Abstract
The incidence and mortality of colorectal cancer (CRC) is higher in African Americans (AAs) than other ethnic groups in the U. S., but reasons for the disparities are unknown. We performed gene expression profiling of sporadic CRCs from AAs vs. European Americans (EAs) to assess the contribution to CRC disparities. We evaluated the gene expression of 43 AA and 43 EA CRC tumors matched by stage and 40 matching normal colorectal tissues using the Agilent human whole genome 4x44K cDNA arrays. Gene and pathway analyses were performed using Significance Analysis of Microarrays (SAM), Ten-fold cross validation, and Ingenuity Pathway Analysis (IPA). SAM revealed that 95 genes were differentially expressed between AA and EA patients at a false discovery rate of ≤5%. Using IPA we determined that most prominent disease and pathway associations of differentially expressed genes were related to inflammation and immune response. Ten-fold cross validation demonstrated that following 10 genes can predict ethnicity with an accuracy of 94%: CRYBB2, PSPH, ADAL, VSIG10L, C17orf81, ANKRD36B, ZNF835, ARHGAP6, TRNT1 and WDR8. Expression of these 10 genes was validated by qRT-PCR in an independent test set of 28 patients (10 AA, 18 EA). Our results are the first to implicate differential gene expression in CRC racial disparities and indicate prominent difference in CRC inflammation between AA and EA patients. Differences in susceptibility to inflammation support the existence of distinct tumor microenvironments in these two patient populations.
Introduction
Colorectal cancer (CRC) remains the most common gastrointestinal cancer in the United States, despite recent improvements in the diagnosis and treatment of the disease. The incidence and mortality rates of CRC for African Americans (AAs) are higher than in the U.S. general population [1], [2]. Many epidemiologic and genetic investigations have focused on AAs [3], [4], [5], [6] with the goal of deciphering the reasons for such disparities. Whereas one cannot discount the contribution of socioeconomic factors, such as a more advanced stage of disease at diagnosis in AAs, other biological factors also contribute to the progression of colon cancer [4]; [7]. However, a biological basis for the existence of a more aggressive CRC in African American patients remains to be further elucidated. Genomic instability is a crucial feature in tumor development and there are at least 3 distinct pathways in CRC pathogenesis: chromosomal instability (CIN), microsatellite instability (MSI), and CpG island methylator phenotype pathways (CIMP) [8], [9]. Any or all of these pathways may contribute to a more aggressive CRC biology in African Americans. Recent genome-wide association studies in CRC have shown not only strong evidence for common single nucleotide polymorphism (SNP) association in a number of genes and chromosomal regions, but also genetic heterogeneity in CRC association in AAs versus EAs [4], [10], [11], [12], [13]. Different incidence of MSI and different level of methylation for functionally very relevant genes were also reported as a possible factors in CRC racial disparities [8], [14], [15].
We hypothesized that the gene expression profiles of CRC in African-American and European-American patients may reveal biological differences between the two populations that could explain the more aggressive cancer phenotype in African-Americans. Thus, we performed genome-wide gene expression profiling in a large set of tumor samples that were matched for selected clinical variables. We analyzed our results on gene and pathway levels to identify key differences in tumor biology between African-American and European-American patients.
Methods
Patients
One hundred and fourteen tumors (86 included in original analysis and 28 for validation study) and 40 normal tissues from de-identified CRC patients were obtained from the Institutional Research Board (IRB) approved University of North Carolina (UNC) Tissue Procurement Facility after UNC School of Medicine IRB approval for this study. Written informed consent was obtained from all patients. All samples were collected between 1999 and 2008 at the time of operation and snap frozen in liquid nitrogen. Patients with known familial adenomatous polyposis and hereditary non-polyposis CRC were excluded. De-identified data including race, tumor, node and metastasis (TNM), grade or differentiation, margin status, and survival were available for the majority of patients.
RNA Isolation and Microarray Hybridization
All RNA isolation and hybridization was performed on Agilent (Agilent Technologies, Santa Clara, CA) human whole genome 4X44 K DNA microarrays at UNC. RNA was extracted from macrodissected snap-frozen tumor samples using All prep Kits (Qiagen, Valencia, CA) and quantified using Nanodrop spectrophotometry (ThermoScientific, Wilmington, DE). RNA quality was assessed with the use of the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA was selected for hybridization using RNA integrity number and by inspection of the 18S and 28S ribosomal RNA. Similar RNA quality was selected across samples. One microgram of RNA was used as a template for cDNA preparation prior to hybridization to Agilent 4X44 K whole human genome arrays. cDNA was labeled with Cy5-dUTP and a reference control (Stratagene; Catalog Number # 740000; Agilent Technologies, Santa Clara, CA; [16] was labeled with Cy3-dUTP using the Agilent low RNA input linear amplification kit and hybridized overnight at 65°C to Agilent 4X44 K whole human genome arrays. Arrays were washed and scanned using an Agilent scanner (Agilent Technologies, Santa Clara, CA).
All microarray data are in MIAME compliant form and raw and processed data has been deposited in the Gene Expression Omnibus (GEO); see http://www.ncbi.nlm.nih.gov/geo/, accession number: GSE28000.
