Abstract
Viruses may very well be the most abundant biological entities on the planet. Yet neither metagenomic studies nor classical phage isolation techniques have shed much light on the identity of the hosts of most viruses. We used a microfluidic digital polymerase chain reaction (PCR) approach to physically link single bacterial cells harvested from a natural environment with a viral marker gene. When we implemented this technique on the microbial community residing in the termite hindgut, we found genus-wide infection patterns displaying remarkable intragenus selectivity. Viral marker allelic diversity revealed restricted mixing of alleles between hosts, indicating limited lateral gene transfer of these alleles despite host proximity. Our approach does not require culturing hosts or viruses and provides a method for examining virus-bacterium interactions in many environments.
Despite the pervasiveness of bacteriophages in nature and their postulated impact on diverse ecosystems (1), we have a poor grasp of the biology of these viruses and their host specificity in the wild. Although substantial progress has been made with certain host-virus systems such as cyanophages (2–5), this is the exception rather than the rule. Conventional plaque assays used to isolate environmental viruses are not applicable to >99% of microbes in nature because the vast preponderance of the microbial diversity on Earth has yet to be cultured in vitro (6). Given the magnitude of the problem, the development of high-throughput, massively parallel sequencing approaches that do not rely on cultivation to identify specific virus-host relations are required. Although metagenomics has revolutionized our understanding of viral diversity on Earth (7–9), that approach has as yet done little to shed light on the nature of specific viral-host interactions, except in restricted cases (10).
Recent advances in microfluidic technology have enabled the isolation and analysis of single cells from nature (11–13). We present an alternative to the classical phage enrichment technique where we use an uncultured virus to capture its hosts from the environment with a microfluidic polymerase chain reaction (PCR) approach called digital multiplex PCR (12, 14). To this end, microbial cells were harvested directly from the environment, diluted, and loaded onto a digital PCR array panel containing 765 PCR chambers operating at single-molecule sensitivity. Samples were diluted such that the majority of chambers were ideally either empty or contained a single bacterium (Fig. 1), achieving a Poisson distribution (15). Because there is no universally conserved gene in viruses (7, 16), we designed degenerate primers (17) to target a subgroup of diverse phagelike elements (18). Concurrently, the small subunit ribosomal RNA (SSU rRNA) gene encoded by each bacterial cell was amplified by using universal “all bacterial” primers (see fig. S1 for experimental design). Possible genuine host-virus associations detectable by this assay are depicted in Fig. 1C. Free phages may also colocalize with hosts; however, these events are not expected to lead to statistically significant colocalizations because of the random nature of these associations (19).
Fig. 1.
End-point fluorescence measured in a panel of a microfluidic digital PCR array. (A) The measured end-point fluorescence from the rRNA channel (right half of each chamber, with the left half masked) and the terminase channel (left half of each chamber, with the right half masked) in a microfluidic array panel. Each panel in the array (1 of 12) consists of 765 reaction chambers 150 µm by 150 µm by 270 µm (6 nl). Retrieved colocalizations are outlined in orange, and positive rRNA chambers randomly selected for retrieval are outlined in gray. FA indicates false alarm (a probable terminase primer-dimer). (B) Normalized amplification curves of all chambers in (A) after linear derivative baseline correction (red, viral; green, rRNA). (C) Specific physical associations between a bacterial cell and the viral marker gene resulting in colocalization include, for example, an attached or assembling virion, injected DNA, an integrated prophage, or a plasmid containing the viral marker gene.
Hunting for phages in the termite hindgut
The system we chose to investigate was the termite hindgut. This microliter-in-scale environment contains ~107 prokaryotic cells per µl (20) with over 250 different species of bacteria (21), making it ideally suited to explore many potential, diverse phage-host interactions. To find a viral marker gene relevant to such an environment, we examined the more abundant candidate viral marker genes present in the sequenced metagenome from a hindgut of a higher termite from Costa Rica collected in 2005 (22) [table S1; search algorithm described in (18)]. We then checked whether any of these viral genes had homologous counterparts in the sequenced genomes of two spirochetes isolated in 1997 from a laboratory colony of genetically and geographically distant termites originally collected in 1986 from Northern California (23, 24). We identified two such genes encoding a large terminase subunit protein (homologous to the T4-associated pfam03237 Terminase_6) and a portal protein (homologous to pfam04860 Phage_portal) exhibiting about 70 to 78% amino acid identity to their closest homologs in the higher termite gut metagenome (table S2). This finding is unexpected given that typically, across biology, portal proteins and terminase proteins from different phages exhibit little overall sequence similarity (25–28). Further analysis revealed that the spirochete viral genes were part of a larger prophage-like element, with the majority of recognizable genes most closely related to Siphoviridae phage genes (19). The association of these genes with prophage-like elements is consistent with the fact that both the Terminase_6 pfam and the Phage_portal pfam describe proteins in known lysogenic and lytic phages.
