Abstract
Commonly observed in colorectal cancer (CRC) is elevated expression of the prostaglandin synthase cyclooxygenase-2 (COX-2). In normal intestinal epithelium, the COX-2 mRNA is targeted for rapid decay through the 3′-untranslated region (3′UTR) adenylate- and uridylate (AU)-rich element (ARE), whereas in tumors ARE-mediated decay is compromised. Here we demonstrate that the COX-2 ARE can mediate degradation through microRNA-mediated regulation. We identified miR-16 to bind the COX-2 3′UTR and inhibit COX-2 expression by promoting rapid mRNA decay. In CRC cells and tumors, miR-16 levels were decreased ~2-fold and miR-16 expression in cancer cells attenuated COX-2 expression and prostaglandin synthesis. The COX-2 ARE is also bound by the RNA-binding protein HuR. In CRC tumors, HuR is overexpressed and localized within the cytoplasm where it promotes ARE-mRNA stabilization. Under conditions of HuR overexpression, miR-16 was unable to promote rapid mRNA decay through the COX-2 ARE. Ribonucleoprotein immunoprecipitation of HuR demonstrated direct association with miR-16 that was reversed when cytoplasmic trafficking of HuR was inhibited. Furthermore, this interaction between HuR and miR-16 promoted the down regulation of miR-16. These new results identify miR-16 as a central post-transcriptional regulator of COX-2 and demonstrate the ability of elevated levels of HuR to antagonize miR-16 function. Along with insight into altered ARE-mediated mRNA decay observed in CRC, these findings provide a new explanation for tumor-derived loss of miR-16.
Keywords: HuR, microRNA-16, COX-2, AU-rich elements, RNA stability, colon cancer
Introduction
Colorectal cancer (CRC) is the third most common form of cancer in both incidence and mortality. Although CRC prevalence has decreased over the past decade due to early detection, it is anticipated that ~141,000 new cases will occur in 2011 (1). In colorectal tumors, various genetic alterations allow for aberrant activation of signaling pathways resulting in enhanced expression of many growth- and inflammation-associated immediate-early response genes. Through their overexpression, these factors can contribute to CRC etiology at virtually all steps of tumorigenesis (2).
A critical point in the regulation of many inflammatory cytokines, growth factors, and proto-oncogenes occurs through post-transcriptional mechanisms that promote rapid mRNA decay (3). A prominent cis-acting RNA element present in a majority of these cancer-associated transcripts is the adenylate- and uridylate (AU)-rich element (ARE) contained within the mRNA 3′-untranslated region (3′UTR). In normal intestinal epithelium, this RNA element serves to target these mRNAs for rapid decay through recognition of ARE-binding proteins and thus limit expression of potentially pathogenic factors (4, 5). However, loss of ARE-mediated post-transcriptional regulation is observed in CRC cells and tumors (6, 7), indicating the significance of this mechanism in carcinogenesis (8).
The cyclooxygenase (COX) enzymes perform the rate-limiting step in prostaglandin synthesis and overexpression of the inducible isoform, COX-2, has been shown to occur at multiple stages of colon carcinogenesis making it a prominent target of chemoprevention (9). In normal cells, COX-2 expression levels are potently regulated through the ARE present in its 3′UTR (10). Whereas, under conditions of neoplastic transformation, the ability of the COX-2 ARE to promote post-transcriptional regulation is compromised (11). This is in part due to overexpression of the ARE-binding protein HuR in CRC cells and tumors, where elevated HuR levels can impede ARE-mediated mRNA decay (6, 7, 12). These findings and others demonstrate increased expression of HuR to occur in a variety of human cancers and promote ARE-containing gene expression (13).
The mechanism by which HuR promotes COX-2 mRNA stabilization appears to be linked to its subcellular localization (11). In non-stressed cells, HuR is predominantly localized to the nucleus (>90%) and can shuttle between the nucleus and cytoplasm (13), whereas in tumor cells and tissues, HuR overexpression is accompanied by its accumulation in the cytoplasm (6, 7, 12). It is hypothesized that the ability of HuR to promote mRNA stabilization requires its translocation to the cytoplasm where it binds target ARE-containing mRNAs and interferes with their rapid decay (14, 15), however the mechanism by which cytoplasmic HuR mediates mRNA stabilization remains to be resolved.
Current work has demonstrated the ability of HuR to control microRNA (miRNA)-mediated post-transcriptional regulation. miRNAs are small non-coding RNAs approximately 21–24 nucleotides in length that primarily bind to the 3′UTR of targeted transcripts through imperfect base-pairing and this interaction is most often associated with regulated expression of the target mRNAs through mRNA degradation and/or translational suppression (16). More recently, HuR has been demonstrated to rescue an ARE-containing mRNA from miRNA-mediated regulation in response to stress (17). In subsequent studies, HuR has further been shown to inhibit translation of specific mRNAs by promoting the recruitment of repressor miRNAs to the targeted 3′UTR (18, 19). Through these contrasting roles, HuR is becoming recognized as a key factor influencing miRNA-mediated regulation (20, 21).
In this report, we demonstrate that the ARE-containing 3′UTR of COX-2 is targeted by the miRNA miR-16 leading to down regulation of COX-2 expression by altering mRNA stability. However, the ability of miR-16 to control COX-2 expression was compromised in the presence of elevated HuR. The ability of HuR to antagonize miR-16 involved a direct HuR/miR-16 interaction that was dependent on the presence of a functional 3′UTR ARE and resulted in decreased miR-16 levels in CRC cells and tumors. These findings offer what we believe are new insights into mechanisms that govern COX-2 regulation in CRC and coordinated crosstalk between ARE-mediated decay and miRNA pathways critical for post-transcriptional regulation of cancer-associated gene expression.
Materials and Methods
Cell culture, DNA transfection, siRNA and miRNA transfection
HeLa, CaCo2, HT-29, LoVo, LS174T, and SKCO1 were purchased from ATCC (Manassas, VA). HCA7 and Moser cells were kindly provided by S. Kirkland (Imperial College, London, UK) and R.D. Beauchamp (Vanderbilt University Medical Center, Nashville, TN), respectively. All cells were maintained in DMEM containing 10% FBS (Hyclone, Logan UT) except SKCO1, HCA7, and CaCo2 cell lines, which were maintained in MEM medium containing 10% (SKCO1, HCA7) or 20% (CaCo2) FBS. HeLa TetOFF/HuR-Flag and HeLa TetOFF/TTP-Flag cell lines were maintained in the presence 2 μg/mL doxycycline (Dox) (Clontech, Mountainview, CA) as described (12); cells were grown in the absence of Dox for 48 hr to induce HuR-Flag or TTP-Flag expression. Where indicated, cells treated with the small molecule inhibitor specific for HuR, MS-444 (20 μM) (22) for the indicated times.
Transient transfections of cells with luciferase reporter constructs containing the COX-2 3′UTR (10) or 1.8-kb COX-2 promoter (23) were accomplished using Lipofectamine Plus (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Where indicated, cells were transfected with the HuR expression vector pcDNA3-HuR-Flag or empty vector (6). After 3 hrs, the media was changed and cells were sequentially transfected with miRNA for 48 hrs. MicroRNA and siRNA transfection of cells using 50 nM hsa-miRNA-16 mature miRNA duplex, random sequence negative control miRNA #2, pre-designed siRNAs for COX-2, HuR, and negative control #1 siRNA (Ambion, Austin, TX) were performed using siQuest (Mirus, Madison, WI) for 48 hrs according to the manufacturer’s instructions. Where indicated, HeLa cells were stimulated with 10 ng/mL IL-1β (R&D Systems) for 24 hr prior to miRNA transfection.
