were absent in the hiPSCs. On the other hand, we also confirmed the higher expression in the hiPSCs of known pluripotency markers such as SOX2, NANOG, OCT4, LIN28 and SALL4. Using the SAM statistical analysis, we found 943 proteins significantly enriched in fibroblasts and 1029 proteins with higher levels in the reprogrammed cells (FDR=1.1%, Supplementary Figure S10B; Figure 3C and D and Supplementary Table S4). When we looked at the GO terms associated with these proteins, we found that the fibroblasts were enriched in terms related to transport, endocytosis, exocytosis and metabolism (Supplementary Figure S11C). On the other hand, the proteins enriched in hiPSCs were enriched in numerous GO categories spanning different biological processes such as nucleic acid metabolism, chromatin organization and cell cycle (Supplementary Figure S11D). To further investigate this, we subjected all the hiPSCs and fibroblast-specific proteins to String analysis (Snel et al, 2000), a bioinformatic tool that reconstructs protein networks based on different features like co-expression of genes, physical interactions and co-citation. Strikingly, we obtained hyper-connected protein networks for both sets of proteins (Supplementary Figure S12). The majority of the proteins showed multiple functional connections with other members, and a protein cluster densely interconnected with thousands of links was clearly observed in both analyses. Therefore, we reasoned that the observed protein networks may constitute the protein backbone that controls pluripotent cells and fully differentiated fibroblast cells.
mRNA and protein correlation
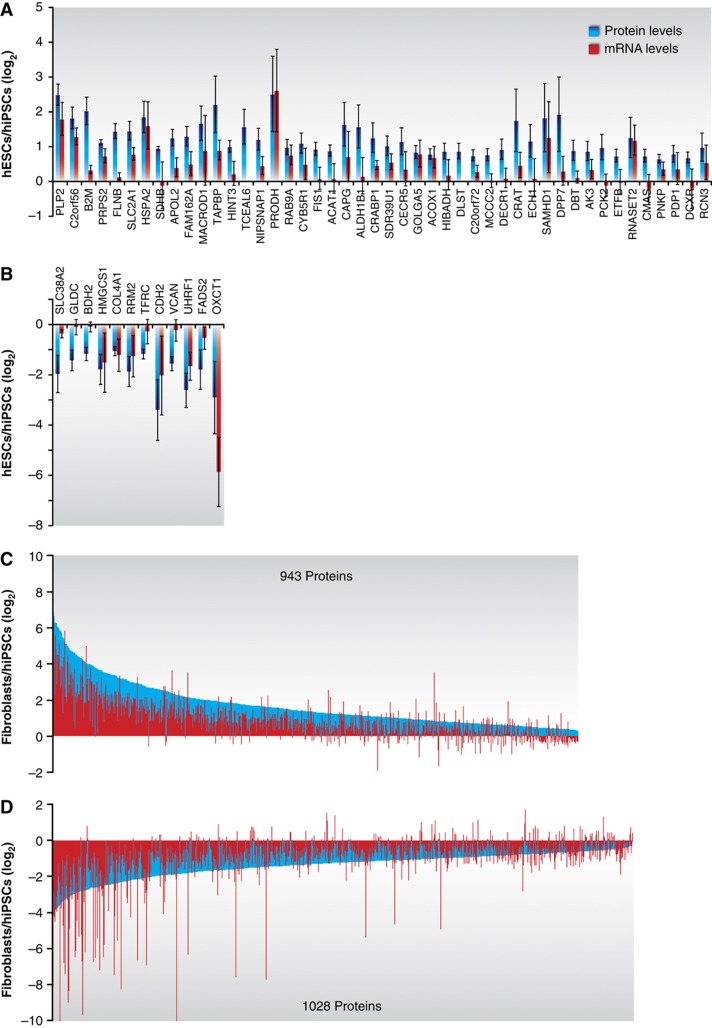
Protein levels are adjusted by an intricate mechanism of gene expression regulation. For instance, recently a poor correlation between protein and mRNA on differentiating mouse ESCs was reported (Lu et al, 2009). To find out if the differences observed in our study were a consequence of transcriptional or translational regulation, we performed paired genome-wide gene expression analyses on the same six samples that were used for the proteomic profiling (Supplementary Methods). Overall, we observed a good correlation between mRNA and protein levels (r∼0.7). Most importantly, when we looked at the transcript levels of the regulated proteins, we found a remarkable agreement (Figure 4). In the hESC/hiPSC comparison, most of the differential proteins were accompanied by a change in the mRNA levels in the same direction (Figure 4A and B). The fibroblast/hiPSC comparison showed the same trend, where most of the genes regulated between these two cell lines were affected at the protein and mRNA levels (Figure 4C and D). These results further authenticated the proteomic measurements and implied a high degree of control at the transcriptional level. Nevertheless, numerous genes were found uncorrelated highlighting the necessity of complementing transcriptomic-based approaches with proteomics.
Figure 4.
