Upstream of N-ras (UNR) is a conserved RNA-binding protein that regulates mRNA translation and stability by binding to sites generally located in untranslated regions (UTRs). In Drosophila, sex-specific binding of UNR to msl2 mRNA and the noncoding RNA roX plays key roles in the control of X-chromosome dosage compensation in both sexes. Here the authors identified its RNA targets in adult male and female flies by high-throughput RNA binding and transcriptome analysis and show that UNR binds to a large set of protein-coding transcripts and to a smaller set of noncoding RNAs in a sex-specific fashion. These results suggest that UNR exploits the transcript diversity generated by alternative processing and alternative promoter usage to bind and regulate target genes in a sex-specific manner.
Keywords: UNR target, sex-specific binding, UTR, alternative processing
Abstract
Upstream of N-ras (UNR) is a conserved RNA-binding protein that regulates mRNA translation and stability by binding to sites generally located in untranslated regions (UTRs). In Drosophila, sex-specific binding of UNR to msl2 mRNA and the noncoding RNA roX is believed to play key roles in the control of X-chromosome dosage compensation in both sexes. To investigate broader sex-specific functions of UNR, we have identified its RNA targets in adult male and female flies by high-throughput RNA binding and transcriptome analysis. Here we show that UNR binds to a large set of protein-coding transcripts and to a smaller set of noncoding RNAs in a sex-specific fashion. The analyses also reveal a strong correlation between sex-specific binding of UNR and sex-specific differential expression of UTRs in target genes. Validation experiments indicate that UNR indeed recognizes sex-specifically processed transcripts. These results suggest that UNR exploits the transcript diversity generated by alternative processing and alternative promoter usage to bind and regulate target genes in a sex-specific manner.
INTRODUCTION
Regulation of gene expression at the post-transcriptional level contributes to generate the molecular and functional diversity that underlies the complexity of living organisms. In addition to alternative promoter usage during transcription, multiple transcripts are produced from a single gene through post-transcriptional mechanisms that include alternative splicing, alternative polyadenylation, and editing. Once processed, mature mRNA isoforms are regulated at the levels of export, localization, stability, and translation. These post-transcriptional mechanisms of control of RNA metabolism are governed by RNA-binding proteins (RBPs). Studies based on high-throughput technologies have revealed not only an enormous diversity at the RNA level, but also a coordinated regulation of RNA by RBPs (for review, see Licatalosi and Darnell 2010). Examples of coordinated regulation by RBPs have led to the RNA operon hypothesis, wherein mRNAs encoding functionally related proteins are coregulated by RBPs in an analogous way in which DNA operons allow coordinated expression in bacteria (Keene 2007; Morris et al. 2009). Because transcripts are usually bound by several RBPs, each transcript can be part of more than one RNA operon, leading to higher-order RNA networks in which RBPs constitute the regulatory nodes.
An example of coordinated regulation is provided by the protein Upstream of N-ras (UNR), a conserved RBP involved in the regulation of mRNA stability and translation (Mihailovich et al. 2010). In Drosophila, UNR contributes to opposite, sex-specific outcomes in X-chromosome dosage compensation via differential binding to RNA. X-chromosome dosage compensation is the process that equalizes the expression of X-linked genes between males (XY) and females (XX), and in Drosophila is achieved by hypertranscription of the single male X chromosome by the dosage compensation complex (DCC), a ribonucleoprotein assembly of at least five proteins (MSL1, MSL2, MSL3, MOF, and MLE) and two noncoding RNAs (roX1 and roX2) (Gelbart and Kuroda 2009). In females, UNR binds to msl2 mRNA and inhibits its translation, contributing to repressing dosage compensation (Abaza et al. 2006; Duncan et al. 2006; Patalano et al. 2009). Binding of UNR to msl2 mRNA depends on the female-specific RBP Sex-lethal (SXL), which, in addition to recruiting UNR to the 3′ UTR of msl2, promotes the retention of a facultative intron in msl2 5′ UTR (for review, see Graindorge et al. 2011). Thus, UNR binds to a sex-specific alternatively processed msl2 transcript in complex with SXL. In males, UNR does not bind to msl2 because SXL is absent. In contrast, UNR binds to the roX RNAs in males, and this binding may contribute to promoting DCC recruitment to the X chromosome (Patalano et al. 2009).
UNR contains five cold-shock domains (CSDs), β-barrel structures that serve to bind single-stranded nucleic acids (Mihailovich et al. 2010). UNR binds to msl2 via a dedicated CSD (Abaza and Gebauer 2008), raising the possibility that each CSD binds RNA independently and suggesting a strong RNA-binding potential for this protein. To gain insight into this potential and to evaluate additional sex-specific roles of UNR, we set to identify the UNR RNA targets in male and female flies. UNR immunoprecipitation from whole adult extracts followed by high-throughput identification of the associated RNAs revealed that UNR binds to a large set of transcripts in a sex-specific fashion. In 42% of the cases, sex-specific binding correlated with sex-specific expression of the target gene. Strikingly, the correlation increased to 83% when differential, sex-specific expression of the UTRs was taken into account. These results indicate that the generation of sex-specific UTRs by alternative splicing, polyadenylation, or alternative promoter usage contributes to sex-specific RNA binding by UNR.
RESULTS
Identification of UNR targets
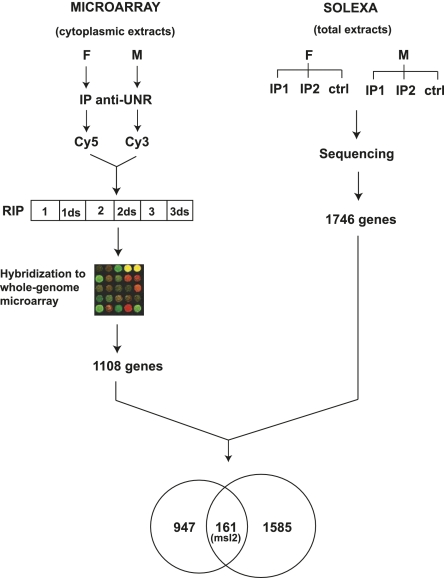
As a first approach to identifying UNR targets, we immunoprecipitated UNR from either male or female adult cytoplasmic extracts and identified the associated mRNAs by microarray analysis (RIP-Chip). Three independent RIP experiments were performed, and two microarrays (direct labeling and dye swap) were hybridized per RIP sample (Fig. 1, left panel). Genes appearing in the upper third of the intensity distribution and present in at least four of the six microarrays were considered. Under these conditions, 1148 probe sets corresponding to 1108 unique genes were identified as UNR targets, including our positive control msl2 (Supplemental Table SI). This number of targets corresponds to 8.1% of the 13,638 genes present in the microarray.