Microarray and statistical analysis
All array data were normalized using LOWESS normalization. Data were excluded for genes with poor spot quality or genes that did not have a mean intensity greater than 10 for one of the two channels (green and red) in at least 70% of the experiments. The log2 ratio of the mean red intensity over mean green intensity was calculated for each gene followed by LOWESS normalization [17]. Missing data were imputed using the k-nearest neighbors imputation (KNN) with k = 10 [18]. Genes that were significantly up- or down-regulated were identified using Significance Analysis of Microarrays (SAM) [19]. SAM assigns a score to each gene on the basis of a change in gene expression relative to the standard deviation of repeated measurements. For genes with scores greater than an adjustable threshold, SAM uses permutations of the repeated measurements to estimate the percentage of genes identified by chance – the false discovery rate (FDR). Analysis parameters (Delta) were set to result in FDR≤5%.
Network and gene ontology analysis
Differentially expressed genes were investigated for network and gene functional interrelation by Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, www.ingenuity.com; [20]. IPA scans the set of input genes to identify networks by using Ingenuity Pathways Knowledge Base for interactions between identified ‘Focus Genes’, in this study, the differentially expressed genes between AA and EA and known and hypothetical interacting genes stored in the knowledge base in IPA software was used to generate a set of networks with a maximum network size of 35 genes/proteins. Networks are displayed graphically as genes/gene products (‘nodes’) and the biological relationships between the nodes (‘edges’). All edges are from canonical information stored in the Ingenuity Pathways Knowledge Base. In addition, IPA computes a score for each network according to the fit of the user's set of significant genes. The score indicates the likelihood of the Focus Genes in a network from Ingenuity's knowledge base being found together due to random chance. A score of 3, as the cutoff for identifying gene networks, indicates that there is only a 1/1000 chance that the focus genes shown in a network are due to random chance. Therefore, a score of 3 or higher indicates a 99.9% confidence level to exclude random chance.
Ten-fold Cross Validation (Ten-f-CV)
Ten-f-CV analysis [21] was used to select smaller representative set of genes for validation study by qRT-PCR. Using Ten-f-CV analysis we identified 10 genes that can predict the ethnicity of the patient for whom the array was done with an error rate of 6%, suggesting that these 10 genes are representative of the entire gene list.
Quantitative real-time PCR
Validation of microarray results was performed on 28 CRC patients (10 AA, and 18 EA). Ten differentially expressed genes (identified by SAM and selected using the Ten-f-CV method) were validated by qRT-PCR. The hydroxymethylbilane synthase (HMBS) gene served as the housekeeping gene [22]. qRT-PCR was performed in duplicates using SYBR Green Gene Expression Assays (Applied Biosystems, Forester City, CA), which include pre-optimized primer sets specific for the genes being validated [23]. The validated genes were: Crystalline, beta B2 (CRYBB2), phosphoserine phosphatase homologue (PSPH), Adenosine deaminase-like (ADAL), V-set and immunoglobulin domain containing 10 like (VSIG10L), Chromosome 17 open reading frame 81 (C17orf81), Ankyrin repeat domain 36B (ANKRD36B). Zinc finger protein 83 (ZNF83), Rho GTPase activating protein 6 (ARHGAP6), WD repeat domain 8 (WDR8), TRNA nucleotidyl transferase, CCA-adding, 1 (TRNT1), and HMBS. Data were collected using the ABI PRISM 7500 sequence detection system (Applied Biosystems, Forster City, CA). qRT-PCR data for each sample were normalized using expression of the housekeeping gene HMBS. Graphs were prepared from normalized data relative to HMBS. Statistical analysis of these data was performed with a two-sided t-test or with a two-sided Wilcoxon rank-sum test if the expression data did not follow normal distribution.
Results
Identification of differentially expressed genes between AA and EA CRC patients
Patient population characteristics for 43 AA and 43 EA patients were matched by TNM staging ( Table 1 ). The two populations were similar in age, gender and tumor localization. Forty non-tumor colon tissues (13 AAs and 27 EAs) were used for genetic comparisons of normal colon gene expression between AAs and EAs.
Table 1. Clinical characteristics of the study population.
| Characteristics | Whites (n = 43) | Blacks (n = 43) | p-Value * | ||
| No. | % | No. | % | ||
| Mean Age (y) | 67 | n/a | 62.7 | n/a | 0.18** |
| Gender | |||||
| Male | 19 | 44.19 | 25 | 58.14 | |
| Female | 24 | 55.81 | 18 | 41.86 | 0.28 |
| Location | |||||
| Right | 17 | 39.53 | 22 | 51.16 | |
| Left | 21 | 48.83 | 21 | 48.83 | |
| Unknown | 5 | 11.62 | 0 | 0.00 | 0.06 |
| Tumor Stage | |||||
| 1 | 4 | 9.30 | 4 | 9.30 | |
| 2 | 16 | 37.21 | 16 | 37.21 | |
| 3 | 8 | 18.60 | 8 | 18.60 | |
| 4 | 15 | 34.88 | 15 | 34.88 | 1.00 |
*p-values based on Fisher's exact test.