As a viral marker gene for this prophage-like element, we chose the large terminase subunit gene. This gene is a component of the DNA packaging and cleaving mechanism present in numerous double-stranded DNA phages (26) and is considered to be a signature of phages (29). We consequently designed degenerate primers on the basis of the collection of 50 metagenome and treponeme-isolate alleles of this gene. The ~820–base pair (bp) amplicon spanned by these primers covered about two-thirds of this gene and about 77% of the predicted N-terminal domain containing the conserved adenosine triphosphatase (ATPase) center (26, 30), the “engine” of this DNA packaging motor (31) (see alignments in figs. S2 and S3). Testing these primers against the RefSeq viral database (32) did not yield any hits (fig. S2). Indeed, the closest homolog of this gene in the RefSeq viral database displayed only 25% amino acid identity (table S2). Thus, although this terminase gene was associated with the Terminase_6 pfam, the termite-related alleles appear to be part of a novel assemblage of terminase genes in this environment and not closely related to previously sequenced phages (fig. S2).
Given that terminase genes of different phages often exhibit less sequence similarity (see above), the fact that we found such closely related terminase genes from such distantly related termites collected from well-separated geographical locations (California and Costa Rica) and from specimens collected almost two decades apart led us to speculate that this family of viral genes and prophage-like elements might be ubiquitous in termites. Indeed, to date we have identified close homologs of the large terminase subunit gene in the gut communities of nine termite species belonging to four families collected from five different geographical locations. We therefore wished to identify the bacterial hosts associated with this viral marker gene. To this end, we collected representatives of a third previously unexamined termite family (Rhinotermitidae; Reticulitermes hesperus, from a third geographical location in Southern California) over a span of 6 months (table S3). We then performed seven independent experiments, where in each case the hindgut contents of three worker termites were pooled, diluted, and loaded onto a digital PCR array, screening in total ~3000 individual hindgut particles (i.e., individual cells or possibly clumps of cells positive for the SSU rRNA gene).
Identification of previously unknown uncultured bacterial hosts
Of the 41 retrieved colocalizations, 28 were associated with just four phylotypes designated phage hosts I, II, III, and IV (compare Fig. 2, Table 1, and the phylogenetic analysis in fig. S4 and tables S4 and S5). Statistically, the reproducible coamplifications were significant and cannot be explained by random colocalization of two unassociated genes (Table 1). Furthermore, these associations were independently reproduced in specimens from different colonies collected 6 months apart (Fig. 2), indicative that relations between specific host bacteria and viral markers were being revealed.
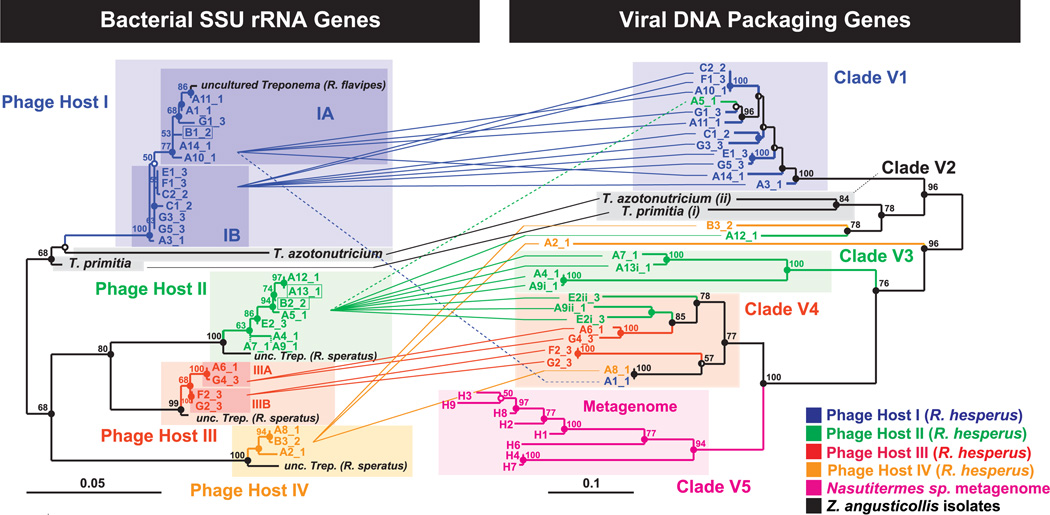
Fig. 2.