FIGURE 1.
Schematic representation of the experimental design to identify UNR targets. Targets were identified by RIP-Chip (microarray, left panel) or RIP-seq (Solexa, right panel). For RIP-Chip, UNR was immunoprecipitated (IP) from cytoplasmic extracts of adult male and female flies, and the RNA in the pellet amplified and labeled for hybridization to microarrays. Three independent IPs were performed, and six microarrays including the dye-swap controls (ds) were hybridized. For RIP-seq, UNR was immunoprecipitated from total extracts of adult males and females in duplicate experiments (IP1 and IP2), carrying a mock IP with rabbit IgG as control (ctrl). The number of targets identified using each technique is indicated, and a Venn diagram is shown to visualize the overlap. The position of msl2 is indicated.
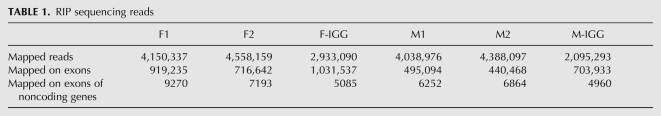
The microarray analysis showed differences in target binding in males and females, but very few targets passed the thresholds to be considered significantly differentially bound (Benjamini-Hochberg FDR cutoff of 10%). This could be due to the limited dynamic range of the microarray, combined with Chip-to-Chip experimental variation. It has been shown that larger differences in gene expression can be observed by deep sequencing as compared with microarrays (Hoen et al. 2008). Thus, to have a more quantitative estimate of the binding of UNR to its targets, we performed RIP-seq experiments. Sequencing allows the unbiased identification of targets, including non-annotated transcripts and noncoding RNAs. Because UNR binds to the nuclear noncoding RNAs roX1 and roX2 (Patalano et al. 2009), in order to identify coding and noncoding poly(A)+ transcripts bound by UNR independently of their subcellular distribution, we used total male and female adult extracts for RIP-seq. A RIP experiment including a duplicate with anti-UNR antibodies and one IgG control was performed per sex, and the RNA isolated from the pellet was sequenced (Fig. 1, right panel). All tags mapping unambiguously on exons of annotated FlyBase transcripts were considered (Table 1). The IgG control was used to correct for serendipitous binding and for nonspecific binding of abundant transcripts (see Halbeisen et al. 2008 and references therein).
TABLE 1.
RIP sequencing reads
To identify significant enrichment in the IP with respect to the IgG control, a 2 × 2 χ2 test similar to that previously introduced for analysis of SAGE data was used (Michiels et al. 1999). This method takes into account both the relative enrichment of each gene in the IP versus the IgG, and the relative abundance of the gene in each sample (number of sequenced tags assigned to each gene vs. the overall number of tags of the experiment). Targets passing a strict Bonferroni-corrected P-value threshold of 5 × 10−6 as well as a fold ratio IP/IgG threshold of 2 in either sex were considered positive (for details, see Materials and Methods). The calculated P-value, assigned to each gene, was used as a direct measure of binding in further analysis. A total of 1746 protein-coding gene targets of UNR were identified in this manner, including our positive control msl2, which showed an IP/IgG ratio of 2.0187 (Supplemental Table SII). This number of targets corresponds to 12.8% of the 13,653 genes annotated in the current FlyBase genome annotation used in our analysis.
A comparison of the targets identified by RIP-Chip and RIP-seq indicated an overlap of ∼15% (Fig. 1). Several factors could contribute to this limited overlap, including experimental variation between independent biological RIP replicates, the intrinsic differences associated with each technique, and the use of a different starting material for RIP-Chip and RIP-seq (cytoplasmic vs. total extracts, respectively). The Pearson correlation between different RIPs in RIP-seq was of 0.94 (females) and 0.96 (males); in RIP-Chip, the correlation ranged from 0.67–0.83 in cross-comparisons of different RIPs, while it increased to 0.90–0.97 for replicates within the same RIP (data not shown). These data indicate that the variation between independent biological replicates is relatively small and thus should contribute little to the limited overlap between the RIP-Chip and RIP-seq data. Other factors such as the use of total versus cytoplasmic RNA have the potential to contribute more decisively, because large variations have been recently reported when these two types of starting material are compared in gene expression studies using microarrays (Trask et al. 2009) (see Discussion).
The modest overlap also suggests that the number of UNR targets is underestimated. Indeed, it is not unexpected that a large set of transcripts associates with a ubiquitous RNA-binding protein in whole organisms.
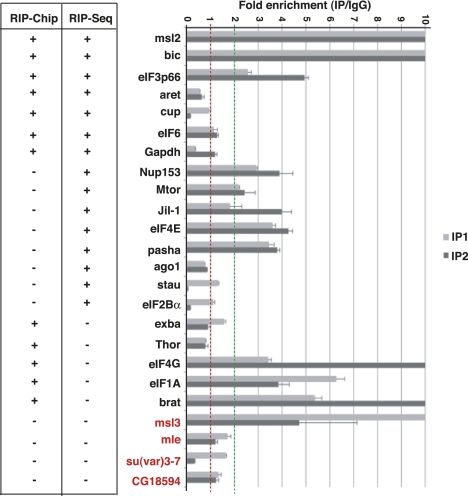
To compare the efficiency of RIP-Chip and RIP-seq in the identification of UNR targets, RT-qPCR validation was performed on a list of 24 genes of interest to our laboratory, whose products have roles in X-chromosome dosage compensation or translational control. The list includes 20 targets that were positive by RIP-Chip, RIP-seq, or both and displayed diverse ranking values, and four genes that were negative by both techniques. Two independent IPs were performed on mixed samples containing the same amounts of male and female extracts, carrying parallel IPs with rabbit IgG as controls. As for RIP-seq, we considered positive in RT-qPCR those targets that fit the threshold IP/IgG ≥ 2. Given this threshold, 12 of the 20 targets were clearly positive by quantitative PCR, some with enrichments above 10-fold (e.g., msl2), while one of the negatives (msl3) turned positive by qPCR (Fig. 2). A comparison of RIP-Chip, RIP-seq, and RT-qPCR indicates a comparable validation rate for both high-throughput techniques.