**p-value based on t-test.
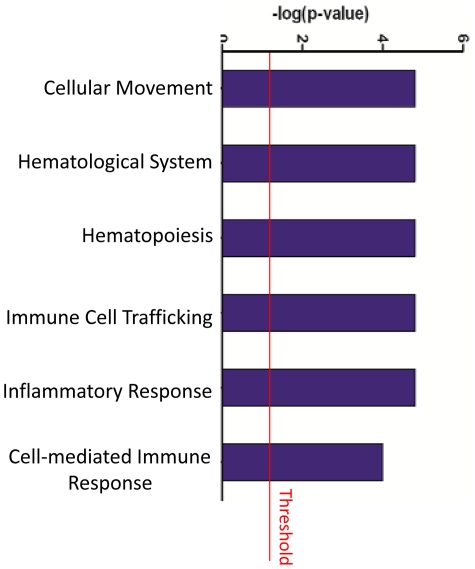
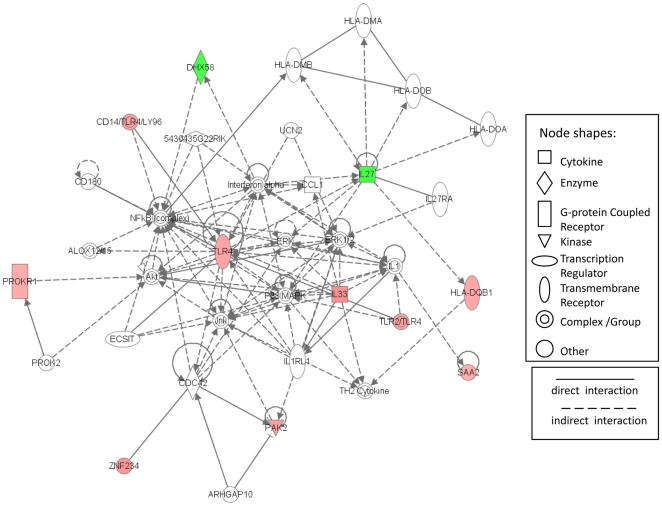
The comparison of gene expression profiles from AA and EA tumors using SAM revealed 95 gene transcripts to be differentially expressed between the two groups at FDRs of ≤5%. Fifty-eight genes were up regulated ( Table 2 ) and 37 down regulated ( Table 3 ) in tumor of AAs. We used Ingenuity Pathway Analysis to assess disease and pathway associations of these 95 genes that were differentially expressed in CRC tumors by race. The disease association analysis revealed associations of differentially expressed genes with genetic pathways that are linked to inflammatory response, hepatic system disease, developmental disorders, genetic disorders and neurologic disease (Table S1). The six top associated pathways for differentially expressed genes are shown in Fig. 1 . Three of these six pathways are related to inflammatory and immune response. Differentially expressed genes in the five highest scoring networks are shown in Table 4 . Top associated network functions for differently expressed genes were: 1) organismal injury and abnormalities, gene expression, cellular development 2) lipid metabolism, small molecule biochemistry, molecular transport 3) cellular assembly and organization, organ development, carbohydrate metabolism 4) antigen presentation and inflammatory response, cellular movement 5) behavior, digestive system development and function, endocrine system development and function. One of these networks (network 4; antigen presentation and inflammatory response) is graphically represented in Fig. 2 . Seven of the nine genes in this network were up regulated in AA patients (HLA-DQB1, IL33, PAK2, PROKR1, SAA2, TLR4, ZNF234), and two genes were down regulated (DHX58, IL27).
Table 2. Up-regulated genes in colorectal tumors of African American patients.
| No. | Gene ID | Gene Name | Gene Title | Chromosome Location | Fold Change |
| 1 | 1415 | CRYBB2 | Crystallin,beta B2 Crystallin,betaB2 | 22q11.2-q12.1|22q11.23 | 3.10 |
| 2 | 5723 | PSPH | Phosphoserinephosphatase | 7p15.2-p15.1 | 3.38 |
| 3 | 116285 | ACSM1 | Acyl-CoAsynthetase medium-chain family member 1 | 16p12.3 | 2.47 |
| 4 | 23008 | KLHDC10 | Kelchdomain containing 10 | 7q32.2 | 1.48 |
| 5 | 90865 | IL33 | Interleukin33 | 9p24.1 | 3.08 |
| 6 | 6231 | RPS26 | Ribosomalprotein S26 | 12q13 | 1.63 |
| 7 | 401081 | FLJ22763 | Hypotheticalgene supported by AK026416 | 3q13.13 | 4.69 |
| 8 | 10780 | ZNF234 | Zincfinger protein 234 | 19q13.31 | 2.09 |
| 9 | 2863 | GPR39 | Gprotein-coupled receptor 39 | 2q21-q22 | 1.58 |
| 10 | 60439 | TTTY2 | Testis-specifictranscript, Y-linked 2 (non-protein coding) | Yp11.2 | 1.55 |
| 11 | 27012 | KCNV1 | Potassiumchannel, subfamily V, member 1 | 8q22.3-q24.1 | 2.58 |
| 12 | 55020 | TTC38 | Tetratricopeptiderepeat domain 38 | 22q13 | 1.67 |
| 13 | 256380 | SCML4 | Sexcomb on midleg-like 4 (Drosophila) | 6q21 | 2.90 |
| 14 | 5062 | PAK2 | P21protein (Cdc42/Rac)-activated kinase 2 | 3q29 | 1.43 |
| 15 | 540 | ATP7B | ATPase,Cu++ transporting, beta polypeptide | 13q14.3 | 1.66 |
| 16 | 23562 | CLDN14 | Claudin14 | 21q22.3 | 2.35 |
| 17 | 10911 | UTS2 | Urotensin2 | 1p36 | 2.45 |