Transcript levels of the differentially expressed proteins between hiPSCs and hESCs. The mRNA levels of the identified proteins were measured on the same samples that were used for the proteomic analyses using microarrays (Affymetrix platform). Gene symbols were used to correlate the measurements from both approaches. The figure shows the transcript levels for all the significantly regulated proteins found for hESCs>hiPSCs (A), hESCs<hiPSCs (B), fibroblasts>hiPSCs (C) and fibroblasts<hiPSCs (D). Protein and mRNA ratios were calculated as the average of the four measurements obtained (IMR90 and 4Skin experiments with two biological replicas each). The corresponding error bars (standard deviations) are shown for each value (error bars in C and D were omitted for a better visualization of the figure).
Discussion
Since the discovery in 2006 that somatic cells can be reprogrammed to an embryonic-like state (Takahashi and Yamanaka, 2006), a fundamental question remains unanswered, i.e., are hiPSCs equivalent to hESCs, their natural counterparts? This is especially relevant as genetic defects may affect hiPSCs during differentiation and/or transplantation (Hanna et al, 2010). The conventional procedure to evaluate the pluripotency of iPSCs is based on biological assays for developmental potency. However, the tetraploid complementation assay, which is considered the gold standard test for pluripotency, is restricted to murine cell lines. Accordingly, hiPSCs need to be examined extensively at the molecular level. This allows the characterization of hiPSCs on the basis of quantitative measurements, which, at the same time, will increase our knowledge on the underlying mechanisms of pluripotency and self-renewal. In the last few years, several studies have reported the analysis of DNA methylation status, histone modification patterns, coding mRNA and non-coding miRNA expression patterns in both hiPSCs and hESCs. The conclusions derived from such studies are still uncertain, and the presence of a recurrent molecular signature from the parental cell line as a consequence of incomplete reprogramming (i.e., epigenetic memory) is currently being debated. However, all the aforementioned levels of gene expression regulation function in an orchestrated manner to tune the actual molecular effectors of cells: proteins. Here, we have compared the proteomes of two different hiPS cell lines, their corresponding somatic cells and one hES cell line.
Faced with the challenges of the enormous dynamic range of proteins in mammalian cells, we extensively fractionated the samples using an SCX-based approach. This allowed us to separate peptides based on their charge state, which subsequently were sequenced using targeted fragmentation schemes (i.e., CID, ETD, HCD) to enhance peptide identification (Frese et al, 2011). Using this approach, we achieved the identification of one the largest proteome coverage in pluripotent cells and somatic cells, spanning six orders of magnitude in protein abundance (Figure 2, Supplementary Tables S2 and S3). The 50 most abundant proteins consisted of cytoskeleton (e.g., ACT1, ACTBL2, TUBB2A), chaperones (e.g., HSPA90AB1, HSPA8, HSPA2), ribosomal (e.g., RPS27A, HNRPC) and histones (e.g., HIST1H4A, HIST1H2AB), the latter group likely reflecting the high nucleus/cytoplasm ratio of pluripotent stem cells (Thomson et al, 1998). On the other hand, among the less abundant, we found numerous transcription factors (e.g., DMTF1, BTBD1, ZNF316), signaling molecules and regulatory proteins (e.g., EFNB2, SOCS7, STK35). Most importantly, proteins known to be associated with pluripotency and self-renewal such as SOX2, NANOG, OCT4, LIN28 and SALL4 were found to have expression levels in the middle range (i.e., ∼1000 times less abundant than the structural components), which confirms the importance of their functions in these cell. Given the fact that mRNA levels poorly predict protein translation rates (Schwanhausser et al, 2011), our data set may be highly valuable for those applications such as FACS and RNAi screenings, in which knowing the absolute levels of proteins could determine the success of the experiment (van der Flier et al, 2009).
The use of cost-effective dimethyl isotopes in our workflow allowed us to accurately quantify relative protein changes between hESCs, hiPSCs and their parental fibroblast cell lines. Furthermore, the overall good correlation with the mRNA levels (obtained from paired microarray analyses) validated the reliability of our proteomic measurements. The comparison of two different hiPS cell lines with the hESCs confirmed, at the protein level, that the reprogramming process successfully activated the expression of pluripotency genes and repressed those related to terminally differentiated fibroblasts. The proteomes of hiPSCs and hESCs were found to be very similar, where 97.8% of the proteins displayed nonsignificant changes. Nevertheless, a small subset of proteins (58) was found differentially expressed in common in the two experiments conducted (Figure 3B and Supplementary Table S4), among them are several components of the immune system. Our results showed that iPSCs have reduced levels (less than 3-fold) of two proteins that are essential for the cell-surface expression of HLA class I and correct antigen presentation: B2M and tapasin (TAPBP). In agreement with our results, it has been shown that the reprogramming process might downregulate, through epigenetic mechanisms, MHC and processing molecules (Suárez-Alvarez et al, 2010). Further experimentation will be necessary to find out if these findings may impact the immunogenicity of hiPSCs, but, interestingly, a recent report has described immune rejection on autologous transplanted murine iPSCs (Zhao et al, 2011).