FIGURE 2.
Validation of UNR targets by RT-qPCR. Two independent IP experiments were performed (light and dark gray bars), carrying parallel IPs with rabbit IgG as negative controls. Targets were considered positive if present in the UNR IP at least twofold over the control (green line). The data were background-normalized for the presence of the 28S rRNA and represented relative to the IgG control (red line). (Red) Genes that were negative by both RIP-Chip and RIP-seq. A comparison with the RIP-Chip and RIP-seq data is shown at the left.
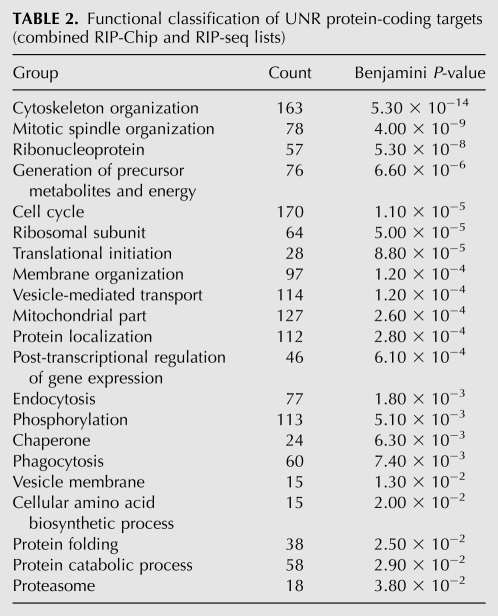
Functional Gene Ontology analysis using DAVID (Huang et al. 2009) revealed that UNR targets were enriched in categories such as cytoskeleton organization, cell cycle, translation and ribosome component, and mitochondria, both if RIP-Chip or RIP-seq data were considered together or separately. A list of GO terms for the combined list is shown in Table 2.
TABLE 2.
Functional classification of UNR protein-coding targets (combined RIP-Chip and RIP-seq lists)
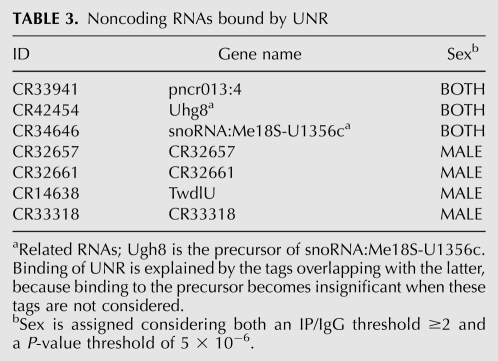
Interestingly, RNA-seq yielded a list of seven noncoding RNAs that were significantly bound by UNR (Table 3). Two of the noncoding RNAs are related, because Ugh8 (UsnRNA host gene 8) is the precursor of Me18S-U1356c, an snoRNA involved in methylation of 18S rRNA (Huang et al. 2005). The function of the remaining noncoding UNR targets is unknown. We expected to find roX1 and roX2 in the list of noncoding UNR targets. Both roX RNAs showed an increased number of tags in the male IP compared with the IgG control but did not pass the P-value threshold used in our analysis (data not shown). These data suggest that the use of strict significance thresholds leads to loss of true positives and support an underestimation in the number of UNR targets.
TABLE 3.
Noncoding RNAs bound by UNR
In sum, the data suggest that UNR binds to a large set of protein-coding transcripts and a reduced group of annotated noncoding RNAs in adult flies.
Extensive sex-specific RNA-binding by UNR
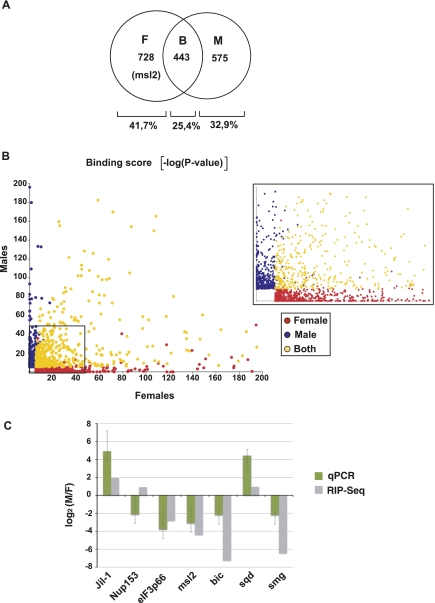
We next evaluated the sex specificity of UNR binding to its targets using the RIP-seq data. The sex-specific association of UNR to its targets (male, female, or both) is indicated in Supplemental Tables SII and III for the coding and noncoding transcripts, respectively. The data indicate that nearly 75% of the protein-coding targets bind to UNR in a sex-specific fashion (Fig. 3A). Because the P-value can be used as a direct measure of binding, we defined the −log2 of the P-value as the binding score. We next plotted the binding scores obtained for the UNR targets in both sexes and colored the genes passing the IP/IgG threshold in females (red), males (blue), or both (yellow) (Fig. 3B). In this way, the sex-binding bias of UNR to its targets can be easily visualized. The results show a wide dispersion of binding scores. Most genes considered targets of UNR in either males or females cluster close to the corresponding graph axes, as expected, while those binding to UNR in both sexes are broadly dispersed (Fig. 3B, cf. blue and red with yellow). We validated the sex-binding bias for seven targets by RT-qPCR in independent RIP experiments and found that the binding bias was largely confirmed (Fig. 3C). These data indicate that UNR binds to targets in a sex-specific manner.
FIGURE 3.