Phylogenetic relationship between cultured and uncultured bacterial host rRNA genes and their associated viral DNA packaging genes. (Left) Maximum likelihood (ML) tree of 898 unambiguous nucleotides of the SSU rRNA gene of ribotypes that repeatedly colocalized with the terminase gene, including the two isolated spirochetes Treponema primitia and Treponema azotonutricium. Shorter sequences (A7, 780 bp, and A9, 806 bp) were added by parsimony (dashed branches). (Right) ML tree of 705 unambiguous nucleotides of the large terminase subunit gene. Connecting lines represent colocalized pairs, revealing restricted mixing of terminase alleles between different bacterial hosts. For association of three additional recombinant sequences (boxed on the left), see fig. S5. Statistically, we estimate that an average of 0.6 colocalizations are false [~2% error (19)]. The sequence error rates (40) for the rRNA and terminase genes were measured to be 0 (n = 8) and <0.6 ± 0.3% SD (n = 9), respectively (18). Alleles are named by array (A to G) and retrieval index followed by an underscore and the colony number (colony 1 being sampled 6 months before colonies 2 and 3). Lowercase roman numerals indicate multiple terminases per chromosome. Scale bars represent substitutions per alignment. For interpretation of node support, refer to (18), and for accession numbers, table S10.
Table 1.
Statistics of repeatedly colocalized SSU rRNA genes. The number of repeated colocalizations and occurrences in the reference library are based on a DOTUR analysis (tables S4 and S5). Reference library frequencies are roughly one-third of the colocalization frequencies, indicating that sampling was unbiased. The statistical test to determine the P value is explained in (19).
| Host | No. of repeated colocalizations (n = 41) |
Occurrence in reference library (n = 118) |
P value (one-tailed, n = 41) |
|---|---|---|---|
| Host I | 13 | 5 | 5.4 × 10−18 |
| Host II | 8 | 2 | 7.6 × 10−13 |
| Host III | 4 | 1 | 5.7 × 10−7 |
| Host IV | 3 | 1 | 3.8 × 10−5 |
All four of the phylotypes were members of the spirochetal genus Treponema and exhibited substantial diversity within this genus (table S4). No reproducible or statistically robust associations involving other bacteria were observed. The terminase alleles that associated with these cells shared ≥69.8% identity (average 81.9 ± 8.3% standard deviation, SD) (33) and were divergent from other currently known terminases (fig. S2), suggesting that the primer set amplifies elements exclusively found associated with termite gut treponemes. Analysis of the retrieved terminase gene sequences revealed that they are under substantial negative selection pressure with ω = β/α = 0.079, where ω is the relative rate of nonsynonymous, β, and synonymous, α, substitutions (18) (see table S6 for additional estimates for individual hosts). Furthermore, none of the terminase sequences in Fig. 2 appeared to encode either errant stop codons or obvious frame shift mutations, and functional motifs appeared to be conserved (fig. S2). Together, the sequence data suggest that these genes have been active in recent evolutionary history and are not degenerating pseudogenes (19).
Because the viral marker gene was present in hosts spanning a swath of species of termite gut treponemes, we were interested to see whether this viral marker exhibited any selectivity within this genus. The relative frequency of free-living Treponema phylotypes was determined by randomly sampling chambers positive for the rRNA gene (18) (Fig. 3 and fig. S4). We found that hosts I through IV were relatively infrequent, comprising 1.3% to 6.4% of the sampled Treponema cells (Table 1) and collectively about 9.8% of the sampled bacterial cells (correcting for reagent contaminants). Interestingly, the three most abundant Treponema phylotypes in the survey, constituting ~30, 10, and 9% of the free-swimming spirochetal cells [Reticulitermes environmental phylotypes (REPs) 1, 2, and 3 in Fig. 3; see also fig. S4 and table S5], were never co-retrieved with the viral marker gene to the extent that this target was spanned by our degenerate primers. Given that the degenerate core region (17) of each primer targets residues that were strictly conserved in gut microbes of highly divergent termite specimens (fig. S2) and that these primers successfully amplified this gene from the guts of many different termite species (see above), it appears that these strains are most likely either insensitive to this virus or that only a small percentage are infected (19). Therefore, we conclude that ~50% of the free-swimming spirochetal cells in the gut were likely not infected with an element encoding the targeted viral marker gene, whereas ~12%were potentially infected hosts (Fig. 3).
Fig. 3.