| 18 | 1416 | CRYBB2P1 | Crystallin,beta B2 pseudogene 1 | 22q11.2-q12.1 | 1.59 |
| 19 | 55282 | LRRC36 | Leucinerich repeat containing 36 | 16q22.1 | 2.45 |
| 20 | 348013 | FAM70B | Familywith sequence similarity 70, member B | 13q34 | 1.48 |
| 21 | 81839 | VANGL1 | Vang-like1 (van gogh, Drosophila) | 1p11-p13.1 | 1.49 |
| 22 | 100190939 | LOC100190939 | HypotheticalLOC100190939 | 13q14.13 | 1.60 |
| 23 | 4552 | MTRR | 5-methyltetrahydrofolate-homocysteinemethyltransferase reductase | 5p15.3-p15.2 | 1.74 |
| 24 | 348751 | LOC348751 | Hypotheticalprotein LOC348751 | 2q33.1 | 1.78 |
| 25 | 7099 | TLR4 | Toll-likereceptor 4 | 9q32-q33 | 1.95 |
| 26 | 5789 | PTPRD | Proteintyrosine phosphatase, receptor type, D | 9p23-p24.3 | 2.17 |
| 27 | 3043 | HBB | Hemoglobin,beta | 11p15.5 | 1.82 |
| 28 | 146456 | TMED6 | Transmembraneemp24 protein transport domain containing 6 | 16q22.1 | 2.61 |
| 29 | 253039 | LOC253039 | HypotheticalLOC253039 | 9q33.2 | 1.32 |
| 30 | 2037 | EPB41L2 | Erythrocytemembrane protein band 4.1-like 2 | 6q23 | 1.81 |
| 31 | 59352 | LGR6 | Leucine-richrepeat-containing G protein-coupled receptor 6 | 1q32.1 | 2.28 |
| 32 | 2689 | GH2 | Growthhormone 2 | 17q24.2 | 1.42 |
| 33 | 4886 | NPY1R | NeuropeptideY receptor Y1 | 4q31.3-q32 | 4.03 |
| 34 | 283345 | RPL13P5 | Ribosomalprotein L13 pseudogene 5 | 12p13.31 | 1.99 |
| 35 | 3119 | HLA-DQB1 | Majorhistocompatibility complex, class II, DQ beta 1 | 6p21.3 | 1.55 |
| 36 | 140881 | DEFB129 | Defensin,beta 129 | 20p13 | 1.44 |
| 37 | 144568 | A2ML1 | Alpha-2-macroglobulin-like1 | 12p13.31 | 1.59 |
| 38 | 10783 | NEK6 | NIMA(never in mitosis gene a)-related kinase 6 | 9q33.3-q34.11 | 1.53 |
| 39 | 130399 | ACVR1C | ActivinA receptor, type IC | 2q24.1 | 2.32 |
| 40 | 27283 | TINAG | Tubulointerstitialnephritis antigen | 6p11.2-p12 | 1.97 |
| 41 | 116511 | MAS1L | MAS1oncogene-like | 6p21 | 1.47 |
| 42 | 8908 | GYG2 | Glycogenin2 | Xp22.3 | 1.73 |
| 43 | 145447 | ABHD12B | Abhydrolasedomain containing 12B | 14q22.1 | 3.20 |
| 44 | 401577 | LOC401577 | Hypotheticalprotein LOC401577 | Xp22.33 | 1.49 |
| 45 | 10887 | PROKR1 | Prokineticinreceptor 1 | 2p13.1 | 1.44 |
| 46 | 4889 | NPY5R | NeuropeptideY receptor Y5 | 4q31-q32 | 1.50 |
| 47 | 327657 | SERPINA9 | Serpinpeptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 | 1.37 | |
| 48 | 4860 | NP | Nucleosidephosphorylase | 14q13.1 | 1.47 |
| 49 | 1056 | CEL | Carboxylester lipase (bile salt-stimulated lipase) | 9q34.3 | 1.49 |
| 50 | 81796 | SLCO5A1 | Solutecarrier organic anion transporter family, member 5A1 | 8q13.3 | 1.34 |
| 51 | 6289 | SAA2 | Serumamyloid A2 | 11p15.1-p14 | 2.29 |
| 52 | 171558 | PTCRA | PreT-cell antigen receptor alpha | 6p21.3 | 1.31 |
| 53 | 8309 | ACOX2 | Acyl-CoenzymeA oxidase 2, branched chain | 3p14.3 | 2.21 |
| 54 | 79857 | FLJ13224 | Hypotheticalprotein FLJ13224 | 12p11.21 | 1.28 |
| 55 | 9376 | SLC22A8 | Solutecarrier family 22 (organic anion transporter), member 8 | 1.33 | |
| 56 | 7712 | ZNF157 | Zincfinger protein 157 | Xp11.2 | 1.40 |
| 57 | 10 | NAT2 | N-acetyltransferase2 (arylamine N-acetyltransferase) | 8p22 | 2.45 |
| 58 | 283422 | C12orf36 | Chromosome12 open reading frame 36 | 12p13.1 | 2.07 |
Table 3. Down-regulated genes in colorectal tumors of African American patients.