Several evidences point out that epigenetic mechanisms underlie some of the differences found in transcriptomic studies between hiPSCs and hESCs, reviewed in Hanna et al (2010). Genome-wide maps of nucleosomes, i.e., activating K4me3 and repressive K27me3 marks, revealed that both pluripotent cell lines are markedly similar (Guenther et al, 2010). However, modifications in the histone tails are reversible changes that cause local formation of heterochromatin, whereas DNA methylation leads to long-term repression (Berger, 2007). Analyses of the ‘DNA methylome’ at different base pair resolutions have shown manifest differences between hiPSCs and hESCs, pointing out that the reprogramming process could fail in repressing certain genes from the donor cells (epigenetic memory). Moreover, aberrant methylation patterns acquired during reprogramming (epigenetic mutation) have been described (Lister et al, 2011). Hence, we checked, in the parental cell lines, the levels of the 12 proteins enriched in hiPSCs. Only one protein, the transferrin receptor 1 showed increased levels in both IMR90 and 4Skin fibroblasts, which thereby may be explained by incomplete repression of somatic genes during reprogramming (CDH2 and COL4A1 were also found highly expressed in the IMR90 fibroblasts, but not in the 4Skin cell line). Nevertheless, the remaining proteins showed a lower expression in the fibroblasts when compared with the reprogrammed hiPSCs.
On the basis of the transcriptomic profiling, several groups have reported that hiPSCs can be distinguished from hESCs by the presence of a recurrent molecular signature (Chin et al, 2009; Marchetto et al, 2009; Ghosh et al, 2010). A more recent study that includes a significant higher number of cell lines indicated that gene expression programs in hESCs and hiPSCs partially overlapped, although there was a significant difference on average hES and hiPS cell lines (Bock et al, 2011). Owing to the relatively low throughput of MS-based proteomics when compared with other ‘-omics’, our study was limited to two different hiPS cell lines, i.e., IMR90_iPS and 4Skin_iPS. Though, we found 12 proteins consistently enriched in hiPSCs and 46 proteins in hESCs (Figure 3 and Supplementary Table S4). This may be explained by the similar nature of our hiPS cell lines: they are both retroviral reprogrammed (Yu et al, 2007), have mesoderm origin and were cultured at late passage, which altogether could contribute to reduce noise from the analysis (Bock et al, 2011). To further investigate these findings, we compared our list of differential proteins with the transcript lists derived from several independent analyses (Yu et al, 2007; Maherali et al, 2008; Chin et al, 2009; Guenther et al, 2010). These studies include a broad spectrum of different hiPSCs: alternative reprogramming methods, different somatic origins and low passage cultures. The comparisons showed some genes in common with the differential transcripts published by Yu et al (2007): 12/58 (e.g., CDH2, TFRC, RRM2 ACAT1, CAPG, SDR39U1) and by Maherali et al (2008): 10/58 (e.g., DCXR, RCN3, CDH2, SLC38A2, ACOX1, HSPA2). Nonetheless, the overlap was found not significant (Fisher test, P>0.05) and we did not find any regulated gene common to all the studies. Consequently, our results are in line with the emerging idea that differences between hiPSCs and hESCs rather reflect experimental conditions than a consistent molecular signature. As expected, the comparison of both hiPS cell lines with their somatic donor fibroblast cells showed massive differences in their proteomes. Bioinformatics analyses on the differentially expressed proteins between these cell lines disclosed functionally interconnected protein networks in the hiPSCs and fibroblasts (Figure 3). The protein network in hiPSCs is especially relevant, as it may constitute the protein core regulating pluripotency. Interestingly, within this network we found many proteins known to interact with SOX2, NANOG and OCT4 in hESCs (Wang et al, 2006; Mallanna et al, 2010; Pardo et al, 2010; van den Berg et al, 2010) such as Requiem, PRC1, Wdr3b and P66b and NAC1. Furthermore, numerous proteins present in this network are also target genes of SOX2, NANOG and OCT4, i.e., the molecular circuitry governing pluripotency (Boyer et al, 2005), including SMARCAD1, RIF1, ARID1B, DPPA4 and TLE3.
This study constitutes an invaluable resource for the stem cell community by adding an essential layer, the protein content, to the systems biology view of pluripotency, highlighting the molecular similarities between these pluripotent cell lines. Therefore, it is our hope that our data will serve as a platform for future investigations, which more targeted experimentation might reveal.
Materials and methods
Culture of hiPSCs
Induced pluripotent stem cell lines IMR90_iPS and 4Skin_iPS were cultured in Matrigel (Becton Dickinson)-coated dishes, supplemented with mTeSR1 media (Stem Cells Technologies) and passaged every 7 days. Briefly, the cells were washed once with phosphate-buffered saline (PBS; Gibco) before enzymatic treatment with dipase (Chemicon) for ∼3 min. After neutralization with media, the cells were triturated into small clumps or single cells and seeded onto new Matrigel-coated at a split ratio of 1:3 to 1:8. The cells were incubated at 37°C in 5% CO2 incubator.