RNA analysis
Total RNA was extracted using Trizol reagent (Invitrogen). Northern blotting was performed as described (10) and probed with P32-labeled DNA probes synthesized for COX-2 and actin (Promega, Madison, WI). Complementary DNA (cDNA) synthesis was performed using 1 μg of total RNA in combination with oligo(dT) and Improm-II reverse transcriptase (Promega, Madison, WI). Reverse-transcribed PCR (RT-PCR) was performed using primers for COX-1 sense 5 ′-ACCTTCATCCGAGAGATGCTC-3′ and antisense, 5′-TGACGCTCCAGATTGTCTCCA-3′ (407 bp product); COX-2 sense 5′-GTCACAAGATGGCAAAATGCTG-3′ and antisense 5′-TAAGATAACACTGCAGTGGCTC-3′ (500 bp product); and GAPDH sense 5′-CCACCCATGGCAAATTCCATGGCA-3 ′ and antisense 5 ′-TCTAGACGGCAGGTCAGGTCCACC-3′ (598 bp product). PCR was performed using 25 cycles of denaturation at 95°C for 45 sec, annealing at 50°C for 1 min, and extension at 72°C for 1 min. qPCR analysis was performed as described (12) using the 7300 PCR Assay System (Applied Biosystems, Foster City, CA) with Taqman probes for COX-2 and GAPDH (PTGS2, GAPDH; Applied Biosystems) and SYBR green PCR master mix (Applied Biosystems). The following primers were used in qPCR reactions: Luciferase; sense 5 ′-ACGGATTACCAGGGATTTCAGTC-3 ′ and antisense 5 ′-AGGCTCCTCAGAAACAGCTCTTC-3 ′, BCL-2; sense 5 ′-ATGTGTGTGGAGAGCGTCAA-3′ and antisense 5′-ACAGTTCCACAAAGGCATCC-3′, VEGF, sense 5 ′-TACCTCCACCATGCCAAGTG-3 ′ and antisense 5 ′-AAGATGTCCACCAGGGTCTC-3 ′, and Cyclin E1; sense 5 ′-ATCCTCCAAAGTTGCACCAG-3′ and antisense 5′-AGGGGACTTAAACGCCATT-3′. GAPDH was used as a control for normalization. mRNA decay experiments were initiated by adding actinomycin D (ActD, 5 μg/mL) (Fischer Scientific, Fair Lawn, NJ) to the growth medium at specified times. For mRNA half-life assays in the presence of both HuR and miR-16, cells were co-transfected with COX-2 3′UTR reporter plasmid and pcDNA3-HuR-Flag for 3 hrs, followed by transfection with miR-16 for 48 hr. ActD treatment was performed as described above.
For endogenous miRNA detection, 10 ng of total RNA was converted to cDNA using the Taqman microRNA reverse transcription kit (Applied Biosystems) with miRNA primers specific for mature hsa-miR-16, hsa-miR-15a, and the small nuclear protein RNU6B (U6) control for normalization (Applied Biosystems). qPCR detection of miRNAs was performed using Taqman probes designed for miR-16, miR-15a, U6, and 18S (Applied Biosystems).
Protein and PGE2 analysis
Western blots were performed as described (12) using antibodies against COX-2 (160112; Cayman Chemical Company, Ann Arbor, MI), COX-1 (H-62; Santa Cruz Biotechnology, Santa Cruz, CA), HuR (3A2; Santa Cruz Biotechnology), nucleoporin (p62, BD Biosciences, San Jose, CA), and α-tubulin (DMA1; Sigma, St. Louis, MO). Membranes were stripped and re-probed using β-actin antibody (Clone C4; MP Biomedicals, Solon, OH). Detection and quantitation of blots were carried out as previously described (10).
Cells transfected with luciferase reporter constructs were lysed in reporter lysis buffer (Promega, Madison, WI) and assayed using the Luciferase Assay System (Promega). Reporter gene activities were normalized to total protein; all results represent the average of triplicate experiments. Where indicated, pGL3-Basic and pGL3-Enhancer vectors were used as controls.
Prostaglandin E2 (PGE2) levels in culture media were analyzed by ELISA (R&D Systems, Minneapolis, MN). Where indicated, cells were pre-treated with 10 μM NS-398 (Cayman) for 1 hr, after which the media was removed and cultures were incubated for 20 min with serum-free media containing 10 μM arachidonic acid (Cayman) in serum- free media. Relative PGE2 levels were normalized to total protein levels and are an average of three experiments.
Ribonucleoprotein immunoprecipitations
Immunoprecipitation of ribonucleoprotein complexes (RNP-IP) was performed as described (24). Briefly, 100 μL of protein A/G Plus agarose beads (Santa Cruz) were washed with NT2 buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40) and then coated with 30 μg anti-HuR antibody (3A2), anti-Argonaute 2 (AGO2) (C34C6; Cell Signaling, Danvers, MA), or control IgG diluted in 200 μL of NT2 buffer for 16 hr. Washed beads were incubated with equal amounts of cytoplasmic lysates obtained from cells lysed in polysome lysis buffer (20 mM Tris-HCl pH 7.6, 5 mM MgCl2, 150 mM NaCl, 1 mM DTT, 0.5% NP-40, 100U/mL RNasin (Promega), 0.2 mM PMSF, 10X Protease Inhibitor Cocktail (Sigma) for 30 minutes on ice followed by centrifugation for 30 minutes at 13,200 rpm, 4°C. NT2 buffer was added to a final volume of 1 mL and IP reactions were incubated for 16 hr at 4°C. Reactions were washed 5X with NT2 buffer and total RNA was isolated from immunoprecipitates using 1 mL Trizol per IP reaction after final wash and then used for cDNA synthesis. Analysis of mRNA and miRNA in RNP-IP samples was performed as described above.
In vitro miRNA binding assay
Direct binding of recombinant HuR to miR-16 was measured in homogeneous solution using 2D-FIDA (Fluorescence Intensity Distribution Analysis) as described previously (22, 25). Briefly, 5′TMR labeled miR-16 (5′TMR-UAGCAGCACGUAAAUAUUGGCG, Microsynth, Balgach, Switzerland) was incubated with increasing concentrations of recombinant full length HuR (prepared as described in (25)) in a buffer of PBS pH 7.2, 5 mM MgCl2, 0.2% (w/v) Pluronic F-127 (Invitrogen), 1mM alpha-methylene ATP for at least 20 min at RT. The fluorescence anisotropy was measured using 2D-FIDA, and the Kd was determined by fitting the fluorescence anisotropy data to an equation describing fluorescence anisotropy in dependence of 1:1 complex formation as derived from the law of mass action (25).
Human and mouse tissue samples
Human colon tumors and histologically normal tissue were obtained from surgical remnants from patients with colorectal cancer through the University of South Carolina Center for Colon Cancer Research Tissue Bank. The protocol was approved by the Institutional Review Board of the University of South Carolina. Tissue was snap-frozen in liquid nitrogen, and total RNA was isolated using Trizol from approximately 50 mg of tissue.