| No. | Gene ID | Gene Name | Gene Title | Chromosome Location | Fold Change |
| 1 | 57730 | ANKRD36B | Ankyrin repeatdomain 36B | 2q11.2 | 0.48 |
| 2 | 23587 | C17orf81 | Chromosome 17open reading frame 81 | 17p13.1 | 0.44 |
| 3 | 415 | ARSE | Arylsulfatase E(chondrodysplasia punctata 1) | Xp22.3 | 0.21 |
| 4 | 395 | ARHGAP6 | Rho GTPaseactivating protein 6 | Xp22.3 | 0.58 |
| 5 | 146547 | PRSS36 | Protease, serine,36 | 16p11.2 | 0.36 |
| 6 | 161823 | ADAL | Adenosine deaminase-like | 15q15.3 | 0.65 |
| 7 | 246778 | IL27 | Interleukin 27 | — | 0.36 |
| 8 | 644246 | LOC644246 | Hypothetical proteinLOC644246 | 17q21.31 | 0.42 |
| 9 | 79132 | DHX58 | DEXH (Asp-Glu-X-His)box polypeptide 58 | 17q21.2 | 0.54 |
| 10 | 51095 | TRNT1 | TRNA nucleotidyltransferase, CCA-adding, 1 | 3p25.1 | 0.60 |
| 11 | 55769 | ZNF83 | Zinc fingerprotein 83 | 19q13.3 | 0.65 |
| 12 | 64093 | SMOC1 | SPARC relatedmodular calcium binding 1 | 14q24.2 | 0.44 |
| 13 | 642946 | LQK1 | Hypothetical LOC642946 | 1q32.3 | 0.47 |
| 14 | 147645 | VSIG10L | V-set andimmunoglobulin domain containing 10 like | 19q13.41 | 0.65 |
| 15 | 26150 | RIBC2 | RIB43A domainwith coiled-coils 2 | 22q13.31 | 0.48 |
| 16 | 57531 | HACE1 | HECT domainand ankyrin repeat containing, E3 ubiquitin protein ligase | 0.60 | |
| 17 | 5303 | PIN4 | Protein (peptidylprolylcis/trans isomerase) NIMA-interacting, 4 (parvulin) | Xq13 | 0.62 |
| 18 | 8820 | HESX1 | HESX homeobox1 | 3p14.3 | 0.56 |
| 19 | 79609 | C14orf138 | Chromosome 14open reading frame 138 | 14q21.3 | 0.62 |
| 20 | 816 | CAMK2B | Calcium/calmodulin-dependent proteinkinase II beta | 22q12|7p14.3-p14.1 | 0.64 |
| 21 | 1953 | MEGF6 | Multiple EGF-like-domains6 | 1p36.3 | 0.55 |
| 22 | 79682 | MLF1IP | MLF1 interactingprotein | 4q35.1 | 0.51 |
| 23 | 51340 | CRNKL1 | Crooked neckpre-mRNA splicing factor-like 1 (Drosophila) | 20p11.2 | 0.61 |
| 24 | 100287616 | LOC100287616 | Hypothetical proteinLOC100287616 | 15q24.1 | 0.54 |
| 25 | 201973 | CCDC111 | Coiled-coil domaincontaining 111 | 4q35.1 | 0.67 |
| 26 | 3712 | IVD | Isovaleryl CoenzymeA dehydrogenase | 15q14-q15 | 0.62 |
| 27 | 4942 | OAT | Ornithine aminotransferase(gyrate atrophy) | 10q26 | 0.60 |
| 28 | 57830 | KRTAP5-8 | Keratin associatedprotein 40306 | 11q13.4 | 0.50 |
| 29 | 388610 | TRNP1 | TMF1-regulated nuclearprotein 1 | 1p36.11 | 0.47 |
| 30 | 26152 | ZNF337 | Zinc fingerprotein 337 | 20p11.1 | 0.62 |
| 31 | 100287572 | LOC100287572 | Similar tohCG1996962 | 18q23 | 0.63 |
| 32 | 1427 | CRYGS | Crystallin, gammaS | 3q25-qter | 0.65 |
| 33 | 4435 | CITED1 | Cbp/p300-interacting transactivator,with Glu/Asp-rich carboxy-terminal domain, 1 | Xq13.1 | 0.55 |
| 34 | 339318 | ZNF181 | Zinc fingerprotein 181 | 19q13.11 | 0.69 |
| 35 | 49856 | WDR8 | WD repeatdomain 8 | 1p36.3 | 0.71 |
| 36 | 221322 | C6orf170 | Chromosome 6open reading frame 170 | 6q22.31 | 0.70 |
| 37 | 284018 | C17orf58 | Chromosome 17open reading frame 58 | 17q24.2 | 0.62 |
Figure 1. Ingenuity analysis of top pathways affected in differentially expressed genes between African Americans and European Americans.
Y-axis is an inverse indication of p-value or significance. The Threshold line marks the p = 0.05. (Note that 3 of 6 top pathways shown are related to inflammation and immune response).
Table 4. Functional association of differentially expressed genes generated by IPA.
| ID | Focus Molecules in Network | Score | Focus Molecules | Top Functions |
| 1 | ARSE, C6ORF170, CITED1, CRYBB2, CRYGS, EPB41L2, HBB (includes EG:3043), HESX1, HLA-DQB1, IVD, NAT2, OAT, PTPRD, RPS26, ZNF83 | 28 | 15 | Organismal Injury and Abnormalities, Gene Expression, Cellular Development |
| 2 | ACVR1C, ATP7B, C17ORF81, GH2, GPR39, MLF1IP, NP, PIN4, PTCRA, SLC22A8, TRNT1, TTC38, VANGL1,WDR8 (includes EG:49856) | 26 | 14 | Lipid Metabolism, Small Molecule Biochemistry, Molecular Transport |
| 3 | ANKRD36B, CRNKL1, GYG2, KCNV1, KLHDC10, MEGF6, MTRR, NEK6, PSPH, RIBC2, ZNF337 | 20 | 11 | Cellular Assembly and Organization, Organ Development, Carbohydrate Metabolism |
| 4 | DHX58, HLA-DQB1, IL27, IL33, PAK2, PROKR1, SAA2, TLR4, ZNF234 | 15 | 9 | Antigen Presentation, Inflammatory Response, Cellular Movement |
| 5 | CAMK2B, CEL, NPY1R, NPY5R, SCML4, UTS2 | 9 | 6 | Behavior, Digestive System Development and Function, Endocrine System Development and Function |
Figure 2. Gene network involved in “Inflammatory Response” generated by IPA for differentially expressed genes between African Americans and European Americans.