Characterization of pluripotency
Flow cytometry analysis (FACS)
The expression levels of the pluripotent markers Oct-4, podocalyxin and Tra-1-60 in iPSC populations were assessed by immunofluorescence using flow cytometry. Cells were harvested as a single-cell suspensions using 0.25% trypsin–EDTA (Gibco), fixed and permeabilized (Caltag Laboratories) before incubation with a mouse monoclonal antibody to Oct-4 (1:20, Santa Cruz), podocalyxin (mAb 84, 5 μg, in-house) and Tra-1-60 (1:50, Chemicon). Cells were then washed with 1% BSA/PBS, and incubated in the dark with goat α-mouse antibody FITC-conjugated (DAKO) at 1:500 dilution. After incubation, the cells were washed and resuspended in 1% BSA/PBS for analysis on a FACScan (Becton Dickinson FACS Calibur). All incubations were performed at room temperature for 15 min. For the negative control, cells were stained with the appropriate isotype control.
Staining of hESC for markers
Staining of iPSC was carried out by incubating the cells with fixative Reagent A (Caltag Laboratories) for 1 h, before blocking with 3% BSA/PBS for another hour. After washing with 0.1% Triton/PBS, the cells were incubated with antibodies to Oct-4 (Santa Cruz), SSEA-4 (DHSB) and Tra-1-60 (Chemicon) for 1 h. The detection of bound antibodies to the pluripotent markers was visualized using DAKO goat α-mouse antibody conjugated with PE (diluted 1:500).
Karyotypic stability
Karyotyping analysis was performed by the Cytogenetics Laboratories at the Department of Obstetrics and Gynaecology, KK Women's and Children's Hospital. Cell samples were incubated with BrdU/colcemid (reagent from hospital) for 16 h in 37°C, 5% CO2 incubator.
In-vivo differentiation assay, SCID mice model and teratoma analysis
Induced PSCs were harvested by collagenase (Sigma) treatment and approximately 4–5 × 106 cells were injected with a sterile 22G needle into the rear leg muscle of 4-week-old female SCID mice. Mice that developed tumors approximately 9–10 weeks after injection were killed and the tumors were dissected and fixed in 10% formalin. Tumors were embedded in paraffin, sectioned and examined histologically after hematoxylin and eosin staining.
Sample preparation for MS
Cells (i.e., IMR90_iPS, 4Skin_iPS, hESCs and IMR90 and 4Skin fibroblasts) were harvested by centrifugation at 2500g for 10 min at 4°C. Cell lysis was performed in a buffer containing 8 M urea, 2 M thiourea in a solution of 25 mM ammonium bicarbonate, pH 8.2, with protease and phosphatase inhibitors (Roche). Proteins (∼1 mg) were first reduced/alkylated and digested for 4 h with Lys-C. The mixture was then diluted 4-fold to 2 M urea and digested overnight with trypsin. Digestion was quenched by acidification with formic acid (final concentration 2%). Resulting peptides were then chemically labeled with stable isotope dimethyl labeling as described previously (Boersema et al, 2009). Briefly, IMR90_iPS and 4Skin_iPS peptides were labeled with a mixture of formaldehyde-H2 and sodium cyanoborohydride (‘light’ reagent). For hESCs and IMR90 and 4Skin fibroblast cells, formaldehyde-D2 with cyanoborohydride (‘intermediate’ reagent) and 13C-D2-formaldehyde with cyanoborodeuteride (‘heavy’ reagent) were used respectively. In a second biological replica experiment, hESC and hiPSC reagents were swapped, whereas ‘heavy’-IMR90/4Skin fibroblast was kept constant. The ‘light’, ‘intermediate’ and ‘heavy’ dimethyl-labeled samples were mixed in 1:1:1 ratio based on total peptide amount, which was determined by running an aliquot of the labeled samples on a regular LC-MS/MS run and comparing overall peptide signal intensities.
Before the mass spectrometic analysis, both replicates were fractionated using SCX systems. For Experiment 1, peptides were fractionated as described elsewhere (Helbig et al, 2010). The SCX system consisted of an Agilent 1100 HPLC system (Agilent Technologies, Waldbronn, Germany) with two C18 Opti-Lynx (Optimized Technologies, OR) trapping cartridges and a polysulfoethyl A SCX column (PolyLC, Columbia, MD; 200 mm × 2.1 mm inner diameter, 5 μm, 200-Å). The labeled peptides were dissolved in 10% FA and loaded onto the trap columns at 100 μl/min and subsequently eluted onto the SCX column with 80% acetonitrile (ACN; Biosolve, The Netherlands) and 0.05% FA. SCX buffer A was made of 5 mM KH2PO4 (Merck, Germany), 30% ACN and 0.05% FA, pH 2.7; SCX buffer B consisted of 350 mM KCl (Merck, Germany), 5 mM KH2PO4, 30% ACN and 0.05% FA, pH 2.7. The gradient was performed as follows: 0% B for 10 min, 0–85% B in 35 min, 85–100% B in 6 min and 100% B for 4 min. A total of 45 fractions were collected for each set and dried in a vacuum centrifuge. The second SCX system (Pinkse et al, 2008) was performed using a Zorbax BioSCX-Series II column (0.8-mm inner diameter × 50-mm length, 3.5 μm). SCX solvent A consists of 0.05% formic acid in 20% ACN, while solvent B was 0.05% formic acid, 0.5 M NaCl in 20% ACN. The SCX salt gradient is as follows: 0–0.01 min (0–2% B); 0.01–8.01 min (2–3% B); 8.01–14.01 min (3–8% B); 14.01–28 min (8–20% B); 28–38 min (20–40% B); 38–48 min (40–90% B); 48–54 min (90% B); 54–60 min (0% B). A total of 50 SCX fractions (1 min each, i.e., 50-μl elution volume) were collected and dried in a vacuum centrifuge. Only the second SCX system was used to fractionate lysates from Experiment 2.