APCMin/+ mice 15 weeks of age obtained from the Center for Colon Cancer Research (University of South Carolina) were sacrificed and tumors were identified in small intestinal tissue. Tumor specimens were excised with scissors and pooled into 10 tumors per mouse while adjacent normal epithelium was scraped from the muscular layer. All tissue samples were snap-frozen and total RNA was isolated using Trizol.
Statistical analysis
The data are expressed as the mean +/− SEM. Student’s t-test was used to determine significant differences. P-values less than 0.05 were considered significant.
Results
miR-16 regulates COX-2 expression
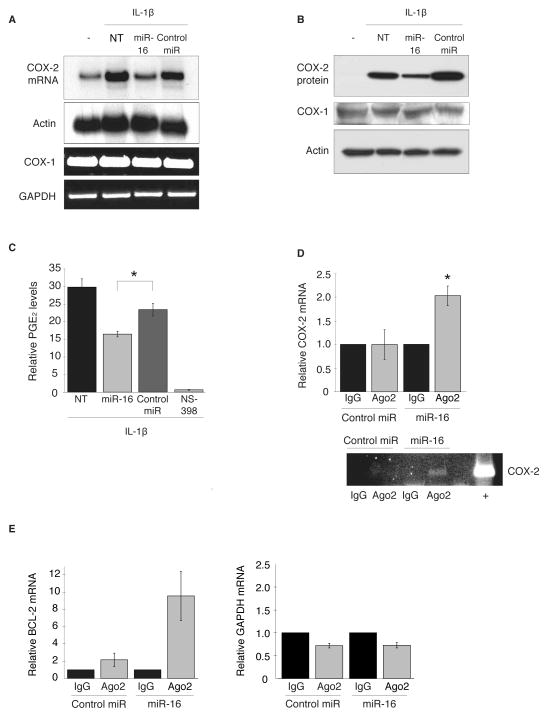
Our prior work had demonstrated the ARE present in the COX-2 3′UTR to be a key cis-acting element regulating expression levels on a post-transcriptional level (10). Based on previous work indicating miR-16 to be a pleiotropic regulator of ARE-containing mRNAs and its ability to target reporter constructs bearing the COX-2 3′UTR (26, 27), we sought to determine if miR-16 may be involved in ARE-mediated regulation of COX-2. The ability of miR-16 to regulate endogenous COX-2 expression was shown using HeLa cells stimulated with IL-1β to induce COX-2 expression for 24 hr and then transfected with mature hsa-miR-16 or a random sequence negative control miRNA for 48 hr. As seen in Fig. 1A, expression of miR-16 in IL-1β-treated HeLa cells reduced COX-2 mRNA approximately 2-fold. This miR-16-dependent loss of COX-2 mRNA was reflected in a similar ~2-fold decrease in COX-2 protein expression and associated prostaglandin E2 (PGE2) synthesis (Fig. 1B and C). The ability of miR-16 to attenuate PGE2 synthesis was consistent with specific targeting of COX-2, since expression of COX-1 mRNA or protein were not impacted by miR-16 (Fig. 1A and B).
Figure 1.
miR-16 regulates COX-2 expression. A. HeLa cells were treated with IL-1β for 24 hr to induce COX-2 expression prior to being transfected with mature miR-16 or control miR for 48 hr; untreated (−) and non-transfected (NT) cells are indicated. COX-2 and COX-1 mRNA was detected by northern blot and RT-PCT, respectively. Actin and GAPDH mRNA are shown as loading controls. Data shown represent 3 experiments. B. Western blot of COX-2 and COX-1 protein expression; actin served as a loading control. C. COX activity measured by PGE2 production in IL-1β-stimulated HeLa cells transfected with miR-16. Pretreatment of cells for 1 hr with 10 μM NS-398 was used as a positive control for COX-2-specific inhibition. Relative PGE2 levels were normalized to total protein levels and are an average of 3 experiments. (*) P = 0.023. D. HeLa cells were stimulated with IL-1β 24 hr prior to transfection with miR-16 for 48 hr. RNP-IP of Ago2 or control IgG was performed to isolate mRNAs associated with the RISC complex. qPCR was used to quantitate COX-2 mRNA levels normalized to the respective IgG control and performed in triplicate. (*) P = 0.008. RT-PCR for COX-2 in RNP-IP reactions in shown in bottom panel. Total RNA from IL-1β-treated HeLa cells was used as a positive RT-PCR control. E. RNP-IP of Ago2 or control IgG from HeLa cells and qPCR of a validated miR-16 target, BCL-2, served as a positive control, and GAPDH served as a negative control.
To establish if the effect of miR-16 on COX-2 expression was mediated through a direct miRNA:mRNA association, ribonucleoprotein immunoprecipitation (RNP-IP) was performed to determine whether COX-2 mRNA would associate with RISC in a miR-16 dependent manner. IL-1β-treated HeLa cells were transfected with miR-16 or control miRNA as described above and immunoprecipitation of cytoplasmic lysates was performed using an antibody against Argonaute 2 (Ago2), a major component of the RISC complex (16). The association of COX-2 mRNA with Ago2 was assayed by qPCR of COX-2 mRNA in immunoprecipitates. As shown in Fig. 1D, COX-2 mRNA was significantly enriched in the Ago2 immunoprecipitation samples where miR-16 was expressed. These results were validated by assaying for an established target of miR-16, BCL-2, in Ago2 immunoprecipitates where miR-16 was expressed (28). Shown in Fig. 1E, BCL-2 mRNA was enriched in miR-16/Ago2 immunoprecipitates, whereas capture of the negative control GAPDH mRNA was unchanged. Taken together, these results indicate that miR-16 facilitates endogenous COX-2 mRNA association with RISC leading to regulation of COX-2 mRNA and protein overexpression.
miR-16 targets the COX-2 3′UTR to promote rapid decay
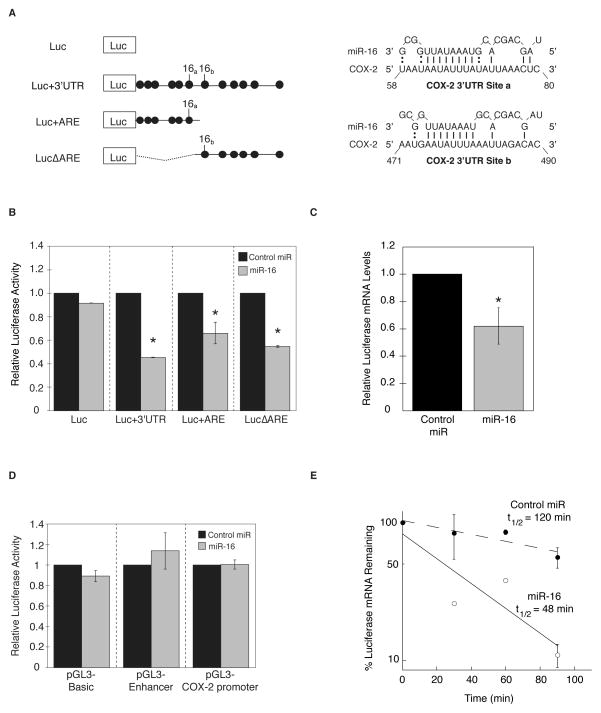
To further investigate the relationship between miR-16 and COX-2 expression, luciferase reporter constructs bearing the full-length 1455 nucleotide COX-2 3′UTR (Luc+3′UTR), the conserved ARE region of the 3′UTR (Luc+ARE), or the ARE region deleted from the 3′UTR (LucΔARE) were utilized (10) (Fig. 2A). Two potential miR-16 binding sites were predicted within the COX-2 3′UTR (denoted miR-16a and miR-16b, Fig. 2A) using the 8 bp ARE-complementary sequence of miR-16 as a prerequisite for targeting AU-rich elements, as single AUUUA motifs are resistant to miR-16 mediated regulation (26). Reporter constructs were transfected in HeLa cells along with miR-16 for 48 hr. Shown in Fig. 2B, miR-16 inhibited luciferase expression ~2-fold in the presence of the full-length COX-2 3′UTR. Similar levels of inhibition were observed in the presence of a single miR-16 site indicating the ability of miR-16 to target both the miR-16a and miR-16b 3′UTR sites with similar efficiencies, and optimal inhibition was observed to occur in the presence of both miR-16 sites suggesting a synergistic effect of multiple miRNA sites. A ~2-fold reduction in luciferase mRNA was observed in the presence of miR-16, consistent with miR-16-dependent reduction of luciferase activity derived from the Luc+3′UTR reporter (Fig. 2C). To ensure that the effects of miR-16 were dependent on the presence of the COX-2 3′UTR, a reporter construct containing a 1.8-kb COX-2 promoter (23) was co-transfected into HeLa cells with either miR-16 or the negative control. Shown in Fig. 2D, overexpression of miR-16 had no effect on COX-2 promoter or control SV40 promoter/enhancer activity, indicating miR-16 targeting of COX-2 is 3′UTR-dependent and not a consequence of indirect transcriptional interference.