Rank abundance curve of free-living Treponema spirochetes in R. hesperus termites identifying putative phage hosts. A library of 118 random chambers positive for the rRNA gene were retrieved, postamplified, and sequenced. Of these, n = 78 were related to the Treponema genus, corresponding to 28 different phylotypes based on an operational taxonomical unit, OTU, cut-off set by DOTUR (41) at 3.1%. We show these 28 phylotypes, designated as REPs, ordered by their abundance. Phylotype abundance is expected to reflect true relative abundances in the gut because single-cell amplification is not susceptible to primer bias or rRNA copy number bias. Phylotypes identified as phage hosts are marked by red bars (with the highly clonal marker associated with host I depicted by green viruses and the divergent marker associated with host II depicted by colored viruses). The most abundant free-living Treponema in the gut—REPs 1, 2, and 3 (blue bars)—were not associated with the viral marker. Remaining bars are gray. Error bars are estimated by the binomial SD. See table S5 for OTU assignment. Note that the isolated spirochetes were not spanned by these REPs (fig. S4).
Phage-host cophylogeny
To elucidate the evolutionary relations between the terminase alleles and their hosts, we examined the phylogeny of the terminase genes associated with each bacterial host. Terminase alleles from R. hesperus formed separate clades from the clades of the two other termite species investigated in this study (clades V2 and V5 in Fig. 2).Within R. hesperus, different bacterial hosts exhibited different patterns of viral allelic diversity. Terminase sequences associated with host I, for example, were highly clonal, with 11 out of 13 terminase alleles sharing 96.7 ± 1.7% SD identity (n = 11, clade V1) (33). Conversely, terminase alleles associated with host II displayed marked diversity (79.1 ± 6.2% identity, n = 11) (33), deep branches, and divergent multiple alleles per bacterium for three out of eight repetitions (with 15 to 31% divergence). The unique features of the terminase alleles associated with host II compared with host I may reflect a more ancient infection or possibly an infection by a phage replicating with a lower fidelity. Alternatively, host II may be a more sensitive bacterial host susceptible to a wider range of phages. Overall, phage terminase alleles associated with different bacterial hosts were significantly divergent with only three exceptions (table S7).
The tandem trees in Fig. 2 reveal multiple possible relations between bacterial hosts and terminase alleles: Whereas host I was associated almost exclusively with a single terminase clade (V1), host II was associated with multiple terminase clades (primarily V3 and V4). Conversely, terminase clade V1 was associated almost exclusively with host I, whereas terminase clade V4 was associated with all bacterial hosts. Overall, the terminase tree was highly structured and displayed specific bacterial host–associated clades (e.g., clades V1 and V3, compare with fig. S5A). Applying the P Test (34) implemented in Fast UniFrac (35) to terminase alleles grouped by bacterial host indeed revealed significant differences between alleles associated with most pairs of hosts (table S8). Grouping terminase alleles by colony, however, did not reveal significant differences between alleles (table S9), indicating that sampling was not a factor in determining the observed host-associated heterogeneity in terminase alleles. The highly nonrandom distribution of host-associated terminase alleles therefore suggests that lateral gene transfer and/or host switching is limited in this system. This result, however, could also reflect the fact that the terminase gene does not appear to shuffle randomly among phages, possibly indicating a connection between DNA packaging and other characteristics of the phage (36). It remains to be seen whether other viral genes follow similar patterns.
The fact that there was little mixing between terminase alleles associated with host I (V1) and the more distantly related hosts II (V3 and V4) and III (V4), whereas alleles of the more closely related hosts II and III (table S4) exhibited a certain degree of mixing (V4), supports the notion that the probability of cross-species transmission or lateral gene transfer decreases with the phylogenetic distance of the hosts (37). The rRNA gene of hosts I through IV also exhibited patterns of micro-diversity that may have physiological relevance (38, 39). These patterns, however, were mirrored only by the terminase alleles of host III. Host I and II terminase alleles appeared to be indifferent to the bacterial host at the subspecies level.
Our results show that, in a marked departure from classical phage enrichment techniques, specific viral-host relations can be revealed in uncultivated cells harvested straight from the environment. We found that variants of a viral packaging gene appear to have infected bacterial hosts across an entire genus of bacteria. Furthermore, despite the substantial potential for lateral gene transfer and/or host switching in this well-mixed, small-volume system, the terminase tree was highly structured and displayed specific bacterial host–associated clades. It will be interesting to continue to monitor the host-virus interactions within this ecosystem as a function of space and time and across the termite community at large, shedding further light on host-virus coevolution in this unique ecosystem. More broadly, the method we have developed enables a highly parallel analysis of host-virus interactions in environmental samples from nearly any environment in nature.