UNR binds to targets in a sex-specific fashion. (A) Venn diagram of the sex-specific distribution of UNR targets identified by RIP-seq. (M) Males; (F) females; (B) both. The position of msl2 is indicated. (B) Representation of the binding scores [−log(P-value)] of UNR targets in males and females. Coloring indicates the genes passing the IP/IgG threshold in females (red), males (blue), or both (yellow). (C) RT-qPCR validation of a set of UNR targets. For each target, three to five independent IPs were performed, the qPCR values were background-normalized for the presence of the 28S rRNA, and the most representative values were taken to obtain this graph (green bars). A comparison with the binding score ratios obtained by RIP-seq (gray bars) is shown.
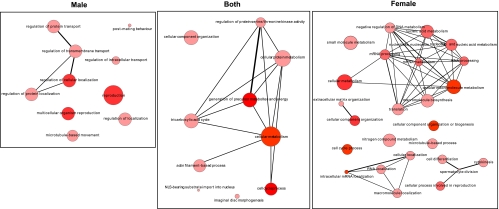
GO analysis of the targets bound by UNR exclusively in one sex using the tools GOrilla and REVIGO (Eden et al. 2009; Supek et al. 2011) revealed different functional groups significantly enriched in either males or females (Fig. 4). In males, processes such as reproduction, transport, and localization were highlighted, whereas in females, regulation of RNA metabolism, cell cycle, and macromolecule biogenesis were enriched. Regulation of cellular metabolism was common to both sexes. These results suggest that RNA binding by UNR may lead to sex-specific functional outcomes for processes other than dosage compensation.
FIGURE 4.
GO analysis of UNR targets bound in males, females, or both. The GOrilla and REVIGO analysis tools were used to generate a network of interconnected circles, where the size of the circle represents the number of genes within the specific GO term, and the intensity of color indicates the P-value significance. The line thickness denotes the degree of similarity between the GO terms.
Sex-specific RNA binding by UNR is not explained by the relative overall expression of target genes
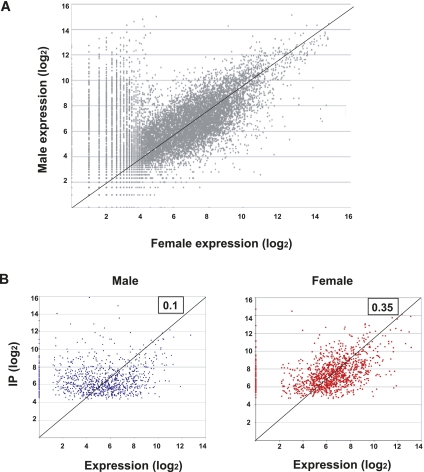
The simplest explanation for the sex-specific binding bias displayed by UNR is that binding just reflects the relative overall expression of targets in males and females. To evaluate this possibility, we sequenced the whole transcriptome of adult male and female flies. A comparison of transcript levels in both sexes showed that most genes were expressed within a threefold range, yet a higher extent of sex-biased expression was detected in males (Fig. 5A). To test for possible correlations between the expression of gene targets and their presence in the UNR immunoprecipitates, we compared the abundance of targets in the IP and the total RNA in each sex, computed as the log2 of the normalized read count (Fig. 5B). The results showed a Pearson correlation coefficient of 0.1 for males and 0.35 for females, indicating that, for most targets, abundance in the IP is not a direct consequence of total RNA abundance.
FIGURE 5.
Assessment of the correlation between the expression of UNR targets and their presence in the UNR immunoprecipitates. (A) Relative expression of genes in male and female adult flies. The transcriptome was sequenced by RNA-seq. The graph represents the normalized tag count for each gene (log2) in either males or females. (B) Poor correlation between expression and RIP. The normalized tag count (log2) obtained for each UNR target in the UNR immunoprecipitates and the total RNA from each sex were compared. (Upper right corner of each plot) Pearson correlation coefficient.
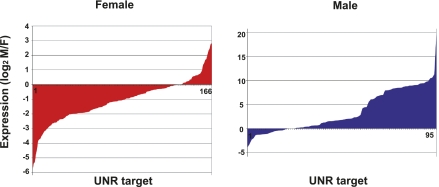
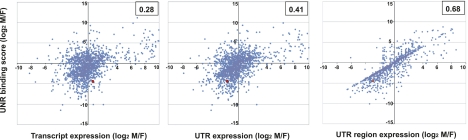
We next compared the relative expression levels of the UNR targets in both sexes with the UNR binding scores defined from the enrichment P-values as indicated before. In case of multiple transcripts annotated for the same gene, to facilitate a correlation we chose the annotated transcript whose expression ratio was closest to the UNR binding score ratio. Of the 1746 UNR targets, 1661 were reliably annotated (i.e., contained at least one transcript with fully annotated UTRs in the FlyBase 5.12 version available at the UCSC Genome Browser, mapped on the BDGP R5/dm3 assembly of the genome) and were used in the analysis. Of these 1661 transcripts, 15.7% (95 targets in males and 166 targets in females) showed the highest enrichment in the corresponding UNR immunoprecipitates (P-value = 0). We classified these targets according to their expression levels in males and females and found that a sizeable proportion were also highly expressed in the corresponding sex, suggesting that many of these targets represent sex-specific transcripts bound by UNR (Fig. 6). A comparison between the UNR binding score ratios and the relative sex-specific expression ratios of the remaining 84.3% transcripts (1400 targets) revealed a poor correlation (Pearson correlation 0.28) (Fig. 7, left panel). Our positive control msl2 (red dot) followed this behavior, because it is bound by UNR in females despite being expressed somewhat better in males (Abaza et al. 2006).
FIGURE 6.
Expression of targets with the strongest UNR binding bias in either sex. UNR targets showing a P-value = 0 in either sex were classified according to their relative sex-specific expression.
FIGURE 7.
UNR binding correlates with sex-specific UTR processing. (Left panel) Representation of the relative UNR binding scores for 1400 UNR gene targets against the expression ratio (normalized tag count) of the transcript closest to the UNR binding ratio. (Middle panel) The relative UNR binding scores were plotted against the expression ratio of the annotated alternative UTR isoform closest to the UNR binding ratio. (Right panel) The UTRs of UNR targets were scanned in a base-by-base manner (see main text for explanation), and the value of the male-to-female coverage ratio closest to the UNR binding ratio was plotted. The position of msl2 (red dot) is highlighted in all graphs. (Upper right corner of each plot) Pearson correlation coefficient.