Mass spectrometric analysis
For Experiment 1, we performed nanoflow LC-MS/MS with an LTQ-Orbitrap XL ETD mass spectrometer (Thermo Electron, Bremen, Germany) coupled to an Agilent 1200 HPLC system (Agilent Technologies). SCX fractions were dried, reconstituted in 10% FA and delivered to a trap column (Aqua™ C18, 5 μm (Phenomenex, Torrance, CA); 20 mm × 100-μm inner diameter, packed in-house) at 5 μl/min in 100% solvent A (0.1 M acetic acid in water). Next, peptides eluted from the trap column onto an analytical column (ReproSil-Pur C18-AQ, 3 μm (Dr Maisch GmbH, Ammerbuch, Germany); 40 cm × 50-μm inner diameter, packed in-house) at approximately 100 nl/min in a 90 min or 3 h gradient from 0 to 40% solvent B (0.1 M acetic acid in 8:2 (v/v) ACN/water). The eluent was sprayed via distal coated emitter tips butt-connected to the analytical column. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS. Full-scan MS spectra (from m/z 300 to 1500) were acquired in the Orbitrap with a resolution of 60 000 at m/z 400 after accumulation to target value of 500 000 in the linear ion trap. For SCX fractions dominated by singly charged and doubly charged peptides, the five most intense ions at a threshold above 5000 were selected for collision-induced fragmentation in the linear ion trap at a normalized collision energy of 35% after accumulation to a target value of 10 000. For highly charged SCX fractions, the five most intense ions at a threshold of above 500 were fragmented in the linear ion trap using electron-transfer dissociation with supplemental activation (ETcaD) at a target value of 50 000. The ETcaD reagent target value was set to 100 000 and the reaction time to 50 ms.
MS analysis for Experiment 2 was performed with the same LC gradient and configuration but using an LTQ-Orbitrap Velos (Thermo Electron). MS data were acquired with a data-dependent decision tree method as described (Frese et al, 2011). Briefly, following the survey scan (30 000 FHMW), the 10 most intense precursor ions were subjected to HCD, ETD-IT or ETD-FT fragmentation. The choice of the most appropriate technique for a selected precursor was determined by a preprogrammed data-dependent decision tree. Essentially, doubly charged peptides were subjected to HCD fragmentation and more highly charged peptides were fragmented using ETD. The normalized collision energy for HCD was set to 35%. ETD was enabled with supplemental activation and the reaction time was set to 50 ms for doubly charged precursors.
Data processing
MS data were processed and quantified with Proteome Discoverer (version 1.3, Thermo Electron) with standardized workflows. This ensures consistent and reproducible quantification for all samples, avoiding possible bias introduced by manual intervention. These workflows are made available as Supplementary Materials. For Experiment 1, peptide identification was performed with Mascot 2.3 (Matrix Science) against a concatenated forward-decoy IPI Human database supplemented with all the frequently observed contaminants in MS (version 3.68, 174 650 entries). The following parameters were used: 50 p.p.m. precursor mass tolerance, 0.5 Da fragment ion tolerance, up to 2 missed cleavages, carbamidomethyl cysteine as fixed modification and oxidized methionine as variable modifications. Dimethyl-based quantitation method was chosen in Proteome Discoverer, with mass precision requirement of 2 p.p.m. for consecutive precursor measurements. Besides, taking into account the isotopic effect of deuterium, we applied 1 min of retention time tolerance for isotope pattern multiplets and allowed spectra with 2 missing channels to be quantified. After identification and quantification, we combined all results originating from the same biological replica and filtered them according to very stringent peptide acceptance criteria. These criteria include (i) mass deviations of ±5 p.p.m., (ii) Mascot Ion Score of at least 20, (iii) a minimum of 7 amino-acid residues per peptide and (iv) position rank 1 in Mascot search. As a result, we obtained peptide FDRs of 0.23 and 0.19% for two respective biological replicas (Supplementary Table S1). Finally, peptide ratios were then normalized against the median (log2). The same criteria were subsequently applied to analyze Experiment 2 except for the following: 0.5 Da fragment ion tolerance for ETD-IT, and 0.02 Da fragment ion tolerance for both HCD and ETD-FT. Peptide FDRs obtained for Experiment 2 were 0.56 and 0.59%, respectively.