Figure 2.
miR-16 targets the COX-2 3′UTR and alters mRNA stability. A. Luciferase reporter constructs without the COX-2 3′UTR (Luc), or fused to the full-length COX-2 3′UTR (Luc+3′UTR), the COX-2 AU-rich element (Luc+ARE), or the ARE deleted from the full-length 3′UTR (LucΔARE). AUUUA sequence elements are denoted by filled circles and putative miR-16 binding sites are indicated. B. HeLa cells were co-transfected with 3′UTR reporter constructs and miR-16 (grey bars) or control miRNA (black bars) for 48 hr. Luciferase activity was normalized to total protein and is the average of 3 experiments. (*) P ≤ 0.05. C. HeLa cells were transfected with Luc+3′UTR reporter construct in addition to miR-16 (grey bars), or control miRNA (black bars). Cells were harvested for total RNA after 48 hr and luciferase mRNA was analyzed by qPCR. (*) P = 0.0444. D. HeLa cells were co-transfected with COX-2 1.8 kb promoter-luciferase reporter construct and miR-16 (grey bars) or control miRNA (black bars) for 48 hr. Control transfections containing the empty vector (pGL3-Basic) and a vector containing the SV40 enhancer (pGL3-Enhancer) are shown. E. HeLa cells were co-transfected with Luc+3′UTR in addition to control miRNA (black circles) or miR-16 (open circles) for 48 hr. ActD (5 μg/mL) was added at the indicated times and Luc mRNA decay was analyzed by qPCR using GAPDH for normalization.
Recent findings have implicated miRNA-mediated mRNA decay to be a predominant mechanism of miRNA-mediated regulation of gene expression (29) and the results shown in Fig. 1 indicate a decrease in COX-2 mRNA levels in the presence of miR-16. To determine whether miR-16-mediated COX-2 mRNA loss was due to elevated mRNA decay, HeLa cells co-transfected with Luc+3′UTR reporter construct and miR-16 were treated with actinomycin D (ActD) to halt transcription and determine mRNA half-life. miR-16 promoted accelerated decay of Luc+3′UTR mRNA, with >2-fold reduction in mRNA half-life as compared to control miRNA (Fig. 2E). The stability of the luciferase control vector was unaltered in the presence of miR-16 or control miR (data not shown). These results substantiate that targeted down regulation of COX-2 by miR-16 occurs via the 3′UTR to promote enhanced mRNA degradation.
Altered expression of miR-16 is observed in CRC allowing for increased COX-2
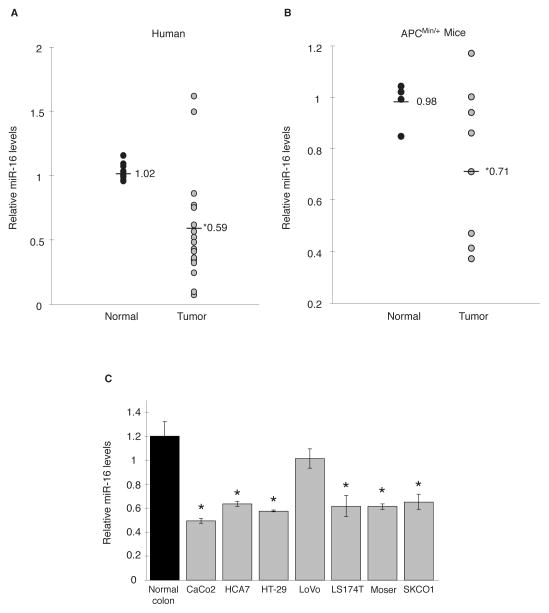
A characteristic feature observed in many human malignancies, including colorectal cancer, is dysregulated miRNA expression (30). Our findings indicating the ability of miR-16 to inhibit COX-2 expression suggested the possibility that miR-16 expression levels are altered during colorectal tumorigenesis and this may contribute to increased COX-2 expression in tumors. This was evaluated by assaying miR-16 expression in normal colon and tumor tissue. In 16 of 18 tumor samples decreased miR-16 levels were observed compared to normal tissue, averaging a 1.7-fold decrease in expression (Fig. 3A). To further validate these results, miR-16 levels were evaluated from normal small intestinal tissue and tumors isolated from an APCMin/+ mouse, a murine model of gastrointestinal cancer in which COX-2 is overexpressed (31). Indicated in Fig. 3B, mouse intestinal tumors display a significant 1.4-fold decrease in miR-16 expression. Decreased miR-16 expression was also observed CRC cell lines compared to normal colon tissue (Fig. 3C). Interestingly, LoVo cells do not exhibit a considerable difference in miR-16 expression, which may account for the limited COX-2 expression observed in this CRC cell line (6, 12).
Figure 3.
miR-16 expression is down regulated in intestinal tumors and CRC cells. A. Total RNA was isolated from human colon tumors and matched normal colonic tissue (n = 18) and assayed for miR-16 expression levels by qPCR. Detection of RNU6B (U6) served as an internal control. Relative miR-16 expression levels for tumors were normalized to their respective matched normal tissue. Average fold change between normal and tumor samples is shown. (*) P = 0.0002. B. Small intestinal tumors and normal tissue isolated from APCMin/+ mice was examined for miR-16 levels by qPCR and normalized to U6. Relative miR-16 levels were normalized to the normal tissue sample closest to the average of all normal samples. (*) P = 0.0381. C. Endogenous miR-16 expression in CRC cell lines CaCo2, HCA7, HT-29, LoVo, LS174T, Moser, and SKCO1 was compared the average of 5 normal colon tissue samples. (*) P ≤ 0.05.