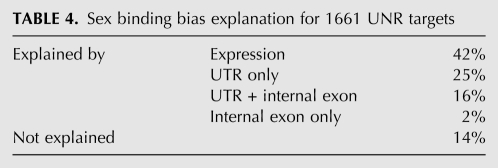
Differential binding of UNR to a target was considered to be explained by expression if the UNR binding score and expression log2 ratios differed by <1.5. Using this threshold, the UNR binding bias could be explained by sex-specific expression of the target in 42% of the 1661 cases (Table 4). These data indicate that the relative abundance of targets in both sexes is not the sole explanation for the UNR binding bias.
TABLE 4.
Sex binding bias explanation for 1661 UNR targets
Generation of alternative UTRs contributes to sex-specific binding of UNR to its targets
UNR has been shown to regulate post-transcriptional gene expression by binding to the 5′ or 3′ UTRs of its targets (Mihailovich et al. 2010). Thus, a possibility to explain the UNR binding bias is that the UTRs of UNR target transcripts are different in males and females. To determine whether UNR binding agrees with sex-specific UTR usage, we computed the number of sequence tags falling on each individual UTR for all annotated transcript isoforms of the 1400 UNR targets indicated above. We next selected, for each target, the UTR whose relative male-to-female enrichment was closer to the UNR binding ratio and plotted its relative expression against the UNR binding score (Fig. 7, middle panel). The results show a modest improvement of the correlation between UNR binding and expression of individual UTR isoforms (Pearson correlation 0.41). msl2 moves to the correlation quarter in this graph (red dot), as expected from the fact that the msl2 transcript recognized by UNR contains a facultative intron in the 5′ UTR that is retained specifically in females (Graindorge et al. 2011).
Surprisingly, only 50% of the UNR target genes (703 targets) had more than one annotated transcript in FlyBase. Recent work has revealed that the fraction of genes with alternatively spliced transcripts is underestimated by a factor of 3 on the annotated portion of the Drosophila genome (Graveley et al. 2010; The modENCODE Consortium 2010). The modENCODE Consortium has detected 14,862 genes and 36,274 transcripts, while the current FlyBase annotation (5.12) available on the UCSC Genome Browser, used in our analysis, includes 13,653 genes and 20,955 transcripts. Thus, incomplete gene annotation may have contributed to the observed modest correlation between UNR binding and UTR expression. To overcome this difficulty and to assess whether the differential binding could also be due to extended 5′ or 3′ UTRs or internal alternatively processed regions, we adopted the following strategy: For each annotated transcript, we computed the normalized base-by-base coverage obtained by RNA-seq in both males and females. In this way, for each annotated exon of target genes, we obtained a base-by-base plot of male versus female usage. This strategy is similar to that used for ChIP-seq, in which a base-pair by base-pair coverage enrichment is computed by comparing two samples (Park 2009). We then selected the regions of target transcripts that had been sequenced at least three times in either sex (i.e., 3× coverage) and contained a position whose coverage ratio was closest to the UNR binding ratio. A plot of the coverage ratio for UTR regions against the UNR binding ratio shows a dramatically improved correlation (Pearson coefficient 0.68) (Fig. 7, right panel). msl2 moves further into the correlation line in this graph, consistent with the fact that the alternatively processed intron represents a relatively small region of the 5′ UTR. These results suggest that alternative UTR processing contributes to the sex-specific binding pattern displayed by UNR.
Differential binding of UNR to a transcript was considered to be explained by a sex-specific UTR region if the UNR binding score and the UTR region expression log2 ratios differed by <1.5. Altogether, while differential UNR binding was explained in 42% of the cases by sex-specific expression of the target, the correlation increased to 83% when sex-specific UTR usage was taken into account. Moreover, when a variation with respect to the overall expression ratio was observed within an internal coding exon, it usually was matched by a similar variation in the coverage ratio within one of the UTRs of the transcript, suggesting that UTR and coding exon processing are tightly linked (Table 4). Within the group of genes that were explained by UTR usage, about the same proportion was explained by the 5′ or the 3′ UTR (42% vs. 58%, respectively), suggesting that the UNR binding site is equally likely to be present at either UTR. This fits well with the known roles of UNR in translation and stability, because UNR has been shown to function as an IRES trans-acting factor by binding to the 5′ UTR, and as a repressor of translation by binding to the 3′ UTR (for review, see Mihailovich et al. 2010). We used Weeder (Pavesi et al. 2004) to search for over-represented motifs on the 5′ UTRs of targets explained by this untranslated region, using the Drosophila mRNA oligo frequencies as background. The highest-scoring motifs consisted of AAAAUAGU, a purine-rich sequence somewhat similar to the human UNR-binding site identified by SELEX (Triqueneaux et al. 1999), and UUUg/uUUUU, a sequence that resembles a SXL-binding site (Singh et al. 2000). A similar search using the 3′ UTRs was difficult because the highest-scoring motifs were variations of the polyadenylation signal.