The MS data associated with this manuscript can be downloaded from ProteomeCommons.org under the following Tranche hash: d/Ci8K23/EI0YIsZ7OQpxQjkHUlCdAMtiBKQT6a+4McomzkpsxZYLnZYcm1vjmMhHv94w5or5jGg/6l42VgFGtFZSC0AAAAAAAAOtw==.
Microarray analysis
RNA isolation
Cells were washed twice in PBS (Gibco), harvested and quantified using the NucleoCounter (Chemometec). RNA was isolated using TRIzol (Invitrogen)/chloroform according to manufacturer's protocol. RNA in the extracted aqueous phase was concentrated by precipitation with an equal volume of isopropanol at −20°C overnight, and further purified using RNeasy Mini Kit (Qiagen). Quantity of RNA was measured by NanoDrop (Thermo Scientific) and quality evaluated by capillary electrophoresis on QIAxcel (Qiagen).
Transcriptome analysis
Affymetrix U133 Plus 2.0 GeneChip Microarrays (Human Genome) were used according to manufacturer's protocol. Briefly, GeneChip 3′ IVT Express Kit was used to generate labeled amplified RNA (aRNA) from 500 ng of total RNA, with 12.5 μg of fragmented aRNA subsequently hybridized onto each microarray. Intact aRNA yield was measured by NanoDrop, and quality of intact and fragmented aRNA evaluated on 2% agarose gel. Hybridizations were carried out at 45°C for 16 h with reagents from GeneChip Hybridization, Wash, and Stain Kit, with subsequent washing and staining of arrays automated and preprogrammed (FS450_0001) on Fluidics Station 450. Arrays were scanned using GeneChip Scanner 3000.
Microarray data analysis
Affymetrix Expression Console software was used to analyze scanned image files. Array data were quantified and normalized using MAS5.0. algorithm, with corresponding detection calls for each probeset determined. Only data from probesets that (i) were flagged as present in at least one set of biological replicates, and (ii) possessed the highest summed intensities among redundant probesets—those annotated with similar gene name or unigene identifier, were used for subsequent comparison with the proteome study. Microarray data are available at the NCBI Gene Expression Omnibus database under the accession numbers GSE26451 and GSE26453.
Bioinformatic analysis
Protein classification (molecular function, biological process, cellular component and protein class) was performed using the PANTHER classification system (Mi et al, 2007). GO enrichment was performed using a Binomial test as described elsewhere (Cho and Campbell, 2000), the entire list of identified proteins was used as the reference data set. Protein and mRNA levels were combined using official gene symbols (HUGO). Protein networks were created with String (Snel et al, 2000).
Statistical analysis
Significance analysis of microarrays (Tusher et al, 2001; Roxas and Li, 2008) was used to identify significantly regulated proteins between hESCs, hiPSCs and fibroblasts. Only those proteins that were quantified in both experiments (i.e., IMR90 and 4Skin) and in both biological replicas were analyzed under statistical criteria. Overall, log2-transformed ratios for 2683 proteins were processed by using the MultiExperiment Viewer software (version 4.7.4) (Saeed et al, 2006). Briefly, one-class test was used (the mean value to test again was 0), 1000 permutations were chosen for a better approximation of FDR values (Saeed et al, 2006), S0 value was selected according to the method proposed by Tusher et al (2001) and q-values were calculated for each protein. Delta values were manually adjusted to ∼1% FDR. SAM graphs for both comparisons hESCs/hiPSCs and Fibroblasts/hiPSCs are shown in Supplementary Figure S10. The results from the analyses are presented in Supplementary Table S4.
Note added in proof
While this paper was in revision, a related proteomic study appeared comparing hESCs and hiPSCs (Phanstiel et al, 2011). Their findings are well in agreement with ours, reporting a high similarity of hESC and hiPSC proteomes with a small subset of proteins differentially expressed between both pluripotent cell lines.