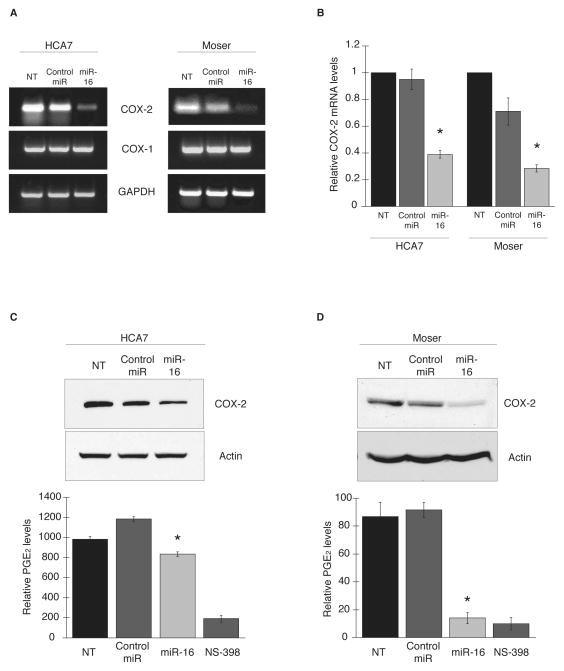
To determine the effects of miR-16 on COX-2 expression in CRC cells, the COX-2 overexpressing HCA7 and Moser lines (12) were transfected with miR-16 or control miRNA for 48 hr and examined for COX-2 mRNA expression. RT-PCR and qPCR analysis showed that COX-2 mRNA levels were attenuated approximately 2-fold in both cell lines in the presence of miR-16 compared to negative control while COX-1 expression levels remained unchanged (Fig. 4A and B). The ability of miR-16 to target COX-2 protein expression and PGE2 synthesis was also evaluated. Expression of miR-16 in HCA7 cells elicited a 2-fold decrease in COX-2 protein levels and a significant decrease in PGE2 levels compared to the negative control (Fig. 4C). Moser cells (Fig. 4D), appeared to be more responsive to miR-16 overexpression in that COX-2 protein levels were diminished in the presence of miR-16 and PGE2 levels were decreased to the extent of treatment of cells with the COX-2 inhibitor NS-398. Similar effects were observed in the CRC cell line HT-29 (data not shown).
Figure 4.
Overexpression of miR-16 targets COX-2 in colon cancer cells. CRC cell lines HCA7 and Moser were transfected with miR-16 or control miRNA for 48 hr. COX-2 and COX-1 mRNA expression was assayed by RT-PCR (A) and qPCR (B) using GAPDH as loading control. (*) P ≤ 0.05. C and D, COX-2 protein expression in miR-16-transfected HCA7 and Moser cells was examined by western blot; actin served as a loading control. Data shown represent 3 experiments. Respective PGE2 levels from culture media was assayed as described above. (*) P = 0.0405 for HCA7, P = 0.0289 for Moser.
Overexpression of HuR antagonizes miR-16 activity
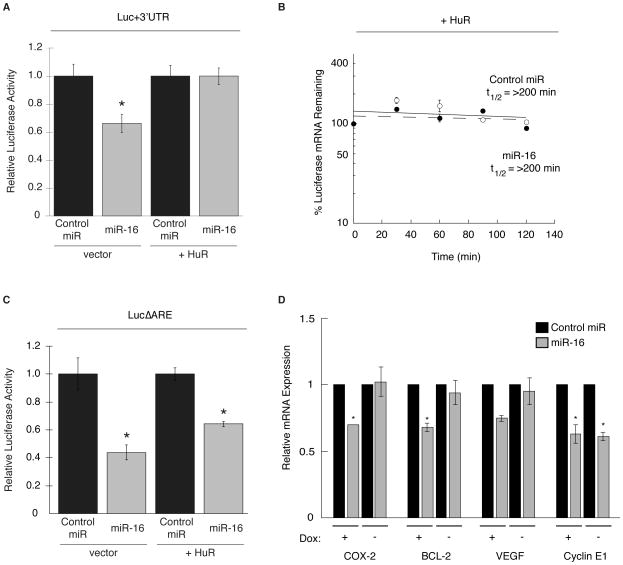
A common feature observed in colorectal tumors is elevated expression of the ARE-binding protein HuR, which is associated with overexpression of COX-2 (6, 7, 12, 32). Based on the proximity between the miR-16 and HuR binding sites within the COX-2 3′UTR, we sought to determine if overexpression of HuR could affect miR-16 targeting of COX-2. The Luc+3′UTR reporter construct was transfected into HeLa cells along with miR-16 and an HuR expression vector or empty vector control for 48 hr. Shown in Fig. 5A, miR-16 inhibited Luc+3′UTR activity to a similar extent as shown above (Fig. 2B). However, in the presence of elevated HuR, the ability of miR-16 to regulate Luc+3′UTR expression was abolished.
Figure 5.
Overexpression of HuR antagonizes the ability of miR-16 to target the COX-2 3′UTR. A. HeLa cells were co-transfected with Luc+3′UTR in addition to a vector expressing Flag-tagged HuR (+HuR) or empty vector, followed by transfection of miR-16 or control miRNA for 48 hr. Luc activity was normalized to the respective control and are the average of 3 experiments. (*) P = 0.0106. B. Luc+3′UTR mRNA decay in HeLa cells co-transfected with HuR expression vector and miR-16 (open circles) or control miRNA (black circles) for 48 hr prior to treatment with ActD at indicated times. Luc mRNA decay was assayed by qPCR and normalized to GAPDH. C. HeLa cells were transfected as described in (A) except the LucΔARE construct was used. Luc activity was normalized to the respective control and are the average of 3 experiments. (*) P ≤ 0.03. D. HeLa-Tet-OFF/HuR-Flag were grown in the absence of Dox for 48 hr to induce expression of HuR. After the first 24 hr of HuR induction, cells were stimulated with IL-1β for 24 hr followed by transfection of miR-16 (grey bars), or control miRNA (black bars). After 48 hr, COX-2, BCL-2, VEGF, and cyclin E1 mRNA levels were analyzed by qPCR using GAPDH for normalization. (*) P ≤ 0.05.
As shown in Fig. 2, miR-16 can accelerate COX-2 mRNA decay through the 3′UTR. To determine if HuR overexpression might interfere with this process, HeLa cells were transfected with Luc+3′UTR, miR-16, and the HuR expression vector. Cells were then treated with ActD, and Luc+3′UTR mRNA half-life was determined by qPCR. Shown in Fig. 5B, overexpression of HuR was shown to stabilize Luc+3′UTR mRNA (t1/2 > 200 min) in the presence of miR-16 or control miRNA. Experiments using Luc control vector lacking the 3′UTR showed no effect on Luc mRNA stability by HuR or miR-16 (data not shown).
To determine if the HuR binding site contained within the functional COX-2 ARE (10) was necessary for HuR-mediated inhibition of miR-16, the LucΔARE reporter construct was transfected into HeLa cells along with miR-16 and HuR expression vector for 48 hr. In contrast to the Luc+3′UTR vector, miR-16 was capable of attenuating LucΔARE activity regardless of HuR expression (Fig. 5C), suggesting that HuR binding to the COX-2 ARE is needed for its miR-16 inhibitory effect.