Supplementary Material
Acknowledgments
We wish to thank D. Baltimore, S. Casjens, D. S. Fisher, R. W. Hendrix, H. J. Lee, M. Lindén, E. G. Matson, R. Milo, V. J. Orphan, S. R. Quake, A. Z. Rosenthal, E. M. Rubin and colleagues at JGI, D. Z. Soghoian, N. D. Wolfe, D. Wu, X. Zhang, and the anonymous referees for their advice and feedback. We also wish to thank E.G.M. and A.Z.R for their assistance in collection of specimens and E.G.M. for ZAS genomic DNA. This project was supported by the NIH Director’s Pioneer Award, NIH American Recovery and Reinvestment Act grant number R01 GM085286-01S, U.S. Department of Energy grant no. DE-FG02-07ER64484, and NSF grant nos. EF-0523267 and CMMI-0758343 and by the Davidow Family Research Fund. GenBank accession numbers are given in table S10.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/333/6038/58/DC1
Materials and Methods
SOM Text
Figs. S1 to S5
Tables S1 to S10
References
References and Notes
- 1.Suttle CA. Nat. Rev. Microbiol. 2007;5:801. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan MB, et al. Environ. Microbiol. 2010;12:3035. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindell D, et al. Nature. 2007;449:83. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 4.Angly FE, et al. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson SJ, et al. PLoS ONE. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugenholtz P. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-reviews0003. reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards RA, Rohwer F. Nat. Rev. Microbiol. 2005;3:504. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 8.Dinsdale EA, et al. Nature. 2008;452:629. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 9.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. Trends Microbiol. 2010;18:11. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson AF, Banfield JF. Science. 2008;320:1047. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 11.Zare RN, Kim S. Annu. Rev. Biomed. Eng. 2010;12:187. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- 12.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 13.Marcy Y, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11889. [Google Scholar]
- 14.Warren L, Bryder D, Weissman IL, Quake SR. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17807. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dube S, Qin J, Ramakrishnan R. PLoS ONE. 2008;3:e2876. doi: 10.1371/journal.pone.0002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohwer F, Edwards R. J. Bacteriol. 2002;184:4529. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose TM, et al. Nucleic Acids Res. 1998;26:1628. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supporting material on Science Online.
- 19.Supporting text is available as supporting material on Science Online.
- 20.Tholen A, Schink B, Brune A. FEMS Microbiol. Ecol. 1997;24:137. [Google Scholar]
- 21.Hongoh Y, Ohkuma M, Kudo T. FEMS Microbiol. Ecol. 2003;44:231. doi: 10.1016/S0168-6496(03)00026-6. [DOI] [PubMed] [Google Scholar]
- 22.Warnecke F, et al. Nature. 2007;450:560. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 23.Leadbetter JR, Schmidt TM, Graber JR, Breznak JA. Science. 1999;283:686. doi: 10.1126/science.283.5402.686. [DOI] [PubMed] [Google Scholar]
- 24.Lilburn TG, et al. Science. 2001;292:2495. doi: 10.1126/science.1060281. [DOI] [PubMed] [Google Scholar]
- 25.Moore SD, Prevelige PE., Jr Curr. Biol. 2002;12:R96. doi: 10.1016/s0960-9822(02)00670-x. [DOI] [PubMed] [Google Scholar]
- 26.Rao VB, Feiss M. Annu. Rev. Genet. 2008;42:647. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 27.Chai S, et al. J. Mol. Biol. 1992;224:87. doi: 10.1016/0022-2836(92)90578-8. [DOI] [PubMed] [Google Scholar]
- 28.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Virology. 1991;183:519. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 29.Casjens S. Mol. Microbiol. 2003;49:277. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Nucleic Acids Res. 2002;30:4009. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, et al. Cell. 2008;135:1251. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2005;33:D501. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Percent identity was measured across 235 unambiguous aligned amino acids.
- 34.Martin AP. Appl. Environ. Microbiol. 2002;68:3673. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamady M, Lozupone C, Knight R. ISME J. 2010;4:17. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casjens SR, et al. J. Bacteriol. 2005;187:1091. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe N, et al. Glob. Change Hum. Health. 2000;1:10. [Google Scholar]
- 38.Moore L, Rocap G, Chisholm S. Nature. 1998;393:465. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JR, et al. Appl. Environ. Microbiol. 2004;70:4103. doi: 10.1128/AEM.70.7.4103-4110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. Appl. Environ. Microbiol. 2005;71:8966. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schloss PD, Handelsman J. Appl. Environ. Microbiol. 2005;71:1501. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.