Supplementary Material
Supplementary Material
Table S1. Experiment information. Information for all the MS analyses performed on two biological replicas for Experiment 1 (hESCs, IMR90_iPS, IMR90_Fibro) and Experiment 2 (hESCs, 4Skin_iPS, 4Skin_Fibro). Simplistic Protein FDR which is calculated by counting the number of Decoy proteins with respect to the total number of identified proteins in each data set
Table S2. Identified and quantified proteins/peptides for Experiment 1 (IMR90). 6,873 and 5,835 unique proteins groups were identified for Biological Replica 1 and 2 respectively. Identifications were filtered at a peptide FDR of < 1% (Mascot Ion Score > 20). Resulting Protein FDR values are shown in Table S1. An expanded view at the peptide level can be obtained by clicking each protein. Prefixes such as CON_ and REV_ represent common contaminants and decoy hits. Protein ratios were first log2-transformed and subsequently normalized according to the median of the data set. In biological replica 1, IMR90_iPS peptides were labelled as ‘Light’, the ES peptides ‘Medium’ while the IMR90 fibroblasts as ‘Heavy’. In the second replica, labels for iPS and ES were swapped while IMR90 remained as ‘Heavy’
Table S3. Identified and quantified proteins/peptides for Experiment 2 (4Skin). 8,548 and 7,154 unique proteins groups were identified for Biological Replica 1 and 2 respectively. Identifications were filtered at a peptide FDR of < 1% (Mascot Ion Score > 20). Resulting Protein FDR values are shown in Table S1. An expanded view at the peptide level can be obtained by clicking each protein. Prefixes such as CON_ and REV_ represent common contaminants and decoy hits. Protein ratios were first log2-transformed and subsequently normalized according to the median of the data set. In biological replica 1, IMR90_iPS peptides were labelled as ‘Light’, the ES peptides ‘Medium’ while the IMR90 fibroblasts as ‘Heavy’. In the second replica, labels for iPS and ES were swapped while IMR90 remained as ‘Heavy’
Table S4. Differential proteins. Significance analysis of microarrays (SAM) (Tusher et al, 2001) was used for the statistical analysis of differentially expressed proteins between hESCs/hiPSCs and Fibroblasts/hiPSCs as proposed by (Roxas et al, 2008). The analysis was performed using the Multi Experiment Viewer software (Saeed et al, 2003). The table contains 4 Excel sheets with both significant and non-significant proteins found for the two comparisons. Significant genes were filtered to approximately 1 % FDR. Upregulated proteins are indicated in red whereas down-regulated proteins are shown in green. Non-significant proteins are in grey. Expected and observed scores are shown for each protein as well as their corresponding q-values (%). The detailed description of the analysis is provided in the Materials section
Acknowledgments
We thank Dr Shabaz Mohammed for mass spectrometry support; Dr Andreas Helbig and Marco Hennrich for SCX; Daniel Navarro and Henk van den Toorn for in-house software development and other members from the Heck Lab for fruitful discussions. We thank Christian Preisinger for critical evaluation of the manuscript. This work was supported by the Netherlands Proteomics Centre, which is part of the Netherlands Genomics Initiative, and by the Agency for Science Technology and Research (A*STAR), Singapore.
Author contributions: All Utrecht University authors performed experiments under the guidance of AJRH, JM and TYL performed the proteomic experiments and analysis. YJK carried out the microarray analyses and VD was involved in the cell culture. JM assembled and analyzed all the data, performed the statistical analyses and wrote the manuscript. AC and AJRH were involved in the conceptual design as well as manuscript writing.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo ABH, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K et al. (2007) Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25: 803–816 [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DEC, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW (2007) Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25: 207–215 [DOI] [PubMed] [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A (2011) Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144: 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4: 484–494 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin ACP, Padmanabhan J, Oh SKW, Choo ABH (2010a) Defined and serum-free media support undifferentiated human embryonic stem cell growth. Stem Cells Dev 19: 753–761 [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA et al. (2009) Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE (2010b) Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell 7: 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ (2000) Transcription, genomes, function. Trends Genetics 16: 409–415 [DOI] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park I-H, Yu J, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K (2009) Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park I-H, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP (2009) Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K (2008) Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells 26: 2467–2474 [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu S-J, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim K-S, Lanza R (2010) Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28: 704–712 [DOI] [PubMed] [Google Scholar]
- Frese CK, Altelaar AFM, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJR, Mohammed S (2011) Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res 10: 2377–2388 [DOI] [PubMed] [Google Scholar]
- Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC (2010) Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE 5: e8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann J, Roschitzki B, Panse C, Fortes C, Barkow-Oesterreicher S, Rutishauser D, Schlapbach R (2010) Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J Proteomics 73: 1740–1746 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA (2010) Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, Lengner CJ, Dausman JA, Jaenisch R (2008) Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R (2010) Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143: 508–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, Mohammed S, Heck AJR (2010) Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteomics 9: 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R (2008) A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3: 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B-Y, Weick JP, Yu J, Ma L-X, Zhang X-Q, Thomson JA, Zhang S-C (2010) Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA 107: 4335–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, Lundin K, Mikkola M, Trokovic R, Peitz M, Brüstle O, Bazett-Jones DP, Alitalo K, Lahesmaa R, Nagy A, Otonkoski T (2011) Copy number variation and selection during reprogramming to pluripotency. Nature 471: 58–62 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel CV, Iseli C, Stevenson BJ, Riggins GJ, Lal A, Mackay A, Harris RA, O'Hare MJ, Neville AM, Simpson AJG, Strausberg RL (2003) Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. Proc Natl Acad Sci USA 100: 4702–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim C-H, Moon J-I, Chung Y-G, Chang M-Y, Han B-S, Ko S, Yang E, Cha KY, Lanza R, Kim K-S (2009) Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J et