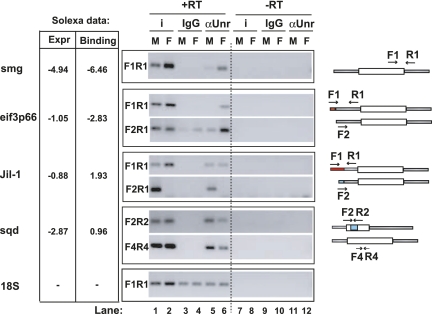
To confirm that UNR binds to specific transcript isoforms in males and females, we performed new RIP experiments and assessed the presence of specific UTRs by RT-PCR on a subset of UNR targets (Fig. 8). To test if UNR binding to a target was linked to the presence of a sex-specific processing event, we designed oligonucleotides that either distinguished mRNAs containing the sex-specific region or detected a common region present in male and female transcripts (Fig. 8, see schematic representation on the right). In all cases, the oligos showed no background in reactions lacking reverse transcriptase (Fig. 8, lanes 7–12). Smaug (smg) binds preferentially to UNR in females, and our in silico analysis indicated that this binding can be explained by sex-specific expression of the transcript (Fig. 8, see Solexa data on the left). Accordingly, the UNR association of smg mirrors its relative abundance in males and females (Fig. 8, first panel, cf. lanes 1,2 with 5,6). Contrary to smg, the strong female binding bias of eIF3p66 cannot be explained by its relative abundance in both sexes but correlates with the presence of an alternative 5′-transcription start site in the UNR-bound transcript (Fig. 8, second panel). Another transcript, Jil-1, binds to UNR preferentially in males despite being more abundant in females. Indeed, RT-qPCR using non-sex-specific primers indicated that Jil-1 is about three times more abundant in females (data not shown). Similar to eIF3p66, Jil-1 binding correlates with the presence of a male-specific transcription start site in the UNR-bound transcript (Fig. 8, third panel). Finally, squid (sqd) was shown to be sex-specifically regulated by alternative splicing: Its second exon was retained preferentially in males, resulting in the production of a truncated protein (Hartmann et al. 2011). We could detect a small difference in the presence of this exon by qPCR when the input samples from males and females were compared, although this difference was not visible by RT-PCR (Fig. 8, fourth panel, F2R2, lanes 1,2; data not shown). However, UNR clearly bound to this transcript preferentially in males (Fig. 8, fourth panel, lanes 5,6), suggesting that alternative splicing contributes to the sex-specific binding bias of sqd. Alternatively, analogous to the recruitment of UNR to msl2 mRNA by SXL, a male-specific factor could promote UNR binding to sqd. UNR could also be preferentially associated with sequences exclusive of another male-specific processing event on the F2R2 transcript. Altogether, these data indicate that alternative splicing and alternative promoter usage contribute to sex-specific RNA binding by UNR.
FIGURE 8.
UNR binds to specific transcript isoforms in males and females. UNR was immunoprecipitated from male (M) or female (F) adult extracts, and the presence of specific RNA isoforms in the pellet was evaluated by RT-PCR. Immunoprecipitations with rabbit IgG, and parallel amplifications without reverse transcriptase (−RT), were carried as negative controls. The amplification of the 18S rRNA is shown as an indication of background. (Left panel) Relative expression and binding data obtained by Solexa deep sequencing [log2(M/F)]. A schematic representation of the RNA targets and the oligos used for amplification is shown on the right. Segments in red and blue indicate alternative annotated regions that we find over-represented in females or males, respectively, as deduced from our Solexa data.
DISCUSSION
Multiple transcripts can be generated from a single gene by differential promoter usage during transcription or by a variety of post-transcriptional mechanisms. Here we show that the conserved RNA-binding protein UNR takes advantage of this diversity to select transcripts in a sex-specific fashion.
Microarray and deep-sequencing analysis suggest that UNR binds to a sizeable set of target transcripts (Fig. 1). This is perhaps not surprising, given that UNR contains five cold-shock domains, each of which could bind to RNA independently (Abaza and Gebauer 2008). The number of targets identified for other RBPs in Drosophila and other organisms ranges from a few tenths to several thousand (Halbeisen et al. 2008). This number is specially high for proteins with multiple RNA-binding domains even considering a single cell type (Hafner et al. 2010). In comparison, the number of targets identified here for UNR in whole adult flies seems nonsaturating. Indeed, the small overlap between the microarray and deep-sequencing data suggests that the number of UNR targets is underestimated. The fact that GO analysis showed significant enrichment for similar categories when targets from RIP-Chip and RIP-seq were analyzed also supports this conclusion. Other factors can also contribute to the limited overlap between the two sets of data, including the differences inherent to each technique and the use of a different starting material for microarray and deep sequencing (cytoplasmic vs. total extracts, respectively). Large differences have been reported in a recent microarray analysis comparing whole-cell versus cytoplasmic RNA (Trask et al. 2009). These investigators found that up to 31% of the differentially expressed genes detected using cytoplasmic RNA can be missed by using total RNA and that the contribution of the nuclear RNA to total RNA variation is far from negligible.
In addition, an incomplete overlap between identified genes was detected when the exact same samples were compared by deep-sequencing and different microarray platforms (Hoen et al. 2008). The overlap could be as small as 15% when differential gene expression was interrogated. Furthermore, techniques with similar principles such as PAR-CLIP and CLIP showed only 58% coincidence in the targets identified for IGF2BP1 (Hafner et al. 2010). These data suggest that different technologies and experimental setups can reveal specific subsets of targets for a given RBP.
Examination of the UNR binding bias indicates that UNR binds to RNA in a sex-specific fashion (Fig. 3). Precedence for sex-specific binding existed from the known UNR targets msl2 and roX (Abaza et al. 2006; Duncan et al. 2006; Patalano et al. 2009). In the case of msl2, UNR is recruited to the 3′ UTR of the transcript by the sex-specific protein SXL (Abaza et al. 2006). Although recruitment by sex-specific binding partners could contribute to the UNR binding profile, this is unlikely to be the sole explanation for the extensive sex-specific binding bias displayed by this protein. Rather, other features such as sex-specific expression and alternative processing of targets can explain most of the UNR binding profile (Fig. 7; Table 4).
A large degree of sex-specific diversity exists at the molecular level (Ellegren and Parsch 2007). In adults of Drosophila, a large proportion of the genes show sex-biased expression (Fig. 5; Parisi et al. 2004; Ayroles et al. 2009; Graveley et al. 2010). Parisi et al. (2004) attributed sex-biased expression to the gonads, where male gonads contribute more sex-specific genes than female gonads. Comparison with those data indicates that only 19% of the UNR target genes are expected to show gonad-biased expression, suggesting that the ovary and testis have little influence on the overall UNR binding pattern (data not shown).