Evaluation of HuR’s suppression of miR-16 function upon other target mRNAs was examined using a Tet-responsive HuR-inducible HeLa cell line (HeLa-Tet-OFF/HuR-Flag) to overexpress HuR (12). In these cells, the removal of Dox from the medium allows for overexpression of HuR that is observed in both the nucleus and the cytoplasm (Supplementary Fig. 1). Cells were grown in the presence or absence of Dox to control HuR overexpression and then stimulated with IL-1β for 24 hr followed by transfection of miR-16 or control miR. As shown in Fig. 5D, miR-16 target mRNAs COX-2, BCL-2 (28), VEGF (33), and Cyclin E1 (34), displayed attenuated mRNA expression in miR-16 transfected cells in the absence of HuR overexpression. When HuR was overexpressed, inhibition of miR-16 function was observed with COX-2, BCL2, and VEGF transcripts but not with cyclin E1. Interestingly, COX-2, BCL2, and VEGF mRNAs all contain 3′UTR ARE motifs, whereas cyclin E1 mRNA does not contain an ARE (35), further supporting that ARE-binding by HuR to the same message in cis is needed for inhibition of miR-16.
Overexpression of HuR attenuates miR-16 expression
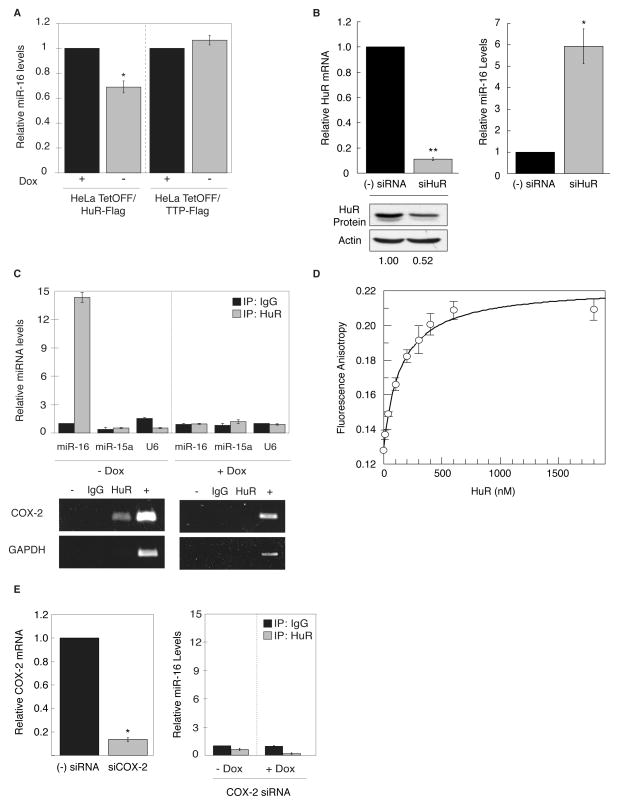
To better understand the basis of the observed inhibition of miR-16 by HuR, we determined whether miR-16 levels were altered by HuR overexpression. HeLa-Tet-OFF/HuR-Flag cells were grown in the absence of Dox to induce expression of HuR and miR-16 levels were quantified by qPCR. As shown in Fig. 6A, overexpression of HuR led to a substantial decrease in miR-16 levels. This loss of miR-16 was not observed with overexpression of the ARE-binding protein TTP using Tet-responsive TTP-inducible HeLa cells (12), supporting a specific effect of HuR in promoting miR-16 reduction. To determine if this effect on miR-16 is observed when HuR expression is attenuated, HCA7 cells were transfected with HuR siRNA for 72 hr and miR-16 levels were quantified by qPCR. As shown in Fig. 6B, a 2-fold reduction in HuR protein levels in response to HuR siRNA resulted in a > 5-fold increase in miR-16 levels. Additionally, various CRC cell lines were examined to determine if endogenous HuR overexpression and cytoplasmic localization associated with decreased miR-16 levels. CaCo2, HCA7, Moser, and HT-29 cells all display nuclear and cytoplasmic localization of HuR consistent with HuR overexpression (12), whereas nuclear localization was observed in LoVo cells (Supplemental Fig. 2). Consistent with HuR’s localization, miR-16 levels were attenuated in all CRC cell lines, except in LoVo cells (Fig. 3C), further supporting that HuR overexpression is needed to regulate miR-16 levels.
Figure 6.
Overexpression of HuR attenuates miR-16 expression. A. HeLa-Tet-OFF/HuR-Flag and HeLa-Tet-OFF/TTP-Flag cells were grown in the absence of Dox for 48 hr to induce expression of HuR or TTP. Relative miR-16 levels were determined by qPCR using U6 as loading control and normalized to miR-16 levels in the absence of HuR or TTP overexpression. Data shown are the average of 3 experiments. (*) P = 0.0028. B. HCA7 cells were transfected with siRNA against HuR (grey bars) or control siRNA (black bars) for 72 hr. Relative HuR mRNA and miR-16 levels were determined by qPCR using 18S for normalization. (**) P = 0.0028, (*) P = 0.0259. Western blot of HuR protein expression after siRNA transfection is shown using loading control actin. C. HeLa Tet-OFF/HuR-Flag cells were grown in the absence of Dox for 48 hr to induce HuR expression. Cytoplasmic extracts were subjected to RNP-IP of HuR or control IgG to isolate miRNAs associated with HuR. qPCR was used to quantitate miR-16, miR-15, and U6 RNA levels using the miR-16/IgG control IP for normalization in each respective condition. Lower panel reflects respective RT-PCR of COX-2 and GAPDH mRNA present in HuR RNP-IP reactions. Positive (+) control is total RNA amplified in RT-PCR reaction. D. 5′TMR-labeled miR-16 was incubated with increasing concentrations of recombinant full length HuR. Fluorescence anisotropy was measured using 2D-FIDA and the affinity was determined by fitting the fluorescence anisotropy data to yield a Kd = 120 ± 9 nM. E. HeLa Tet-OFF/HuR-Flag cells were grown in the absence of Dox and transfected with COX-2 or control siRNA for 48 hr. COX-2 mRNA was assayed by qPCR using GAPDH for normalization. (*) P = 0.0001. Cytoplasmic extracts were subjected to RNP-IP of HuR or control IgG and qPCR was used to quantitate miR-16 present in immunoprecipitates; IgG control IPs were used for normalization.
Based on these results, we speculated whether miR-16 down regulation might involve a direct HuR:miR-16 interaction. To test this, cytoplasmic lysates were generated from Tet-responsive HuR-inducible cells grown in the presence or absence of Dox for 48 hrs and subjected to RNP-IP using anti-HuR antibody or control IgG to determine if HuR is bound to miR-16 when overexpressed and present in the cytoplasm. As shown in Fig. 6C, HuR associated with miR-16 only in immunocomplexes when HuR expression was induced (Dox (−) conditions). As a positive control, COX-2 mRNA was also detected by RT-PCR in HuR immunoprecipitates only when HuR was overexpressed (Fig. 6C). To evaluate the specificity of the HuR:miR-16 interaction, qPCR reactions were performed to detect control U6 snRNA and miR-15a (Fig. 6C). miR-15a is transcribed co-cistronically with miR-16 (26) and was not immunoprecipitated with HuR under any conditions indicating that HuR does not interact with the unprocessed pri-miR-16-1/15a transcript. Pre-miR-16-1 was also analyzed in immunoprecipitates by qPCR analysis using precursor-miRNA-specific forward and reverse primers targeting the stem-loop sequence present in pre-miR-16-1, and was not detected in HuR immunoprecipitates, indicating that the co-immunoprecipitation reflects an interaction of HuR with mature miR-16 (data not shown). This interaction between HuR and miR-16 was also shown to occur in vitro using a fluorescence intensity distribution analysis (2D-FIDA) assay (22, 25), with a Kd of 120 nM observed between recombinant full length HuR and labeled miR-16 (Fig. 6D).