Red symbols are assigned for up-regulated and green for down-regulated genes. Node shape corresponds to the functional role of molecules as shown in the legend. Direct or indirect interactions are shown by complete or dashed lines.
We also performed SAM analysis using non-tumor colon tissues from AA and EA patients and did not see differential gene expression (data not shown), suggesting that the changes we identified are tumor microenvironment specific.
Validation of microarray results
In order to select a representative group of genes for qRT-PCR validation of differentially expressed genes between AA and EA CRC patients, we performed a 10-fold cross validation analysis that resulted in the selection of following ten genes: CRYBB2, PSPH, ADAL, VSIG10L, C17orf81, ANKRD36B, ZNF835, ARHGAP6, TRNT1 and WDR8.
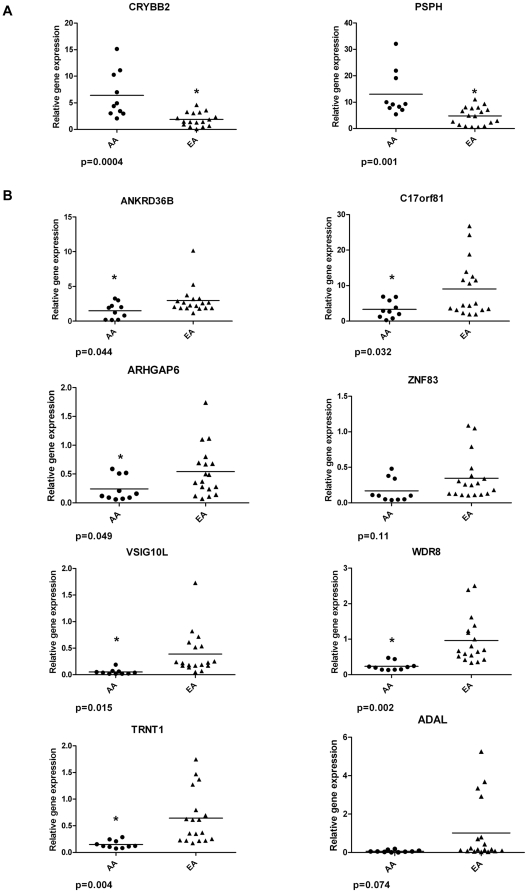
Expression of these ten genes was validated by qRT-PCR on an independent test set of 28 CRC patients (10 AA and 18 EA). The qRT-PCR results are shown in Fig. 3 . Two of the 10 differentially expressed genes were up-regulated in AA vs. EA CRC patients; CRYBB2, p = 0.0004 and PHSP, p = 0.001 ( Fig. 3 ; Panel A.). Eight were down-regulated in AA vs. EA CRC patients; VSIG10L, p = 0.015; C17orf81, p = 0.032; WDR8, p = 0.002; TRNT1, p = 0.004; ANKRD36B, p = 0.044; ARHGAP6, p = 0.049; ADAL, p = 0.074; ZNF83, p = 0.11; ( Fig. 3 ; Panel B.). The direction of change (up or down regulation) for the qRT-PCR validated genes was in agreement with SAM results. Eight of ten genes in the qRT-PCR validation study reached a statistically significant level (p<0.05; CRYBB2, PSPH, C17orf81, ANKRD36B, VSIG10L, WDR8, TRNT1 and ARHGAP6).
Figure 3. qRT-PCR validation analysis of expression of ten selected genes between African American and European American CRC patients.
Panel A. Expression of two up-regulated genes in African American colorectal cancer patients: CRYBB2, PSPH. Panel B. Expression of eight down-regulated genes in African American colorectal cancer patients: ARHGAP6, VSIG10L, C17orf81, ZNF83, ANKRD36B, ADAL, WDE8 and TNRT1. Points are relative Ct values for the individual samples; horizontal lines are mean values for the sample set. The significantly differentially expressed genes (p<0.05) between African-American (n = 10) vs. European-American (n = 18) CRC tumors are labeled by a star. Graphs were prepared from normalized genes expression data relative to the housekeeping (HMBS) gene.