UNR binding strongly correlates with the presence of sex-specific UTR segments (Figs. 7, 8). From the data presented in Figure 7 and from the visual examination of a few random cases, often these segments reflect putative non-annotated processing events. Recent RNA-seq data from the Graveley laboratory and the modENCODE Consortium indicate that the number of transcripts on the annotated portion of the Drosophila genome is heavily underestimated (Graveley et al. 2010; The modENCODE Consortium 2010). Work from these investigators has nearly tripled the fraction of genes with alternatively spliced transcripts and increased the number of edited genes by one order of magnitude. In addition, Hartmann et al. (2011) detected sex-specific changes in 26% of splice junctions that were annotated as constitutive. These data indicate that the sex-specific transcriptome is more complex than previously anticipated. We hypothesize that UNR takes advantage of this complexity to enforce sex-specific gene expression. Furthermore, UNR could contribute to the generation of sex-specific complexity by influencing alternative processing or mRNA stability.
UTRs provide a wealthy source of regulatory diversity. In germ cells of Caenorhabditis elegans, promoters are active by default while post-transcriptional regulation via 3′ UTRs confers cell-type specific expression, primarily by providing recognition sites for inhibitory RBPs (Merritt et al. 2008). Alternative UTR generation by transcriptional and post-transcriptional mechanisms may contribute to sex-specific regulation by UNR and other RBPs. For example, it has been shown that sex-specific qualitative and quantitative differences in the expression of RBPs contribute to shape the sex-specific transcriptome (Hartmann et al. 2011). One such protein is hrp40/squid, which is one of the UNR targets identified in our study. A comparison between a list of 260 RBPs in Drosophila and the list of UNR targets reported here indicates that 19% of the RBPs are encoded by UNR targets (Supplemental Table SIII). These results suggest that UNR contributes to sex-specific gene expression in part by regulating the expression of other RBPs, and highlight the existence of extensive cross-regulation between RBP networks.
MATERIALS AND METHODS
Extract preparation
Adult male and female flies of mixed ages were manually separated, snap-frozen, and ground in liquid nitrogen. Cytoplasmic extract preparations were obtained as previously described (Wilhelm et al. 2000). Total extract preparations were obtained by resuspending the fly powder in one volume of hypotonic buffer (HB) (10 mM HEPES-KOH at pH 7.4; 0.5 mM DTT; 20 U/mL RNasin; 1× Complete Protease inhibitor cocktail; 5 μg/mL pepstatin, leupeptin, and aprotinin) and homogenizing the suspension with a DIAX 900 homogenizer (Heidolph). One additional volume of HB was added, and the mix was incubated for 10 min on ice and further homogenized for 1 min. The homogenate was centrifuged for 20 min at 13 Krpm in the microfuge, and the supernatant was recovered. Salts were added to 150 mM KCl and 5 mM Mg(Cl)2, and the extract was used for immunoprecipitation.
RNA-immunoprecipitation (RIP)
Immunoprecipitation of UNR was performed from saturating amounts of either cytoplasmic or total Drosophila adult extracts using purified IgG from serum generated against a C-terminal fragment of UNR (Abaza et al. 2006). Immunoprecipitation from cytoplasmic extracts was performed as previously described (Abaza et al. 2006). Immunoprecipitation from total extracts was performed using protein A–magnetic beads (Invitrogen). After extensive washing with 1xNET (50 mM TRIS-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40), beads were resuspended in elution buffer (50 mM Tris-HCl at pH 7.4, 5 mM EDTA, 10 mM DTT, 1% SDS) and treated with 40 μg of proteinase K. RNA was then extracted from the supernatant using TRIzol, precipitated, and treated with DNase (TURBO-DNA free kit; Ambion). A similar protocol was used to obtain RNA for RT-qPCR validation studies, except that the RNA was obtained by direct extraction from the magnetic beads using TRIzol. The amount of RNA was assessed by NanoDrop measurement. The quality of the RNA used in RIP-Chip and RIP-seq experiments was assessed in the Bioanalyzer. The efficiency of immunoprecipitation and the integrity of UNR were monitored by Western blot.
Microarray
RNA obtained from RIP of cytoplasmic extracts was used to generate probes for microarray hybridization using the pico version of the ExpressArt mRNA Amplification Kit (Artus) following the manufacturer's instructions. Identical amounts of male and female probes were hybridized against each other in custom-made microarrays printed at the Transcriptome Platform of Barcelona University (Beltran et al. 2007). These microarrays contain the Drosophila Genome Oligo Set version 1.1 (Operon Biotechnologies, Inc.), a collection of 14,593 70-mer oligos representing 13,577 Drosophila genes, in addition to negative and spike-in controls (for a complete description, see Beltran et al. 2007). Microarrays were hybridized at the CRG Genomics Facility following the Corning protocol for UltraGaps slides and scanned using the Agilent scanner, and the images were processed with GenePix image analysis software v6.0. All GenePix Results (GPR) data files were analyzed with R and the Bioconductor modules for the analysis of high-throughput data (Gentleman et al. 2004; Smyth 2004). Spots not fulfilling the quality thresholds were eliminated from the analysis, and the data were background-corrected with the normexp method (Ritchie et al. 2007) and normalized using OLIN (Futschik and Crompton 2005). The list of UNR targets was obtained by considering only genes detected in at least four arrays and falling in the upper third of intensity distribution. UNR sex-specific targets were estimated by computing differential expression analysis using the Bioconductor limma R package (Smyth 2004) with an FDR corrected P-value of <0.1.
Solexa
Sequencing of UNR targets by Solexa was performed at the CRG Genomics Facility using the Illumina NlaIII protocol. Analysis of the raw data indicated a similar number of total reads per sample after eliminating the linker sequences (10 × 106 to 15 × 106 reads), indicating a comparable sample size. Tags were then mapped against the Drosophila genome (BDGP release 5) available at the UCSC Genome Browser using the seqmap tool (Jiang and Wong 2008), allowing at most two mismatches per tag and considering only tags mapping unambiguously on the genome for further analysis. Details on the number of tags uniquely mapped to the Drosophila genome are indicated in Table 1. All tags falling in the same strand within exons of Drosophila transcripts were considered. To compare fold ratios between different experiments, the number of tags per gene computed for each sample was normalized for the total number of tags produced in each experiment. Furthermore, a P-value was assigned to each tag count based on a 2 × 2 χ2 test similar to that previously used for SAGE experiments (Michiels et al. 1999). This method considers simultaneously the enrichment in the IP versus the IgG control and the overall number of tags in both samples. The two IP replicates per sex were considered separately for the P-value calculation. A strict Bonferroni-corrected P-value threshold of 5 × 10−6 was established in order to have an FDR of 0.05. A target was considered positive if the P-value associated with the tag count was lower than this threshold and if the fold ratio IP/IgG ≥ 2.