To determine if the presence of the ARE-containing COX-2 mRNA was a necessary component facilitating HuR:miR-16 interaction, HeLa-Tet-OFF/HuR-Flag cells were grown to induce HuR overexpression and transfected with a COX-2 or control siRNA, then assayed for the presence of miR-16 in HuR immunoprecipitates. As shown in Fig. 6E, the association between HuR and miR-16 was lost in samples where COX-2 mRNA was attenuated.
HuR:miR-16 interaction is dependent on HuR cytoplasmic localization
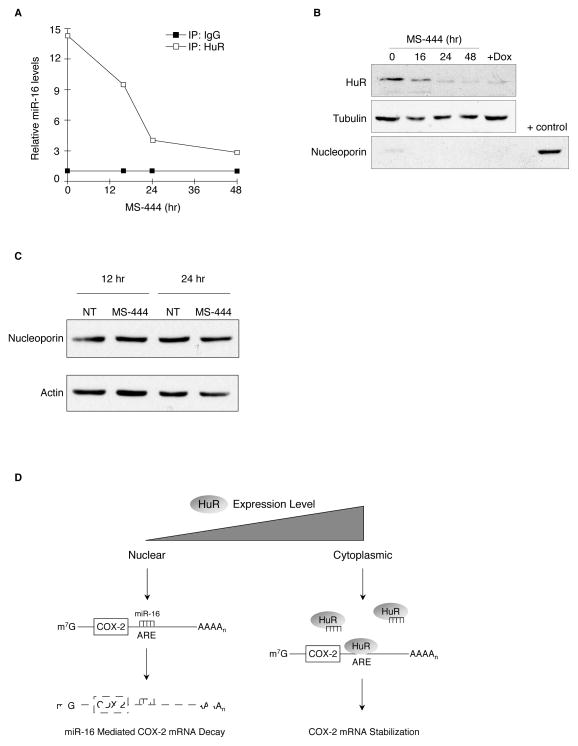
Since the interaction between HuR and miR-16 was observed only under conditions where HuR levels were elevated, we explored whether this interaction was dependent on HuR cytoplasmic localization. To accomplish this, HeLa Tet-OFF/HuR-Flag cells were grown in the absence of Dox to induce HuR expression, after which they were treated with a low-molecular weight inhibitor of HuR, MS-444. This compound has been previously shown to prevent the cytoplasmic trafficking of HuR by interfering with its homodimerization (22). Cytoplasmic lysates were obtained and subjected to HuR RNP-IP followed by qPCR analysis for miR-16. As depicted in Fig. 7A, treatment of cells with MS-444 resulted in a time-dependent decrease in the amount of miR-16 associated with HuR. Cellular fractionation further confirmed that the amount of HuR present in the cytoplasm decreased over time of compound treatment (Fig. 7B) and correlated with loss of miR-16 binding. As a control, total HuR and nucleoporin levels were assayed in cells treated with MS-444 to validate the observed changes in HuR cytoplasmic localization were not due to the loss of either HuR or nucleoporin (in preparation and Fig. 7C). These results indicate that heightened levels of cytoplasmic HuR can interact with mature miR-16 to promote its down regulation and this interaction can be reversed by the addition of an HuR inhibitor that prevents its cytoplasmic localization.
Figure 7.
HuR:miR-16 interaction is dependent on HuR cytoplasmic localization. A. HeLa Tet-OFF/HuR-Flag cells grown in the absence of Dox for 48 hr to induce HuR expression were incubated with 20 μM MS-444 at indicated times. RNP-IP of HuR or control IgG was performed. qPCR was used to quantitate HuR-associated miR-16 (solid squares) normalized to the respective control IP at each time point (open squares). B. Western blot of HuR present in cytoplasmic extracts of HeLa Tet-OFF/HuR-Flag cells after treatment with MS-444 as described in panel A. Tubulin and nucleoporin served as cytoplasmic and nuclear controls, respectively. C. Western blot of nucleoporin present in lysates of HeLa cells after treatment with 20 μM MS-444 for 24sand 48 hrs. Actin served as a loading control. D. Model of HuR inhibition of miR-16 targeting COX-2 mRNA. In normal cells, low levels of HuR are localized to the nucleus allowing for miR-16 targeting of the COX-2 ARE. In tumor cells, HuR is overexpressed and present in the cytoplasm where HuR direct binding of miR-16 prevents its interaction with the ARE. See text for details.
Discussion
Data obtained from several sources clearly demonstrates COX-2 overexpression to be a critical step contributing to various facets of CRC tumorigenesis (9), and growing evidence indicates that this overexpression is facilitated through loss of rapid ARE-mediated mRNA decay (11). Previously, we showed that elevated expression of the mRNA stability factor HuR in colon cancer promoted COX-2 mRNA stabilization by interfering with ARE-mediated decay (6, 12). The findings presented here identify miR-16 to be a key component in targeting COX-2 mRNA for rapid decay and overexpression of HuR directly interferes with this process. This effect was dependent on HuR being overexpressed and present in the cytoplasm, where it maintained the ability to bind both, the COX-2 mRNA and miR-16 and promote miR-16 down regulation as observed in CRC cells and tumors. Additionally, HuR appears to suppress miR-16 function in a mRNA ligand-dependent manner, in that a functional ARE was needed for interference of miR-16 function.
Normal cell growth is associated with rapid decay of ARE-containing mRNAs and selective targeting of these transcripts, which compose 5% to 8% of the human transcriptome (36), is an essential way of controlling their expression. However, a number of observations have indicated deficiencies in ARE-mediated decay occurs during neoplastic transformation of cells (8, 37) and current findings have demonstrated a 3- to 4-fold enrichment in ARE-containing genes to occur during CRC tumorigenesis (38). These observations can be explained in part by the results presented here and indicate ARE-mediated post-transcriptional regulation is an important regulatory mechanism lost during the early stages of CRC tumorigenesis.
An objective of this study was to further elucidate defects in COX-2 post-transcriptional regulation present in CRC in the context of microRNA-mediated regulation. miR-16 was previously determined to promote degradation of ARE-containing transcripts through complementarity to ARE motifs that was independent of canonical miRNA 5′ seed sequence commonly observed in many examples of miRNA-mediated regulation (26, 39, 40). Within the COX-2 3′UTR, there are 2 miR-16 binding sites that contain 8 bp homology to the miR-16 sequence UAAAUAUU previously determined to provide efficient targeting by miR-16 (26). Using 3′UTR reporter constructs, the two regions containing either miR-16 site maintained similar efficiencies in attenuating reporter expression by accelerating rapid mRNA decay, in agreement with work examining COX-2 3′UTR-mediated post-transcriptional regulation by miR-16 in response to diabetic stimuli in leukocytes (27). Furthermore, miR-16 was efficient in controlling endogenous COX-2 expression at both the mRNA and protein level, which subsequently resulted in reduced PGE2 levels in both IL-1β-stimulated HeLa and colon cancer cells. Interestingly, HCA7 cells appeared to be marginally resistant to miR-16 down regulation of COX-2 compared to other colon cancer cells. Previous work has shown that HCA7 cells possess a truncated COX-2 3′UTR variant resulting from alternative polyadenylation and lacks one of the two miR-16 binding sites (miR-16b) (41). Thus, the moderate sensitivity of HCA7 cells to miR-16 can be attributed to the presence of the truncated mRNA transcript, which was reported to be inherently more stable than the full-length transcript (41). These results demonstrate a central role for miR-16 in controlling COX-2 expression in varied physiological and pathological conditions.