al. (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, Ku S, Martynova M, Semechkin R, Galat V, Gottesfeld J, Izpisua Belmonte JC, Murry C, Keirstead HS, Park H-S, Schmidt U et al. (2011) Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8: 106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-H, Agarwal S, Park I-H, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ (2009) Generation of induced pluripotent stem cells from human blood. Blood 113: 5476–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma/'ayan A, Boyer LA, Troyanskaya OG, Whetton AD, Lemischka IR (2009) Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K (2008) A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3: 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, Washburn MP, Rizzino A (2010) Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells 28: 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Marchetto MCN, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR (2009) Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 4: e7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti J-C, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N (2010) Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7: 521–531 [DOI] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD (2007) PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res 35: D247–D252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8: 633–638 [DOI] [PubMed] [Google Scholar]
- Newman AM, Cooper JB (2010) Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 7: 258–262 [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, Yu J, Hebrok M, Hochedlinger K, Costello JF, Song JS, Ramalho-Santos M (2011) Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13: 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J (2010) An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-H, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ (2008a) Disease-specific induced pluripotent stem cells. Cell 134: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ (2008b) Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146 [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, Thomson JA, Coon JJ (2011) Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Meth 8: 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse MWH, Mohammed S, Gouw JW, van Breukelen B, Vos HR, Heck AJR (2008) Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J Proteome Res 7: 687–697 [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K (2010) Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KTG, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4: rs3. [DOI] [PubMed] [Google Scholar]
- Roxas BAP, Li Q (2008) Significance analysis of microarray for relative quantitation of LC/MS data in proteomics. BMC Bioinformatics 9: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Meth Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung H-K, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7: 64–77 [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Snel B, Lehmann G, Bork P, Huynen MA (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 28: 3442–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R (2009) Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136: 964–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Alvarez B, Rodriguez RM, Calvanese V, Blanco-Gelaz MA, Suhr ST, Ortega F, Otero J, Cibelli JB, Moore H, Fraga MF, López-Larrea C (2010) Epigenetic mechanisms regulate MHC and antigen processing molecules in human embryonic and induced pluripotent stem cells. PLoS ONE 5: e10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Thomson JA, Coon JJ (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA 106: 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DLC, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H (2009) Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Muñoz J, Braam SR, Pinkse MWH, Linding R, Heck AJR, Mummery CL, Krijgsveld J (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5: 214–226 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Pinkse MWH, Oostwaard DW-V, Mummery CL, Heck AJR, Krijgsveld J (2007) An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat Methods 4: 677–678 [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC (2009) MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev 18: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung H-K, Nagy A (2009) piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin I, Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L (1995) Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev 9: 2635–2645 [DOI] [PubMed] [Google Scholar]
- Zhao T, Zhang Z-N, Rong Z, Xu Y (2011) Immunogenicity of induced pluripotent stem cells. Nature 474: 212–215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1. Experiment information. Information for all the MS analyses performed on two biological replicas for Experiment 1 (hESCs, IMR90_iPS, IMR90_Fibro) and Experiment 2 (hESCs, 4Skin_iPS, 4Skin_Fibro). Simplistic Protein FDR which is calculated by counting the number of Decoy proteins with respect to the total number of identified proteins in each data set
Table S2. Identified and quantified proteins/peptides for Experiment 1 (IMR90). 6,873 and 5,835 unique proteins groups were identified for Biological Replica 1 and 2 respectively. Identifications were filtered at a peptide FDR of < 1% (Mascot Ion Score > 20). Resulting Protein FDR values are shown in Table S1. An expanded view at the peptide level can be obtained by clicking each protein. Prefixes such as CON_ and REV_ represent common contaminants and decoy hits. Protein ratios were first log2-transformed and subsequently normalized according to the median of the data set. In biological replica 1, IMR90_iPS peptides were labelled as ‘Light’, the ES peptides ‘Medium’ while the IMR90 fibroblasts as ‘Heavy’. In the second replica, labels for iPS and ES were swapped while IMR90 remained as ‘Heavy’
Table S3. Identified and quantified proteins/peptides for Experiment 2 (4Skin). 8,548 and 7,154 unique proteins groups were identified for Biological Replica 1 and 2 respectively. Identifications were filtered at a peptide FDR of < 1% (Mascot Ion Score > 20). Resulting Protein FDR values are shown in Table S1. An expanded view at the peptide level can be obtained by clicking each protein. Prefixes such as CON_ and REV_ represent common contaminants and decoy hits. Protein ratios were first log2-transformed and subsequently normalized according to the median of the data set. In biological replica 1, IMR90_iPS peptides were labelled as ‘Light’, the ES peptides ‘Medium’ while the IMR90 fibroblasts as ‘Heavy’. In the second replica, labels for iPS and ES were swapped while IMR90 remained as ‘Heavy’
Table S4. Differential proteins. Significance analysis of microarrays (SAM) (Tusher et al, 2001) was used for the statistical analysis of differentially expressed proteins between hESCs/hiPSCs and Fibroblasts/hiPSCs as proposed by (Roxas et al, 2008). The analysis was performed using the Multi Experiment Viewer software (Saeed et al, 2003). The table contains 4 Excel sheets with both significant and non-significant proteins found for the two comparisons. Significant genes were filtered to approximately 1 % FDR. Upregulated proteins are indicated in red whereas down-regulated proteins are shown in green. Non-significant proteins are in grey. Expected and observed scores are shown for each protein as well as their corresponding q-values (%). The detailed description of the analysis is provided in the Materials section