Discussion
The causes of the CRC disparity that exists between AA and EA patients remain to be fully elucidated. Although most of the research on this disparity has focused on socioeconomic factors, recent findings strongly support the role of genetic and biological factors. Genetic differences between AA and EA CRC patients were reported for SNP association, for incidence of MSI and level of gene methylation [4], [14], [15]. Any of these differences can result in differential gene expression between AA and EA CRC patients. In this study we analyzed the gene expression profiles of 86 tumors from 43 AA and 43 EA patients. Significant differences in the expression of genes related to immune response and inflammation within the tumor micro-environment were identified between these two groups. This interpretation was supported by both disease association and pathway analyses. Most of the immune-related genes had higher expression in tumors from African-American patients than in those from European-American patients. Although preliminary, these findings are novel and could have implications for cancer therapy. From the present study, we do not know why CRC from African-American patients would have a different immunologic profile than tumors from European-American patients. We hypothesize that the causes of these differences are multifactorial. Chronic inflammation is thought to be a causative factor in colorectal carcinogenesis [24], [25]. It was shown that an immune response signature in the liver of cancer patients predicts metastasis and recurrence of hepato-cellular carcinoma [26]. Thus, future studies should evaluate whether the immunologic profile of CRC in African-American patients is a predisposing factor for tumor progression and metastasis. Previous investigations identified a two-gene tumor signature (CRYBB2 and PSPHL) that accurately differentiated between African-American and European-American prostate cancer patients [27]. Those two genes were also differentially expressed between African-American and European American breast cancer patients [28] In this study we found up-regulation of CRYBB2 and PSPH gene in CRC of African-American patients. Mutations in the CRYBB2 gene are also responsible for familial cataract [29]. PSPHL is a homolog of PSPH. Interestingly, PSPH is located on chromosome 7p15.2, a chromosomal region known to have gain of function related to advanced tumor stage in non-small-cell lung adenocarcinoma [30]. It was shown that increased expression of PSPH in non-small-cell lung cancer corresponds to clinical response to treatment with erlotinib [31]. Thus, it is possible that higher expression of PSPH contributes to CRC susceptibility in AAs and that the levels of PSPH expression may be correlated with response to anti-EGFR treatment. These possibilities will have to be tested in future studies. Considering down-regulated genes in AAs we found lower expression of the C17orf81 gene. Down regulation of this gene was associated with colon cancer [32], suggesting that lower expression of this gene can contribute to more aggressive CRC in AAs. Other down regulated genes in AAs include: TRNT1 (involve in RNA processing; [33]); ARHGAP6 (promotes actin remodeling: [34]); WDR8 (facilitates formation of multiprotein complexes: [35]). Considering the cellular functions of these genes it is not hard to envision how their expression may influence aggressiveness of CRC.
Whole-genome gene expression analysis experiments can be prone to findings that are either unique to a selected patient population or are artificially created by the applied technology. To exclude the possibility of an artifact, two different approaches were used to cross validate our gene expression data. First, we compared our results of the differentially expressed genes between 86 tumors and 40 surrounding non-tumor tissues with those from a published meta-analysis of five CRC gene expression datasets in Oncomine [36], [37], [38], [39], [40]; Table S2. We found a very good agreement between our results and the results of the other 5 meta-analyses. Of the top 20 over-expressed genes in CRC tumors across the 5 other meta-analyses (Oncomine; https://www.oncomine.org), 15 were also found to be significantly up-regulated (FDR, <5%) in our study. Of the top 20 under-expressed genes in CRC tumors across the 5 other meta-analyses, 17 were significantly down-regulated (FDR, <5%) in our study.
Second, we validated the expression of ten key genes via qRT-PCR and confirmed differences in gene expression between CRCs of AAs and EAs for eight of them.
In conclusion, the gene expression profile of CRC corresponds to differences in tumor biology between African-American and European-American patients. The implications of these differences in disease aggressiveness and response to therapy should be evaluated in future studies.
Supporting Information
Ingenuity Pathway Analysis of association of differentially expressed genes with top bio functions.
(DOCX)
Comparison of top 40 differently expressed genes in five other CRC microarray studies and this study.
(DOCX)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grant funding for Gastrointestinal Specialized Program of Research Excellence (GI SPORE, P50 106991) and by the University of North Carolina Center for Gastrointestinal Biology and Disease (CGIBD, P30 DK 34987). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alexander DD, Waterbor J, Hughes T, Funkhouser E, Grizzle W, et al. African-American and Caucasian disparities in colorectal cancer mortality and survival by data source: an epidemiologic review. Cancer Biomark. 2007;3:301–313. doi: 10.3233/cbm-2007-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Devaraj B, Lee A, Cabrera BL, Miyai K, Luo L, et al. Relationship of EMAST and microsatellite instability among patients with rectal cancer. J Gastrointest Surg. 2011;14:1521–1528. doi: 10.1007/s11605-010-1340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kupfer SS, Anderson JR, Hooker S, Skol A, Kittles RA, et al. Genetic heterogeneity in colorectal cancer associations between African and European americans. Gastroenterology. 2011;139:1685e1671–1678. doi: 10.1053/j.gastro.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ollberding NJ, Nomura AM, Wilkens LR, Henderson BE, Kolonel LN. Racial/ethnic differences in colorectal cancer risk: The multiethnic cohort study. Int J Cancer. 