Sequencing of the Drosophila transcriptome was performed from total RNA of adult male and female flies following the NlaIII and RNA-seq protocols. A comparison of both protocols for genes producing at least 20 tags in each sex shows a Pearson correlation of 0.9, indicating that they are comparable (Supplemental Fig. S1). Because the RNA-seq protocol produces more tags, we used the results of RNA-seq throughout this work. Moreover, RNA-seq permits us to quantify not only the overall level of expression of genes, but also of alternative transcript isoforms, thus giving a more accurate view of the transcriptional landscape.
RT-qPCR
Similar amounts of male and female extracts (confirmed by Western blot against tubulin and UNR) were either mixed or used separately for the immunoprecipitation experiments of Figures 2 and 3C, respectively. Parallel experiments with rabbit IgG were performed as negative controls. The same volumes of immunoprecipitated material were reverse-transcribed with SuperScript II (Invitrogen) using a mixture of oligo(T) and random hexamers. Quantitative PCR was performed in a LightCycler 480 (Roche) using LightCycler 480 SYBR Green I Master Mix. Values were corrected for the 28S rRNA, which was amplified as a background control. For each target, the anti-UNR value was expressed relative to the IgG control and normalized to 28S rRNA enrichment.
DATA DEPOSITION
The microarray data set has been submitted to GEO under ID number GSE30963. All Solexa sequencing data have been deposited in GEO under accession number GSE33178.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Britta Hartmann and Belén Miñana for advice, and Juan Valcárcel for suggestions and critical reading of this manuscript. Deep-sequencing and microarray hybridizations were performed at the CRG Genomics Facility. M.M. was supported by a Juan de la Cierva contract from the Spanish Ministry of Science and Innovation (MICINN). L.W. was supported by the National Research Fund, Luxembourg, and cofunded under the Marie Curie Actions of European Commission (FP7-COFUND). C.M. was supported by a fellowship from La Caixa. G.P. and F.Z. were supported by the Italian Ministry of Education and Scientific Research (MIUR) within the FIRB project “Laboratorio Internazionale di Bioinformatica” (LIBI). Work in the F. Gebauer laboratory is supported by grants BFU2009-08243 and Consolider CSD2009-00080 from MICINN.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029603.111.
REFERENCES
- Abaza I, Gebauer F 2008. Functional domains of Drosophila UNR in translational control. RNA 14: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaza I, Coll O, Patalano S, Gebauer F 2006. Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes Dev 20: 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RR, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran S, Angulo M, Pignatelli M, Serras F, Corominas M 2007. Functional dissection of the ash2 and ash1 transcriptomes provides insights into the transcriptional basis of wing phenotypes and reveals conserved protein interactions. Genome Biol 8: R67 doi: 10.1186/gb-2007-8-4-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M, Hentze MW 2006. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: Translational repression for dosage compensation. Genes Dev 20: 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z 2009. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet 8: 689–698 [DOI] [PubMed] [Google Scholar]
- Futschik ME, Crompton T 2005. OLIN: Optimized normalization, visualization and quality testing of two-channel microarray data. Bioinformatics 21: 1724–1726 [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI 2009. Drosophila dosage compensation: A complex voyage to the X chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge A, Militti C, Gebauer F 2011. Post-transcriptional control of X-chromosome dosage compensation. Wiley Interdiscip Rev RNA 2: 534–545 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2010. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP 2008. Post-transcriptional gene regulation: From genome-wide studies to principles. Cell Mol Life Sci 65: 798–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B, Castelo R, Miñana B, Peden E, Blanchette M, Rio DC, Singh R, Valcárcel J 2011. Distinct regulatory programs establish widespread sex-specific alternative splicing in Drosophila melanogaster. RNA 17: 453–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT 2008. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36: e141 doi: 10.1093/nar/gkn705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH 2005. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA 11: 1303–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jiang H, Wong WH 2008. SeqMap: Mapping massive amount of oligonucleotides to the genome. Bioinformatics 24: 2395–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels EM, Oussoren E, Van Groenigen M, Pauws E, Bossuyt PM, Voûte PA, Baas F 1999. Genes differentially expressed in medulloblastoma and fetal brain. Physiol Genomics 1: 83–91 [DOI] [PubMed] [Google Scholar]
- Mihailovich M, Militti C, Gabaldon T, Gebauer F 2010. Eukaryotic cold shock domain proteins: Highly versatile regulators of gene expression. Bioessays 32: 109–118 [DOI] [PubMed] [Google Scholar]
- The modENCODE consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD 2009. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip Rev Syst Biol Med 2: 162–180 [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lü J, Doctolero M, Vainer M, Chan C, Malley J, et al. 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol 5: R40 doi: 10.1186/gb-2004-5-6-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ 2009. ChIP-Seq: Advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patalano S, Mihailovich M, Belacortu Y, Paricio N, Gebauer F 2009. Dual sex-specific functions of Drosophila Upstream of N-ras in the control of X chromosome dosage compensation. Development 136: 689–698 [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G 2004. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res 32: W199–W203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Singh R, Banerjee H, Green MR 2000. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor Sex-lethal. RNA 6: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T 2011. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6: e21800 doi: 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask HW, Cowper-Sal-lari R, Sartor MA, Gui J, Heath CV, Renuka J, Higgins AJ, Andrews P, Korc M, Moore JH, et al. 2009. Microarray analysis of cytoplasmic versus whole cell RNA reveals a considerable number of missed and false positive mRNAs. RNA 15: 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triqueneaux G, Velten M, Franzon P, Dautry F, Jacquemin-Sablon H 1999. RNA binding specificity of Unr, a protein with five cold shock domains. Nucleic Acids Res 27: 1926–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD 2000. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol 148: 427–440 [DOI] [PMC free article] [PubMed] [Google Scholar]