A fundamental question regarding the role of miR-16 in CRC etiology pertains to identifying and understanding the factors that contribute to its loss in tumors. The results presented here demonstrate decreased miR-16 levels to be present in both human colorectal tumors and cell lines, along with loss in murine adenomas derived from the APCMin/+ mouse. These results are in agreement with those showing reduced miR-16 levels present in the colorectal microRNAome (42) and detection of lower miR-16 expression observed in the stool of advanced stage CRC patients (43). Based on these findings, we hypothesize that tumor-derived loss of miR-16 expression contributes to COX-2 overexpression observed during CRC tumorigenesis.
Various mechanisms have been reported that influence miR-16 expression in tumors. miR-16 is part of a polycistronic microRNA also encoding miR-15a located on chromosome 13q14. While genomic loss of this region in chronic lymphocytic leukemia results in miR-16 loss (44), this region is not an observed site of genomic alterations in CRC (45). Expression of miR-16 initiates from transcription of the DLEU2 promoter and overexpression of c-Myc can suppress miR-16 expression through repression of the DLEU2 promoter (46). Finally, miR-16 maturation can be modulated by p53 through its interaction with the microRNA processing complex involving Drosha and p68 (47), indicating that loss of p53 in CRC tumors may contribute to decreased mature miR-16 levels owing to altered miRNA processing.
The mRNA stability factor HuR is overexpressed in a variety of cancers, particularly CRC, and this overexpression is characterized with increased cytoplasmic localization (12, 13). In this context, HuR can interfere with ARE-mediated decay, however the manner in which HuR facilitates this process is still not understood. The findings presented here identify a new facet of this process in that HuR can antagonize miR-16 targeting of the COX-2 3′UTR and promote mRNA stabilization. Our data indicate a mechanism where HuR inhibition of miR-16 occurs by HuR directly binding to miR-16 and preventing its interaction with the COX-2 3′UTR (Fig. 7D). This association between HuR and miR-16 appears facilitated by nucleation at the COX-2 ARE, since deletion of the ARE allowed for miR-16 function through the site adjacent to the ARE (miR-16b) and HuR:miR-16 interaction was not detected when the COX-2 transcript was down regulated by siRNA. In support of this, HuR was observed to interfere with miR-16 targeting of other ARE-containing transcripts, whereas miR-16 was capable of regulating non-ARE-containing target cyclin E1 mRNA regardless of HuR expression status. More importantly, the ability of HuR to inhibit miR-16 was only apparent under conditions observed in CRC cells and tumors where HuR was overexpressed and located in the cytoplasm. Furthermore, inhibition of HuR cytoplasmic trafficking by MS-444 diminished its association with miR-16. Based on these observations, we hypothesize that a central event occurring during cellular transformation or in response to stress is the altered nucleocytoplasmic localization of HuR; the cytoplasmic presence of HuR allows it access to mature miR-16 at the COX-2 ARE ultimately leading to pathogenic mRNA stabilization.
Unexpected in this study were the results indicating HuR’s ability to promote miR-16 down regulation and that siRNA knockdown of HuR in CRC cells allows for increased miR-16 levels. While this suggests a much more active role of HuR in RNA and specifically miRNA metabolism, it remains to be seen how this newly found activity of HuR might connect to its recently discovered RNA 3′-modifying enzymatic activity (48). Whether through a direct enzymatic action or not, our data strongly suggest a novel mechanism describing how HuR overexpression interferes with ARE-mediated mRNA decay. By virtue of its cytoplasmic localization in tumor cells, HuR actively contributes to miR-16 elimination, allowing for unrestricted HuR binding to the COX-2 ARE. This ability of HuR to regulate miRNA levels when overexpressed extends to other miRNAs such as miR-30 and let-7a (unpublished observations), which may in part explain the observed loss of specific miRNAs in tumors. The findings reported in this study shed a new light on the role of HuR in connecting miRNA and ARE-mediated post-transcriptional regulation and indicate it to be a viable therapeutic target that can impact both mRNA and miRNA expression levels.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by the NIH grant R01CA134609 (D. Dixon) and the American Cancer Society Research Scholar grant RSG-06-122-01-CNE (D. Dixon).
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–14. e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 3.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 4.Anant S, Houchen CW, Pawar V, Ramalingam S. Role of RNA-binding proteins in colorectal carcinogenesis. Curr Colorectal Cancer Rep. 2010;6:68–73. doi: 10.1007/s11888-010-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon DA, Tolley ND, King PH, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez de Silanes I, Fan J, Yang X, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–54. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 8.Benjamin D, Moroni C. mRNA stability and cancer: an emerging link? Expert Opin Biol Ther. 2007;7:1515–29. doi: 10.1517/14712598.7.10.1515. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–8. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J Biol Chem. 2000;275:11750–7. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 11.Young LE, Dixon DA. Posttranscriptional regulation of cyclooxygenase 2 expression in colorectal cancer. Curr Colorectal Cancer Rep. 2010;6:60–7. doi: 10.1007/s11888-010-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–79. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdisciplinary Reviews: RNA. 2010;1:214–29. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keene J. Why is Hu where? Shuttling of early-response messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 18.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–94. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meisner NC, Filipowicz W. Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. Adv Exp Med Biol. 2010;700:106–23. [PubMed] [Google Scholar]
- 22.Meisner NC, Hintersteiner M, Mueller K, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3:508–15. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 23.Dixon DA, Tolley ND, Bemis-Standoli K, et al. Expression of COX-2 in platelet-monocyte interactions occurs via combinatorial regulation involving adhesion and cytokine signaling. J Clin Invest. 2006;116:2727–38. doi: 10.1172/JCI27209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. Embo J. 2004;23:3092–102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M, Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5:1432–47. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 26.Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–34. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 27.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J Biol Chem. 2008;283:36221–33. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1:11–21. doi: 10.1038/35094017. [DOI] [PubMed] [Google Scholar]
- 32.Denkert C, Koch I, von Keyserlingk N, et al. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol. 2006;9:1261–9. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 33.Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Q, Fu H, Sun F, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 2010;39:D66–9. doi: 10.1093/nar/gkq990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–4. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanies CL, Smith JJ, Kis C, et al. Oncogenic Ras and transforming growth factor-beta synergistically regulate AU-rich element-containing mRNAs during epithelial to mesenchymal transition. Mol Cancer Res. 2008;6:1124–36. doi: 10.1158/1541-7786.MCR-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 40.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetrapolin binds to the 3′ untranslated region of cyclooxygenase-2 mRNA: A polyadenylation variant in a cancer cell line lacks the binding site. J Biol Chem. 2003;278:13928–35. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 42.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed FE, Jeffries CD, Vos PW, et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6:281–95. [PubMed] [Google Scholar]
- 44.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staub E, Grone J, Mennerich D, et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol Cancer. 2006;5:37. doi: 10.1186/1476-4598-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 48.Meisner NC, Hintersteiner M, Seifert JM, et al. Terminal adenosyl transferase activity of posttranscriptional regulator HuR revealed by confocal on-bead screening. J Mol Biol. 2009;386:435–50. doi: 10.1016/j.jmb.2008.12.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.