2010 doi: 10.1002/ijc.25822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinikoor LC, Satia JA, Schroeder JC, Millikan RC, Martin CF, et al. Associations between trans fatty acid consumption and colon cancer among Whites and African Americans in the North Carolina colon cancer study I. Nutr Cancer. 2009;61:427–436. doi: 10.1080/01635580802710725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams CD, Satia JA, Adair LS, Stevens J, Galanko J, et al. Antioxidant and DNA methylation-related nutrients and risk of distal colorectal cancer. Cancer Causes Control. 2010;21:1171–1181. doi: 10.1007/s10552-010-9544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton AM, Sandler R, Carethers JM, Millikan RC, Galanko J, et al. 5,10-methylenetetrahydrofolate reductase 677 and 1298 polymorphisms, folate intake, and microsatellite instability in colon cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2023–2029. doi: 10.1158/1055-9965.EPI-05-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones BA, Christensen AR, Wise JP, Sr, Yu H. Glutathione S-transferase polymorphisms and survival in African-American and white colorectal cancer patients. Cancer Epidemiol. 2009;33:249–256. doi: 10.1016/j.canep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Katkoori VR, Jia X, Shanmugam C, Wan W, Meleth S, et al. Prognostic significance of p53 codon 72 polymorphism differs with race in colorectal adenocarcinoma. Clin Cancer Res. 2009;15:2406–2416. doi: 10.1158/1078-0432.CCR-08-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kupfer SS, Anderson JR, Ludvik AE, Hooker S, Skol A, et al. Genetic associations in the vitamin d receptor and colorectal cancer in african americans and Caucasians. PLoS One. 2011;6:e26123. doi: 10.1371/journal.pone.0026123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roff A, Wilson RT. A novel SNP in a vitamin D response element of the CYP24A1 promoter reduces protein binding, transactivation, and gene expression. J Steroid Biochem Mol Biol. 2008;112:47–54. doi: 10.1016/j.jsbmb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashktorab H, Smoot DT, Carethers JM, Rahmanian M, Kittles R, et al. High incidence of microsatellite instability in colorectal cancer from African Americans. Clin Cancer Res. 2003;9:1112–1117. [PubMed] [Google Scholar]
- 15.Brim H, Kumar K, Nazarian J, Hathout Y, Jafarian A, et al. SLC5A8 gene, a transporter of butyrate: a gut flora metabolite, is frequently methylated in African American colon adenomas. PLoS One. 2011;6:e20216. doi: 10.1371/journal.pone.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dybkaer K, Zhou G, Iqbal J, Kelly D, Xiao L, et al. Suitability of stratagene reference RNA for analysis of lymphoid tissues. Biotechniques. 2004;37:474. doi: 10.2144/04373RT03. [DOI] [PubMed] [Google Scholar]
- 17.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori R, Xiong S, Wang Q, Tarabolous C, Shimada H, et al. Gene profiling and pathway analysis of neuroendocrine transdifferentiated prostate cancer cells. Prostate. 2009;69:12–23. doi: 10.1002/pros.20851. [DOI] [PubMed] [Google Scholar]
- 21.Camp JT, Elloumi F, Roman-Perez E, Rein J, Stewart DA, et al. Interactions with fibroblasts are distinct in Basal-like and luminal breast cancers. Mol Cancer Res. 2011;9:3–13. doi: 10.1158/1541-7786.MCR-10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Round JL, Humphries LA, Tomassian T, Mittelstadt P, Zhang M, et al. Scaffold protein Dlgh1 coordinates alternative p38 kinase activation, directing T cell receptor signals toward NFAT but not NF-kappaB transcription factors. Nat Immunol. 2007;8:154–161. doi: 10.1038/ni1422. [DOI] [PubMed] [Google Scholar]
- 24.Secher T, Gaillot O, Ryffel B, Chamaillard M. Remote control of intestinal tumorigenesis by innate immunity. Cancer Res. 2010;70:1749–1752. doi: 10.1158/0008-5472.CAN-09-3401. [DOI] [PubMed] [Google Scholar]
- 25.Westbrook AM, Szakmary A, Schiestl RH. Mechanisms of intestinal inflammation and development of associated cancers: lessons learned from mouse models. Mutat Res. 2010;705:40–59. doi: 10.1016/j.mrrev.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 27.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 28.Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4:e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, et al. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–668. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 30.Choi JS, Zheng LT, Ha E, Lim YJ, Kim YH, et al. Comparative genomic hybridization array analysis and real-time PCR reveals genomic copy number alteration for lung adenocarcinomas. Lung. 2006;184:355–362. doi: 10.1007/s00408-006-0009-0. [DOI] [PubMed] [Google Scholar]
- 31.Tan EH, Ramlau R, Pluzanska A, Kuo HP, Reck M, et al. A multicentre phase II gene expression profiling study of putative relationships between tumour biomarkers and clinical response with erlotinib in non-small-cell lung cancer. Ann Oncol. 2010;21:217–222. doi: 10.1093/annonc/mdp520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saaf AM, Halbleib JM, Chen X, Yuen ST, Leung SY, et al. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell. 2007;18:4245–4260. doi: 10.1091/mbc.E07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagaike T, Suzuki T, Tomari Y, Takemoto-Hori C, Negayama F, et al. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J Biol Chem. 2001;276:40041–40049. doi: 10.1074/jbc.M106202200. [DOI] [PubMed] [Google Scholar]
- 34.Prakash SK, Paylor R, Jenna S, Lamarche-Vane N, Armstrong DL, et al. Functional analysis of ARHGAP6, a novel GTPase-activating protein for RhoA. Hum Mol Genet. 2000;9:477–488. doi: 10.1093/hmg/9.4.477. [DOI] [PubMed] [Google Scholar]
- 35.Ewing RM, Chu P, Elisma F, Li H, Taylor P, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci U S A. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 39.Kurashina K, Yamashita Y, Ueno T, Koinuma K, Ohashi J, et al. Chromosome copy number analysis in screening for prognosis-related genomic regions in colorectal carcinoma. Cancer Sci. 2008;99:1835–1840. doi: 10.1111/j.1349-7006.2008.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ingenuity Pathway Analysis of association of differentially expressed genes with top bio functions.
(DOCX)
Comparison of top 40 differently expressed genes in five other CRC microarray studies and this study.
(DOCX)