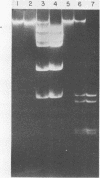
Abstract
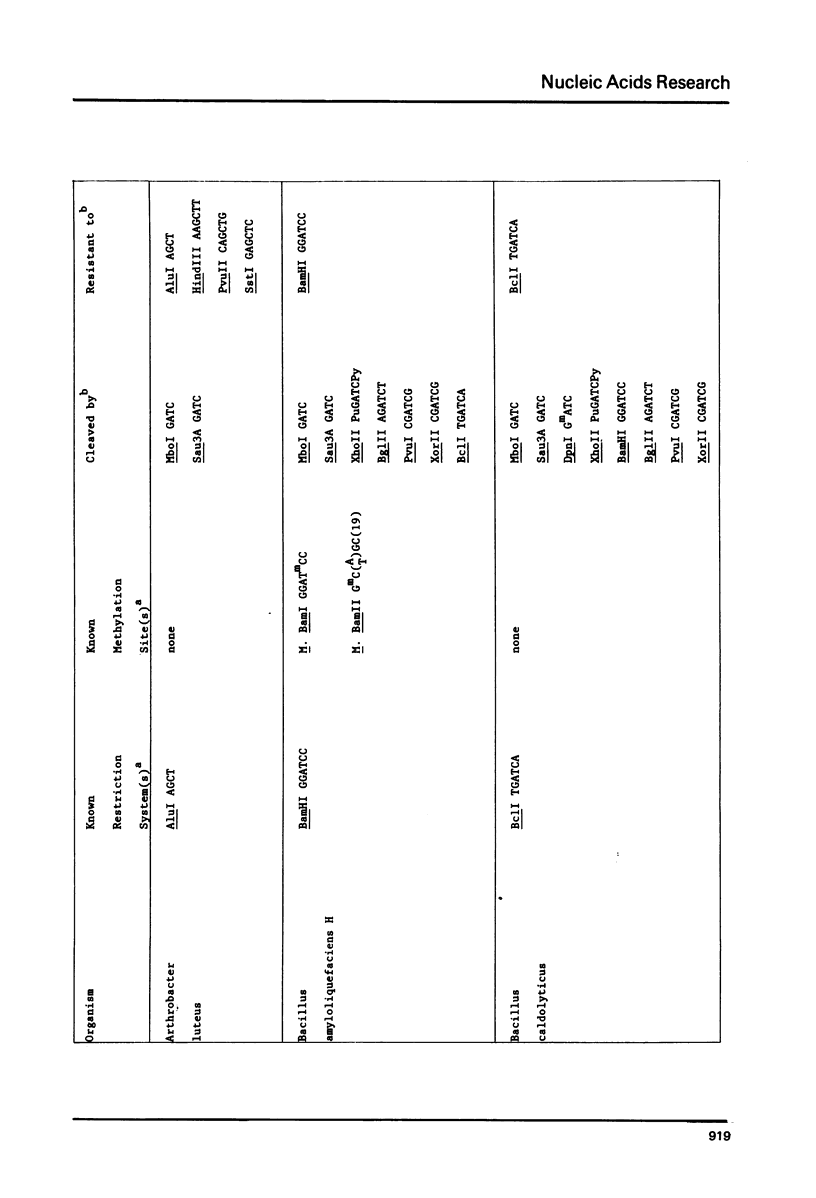
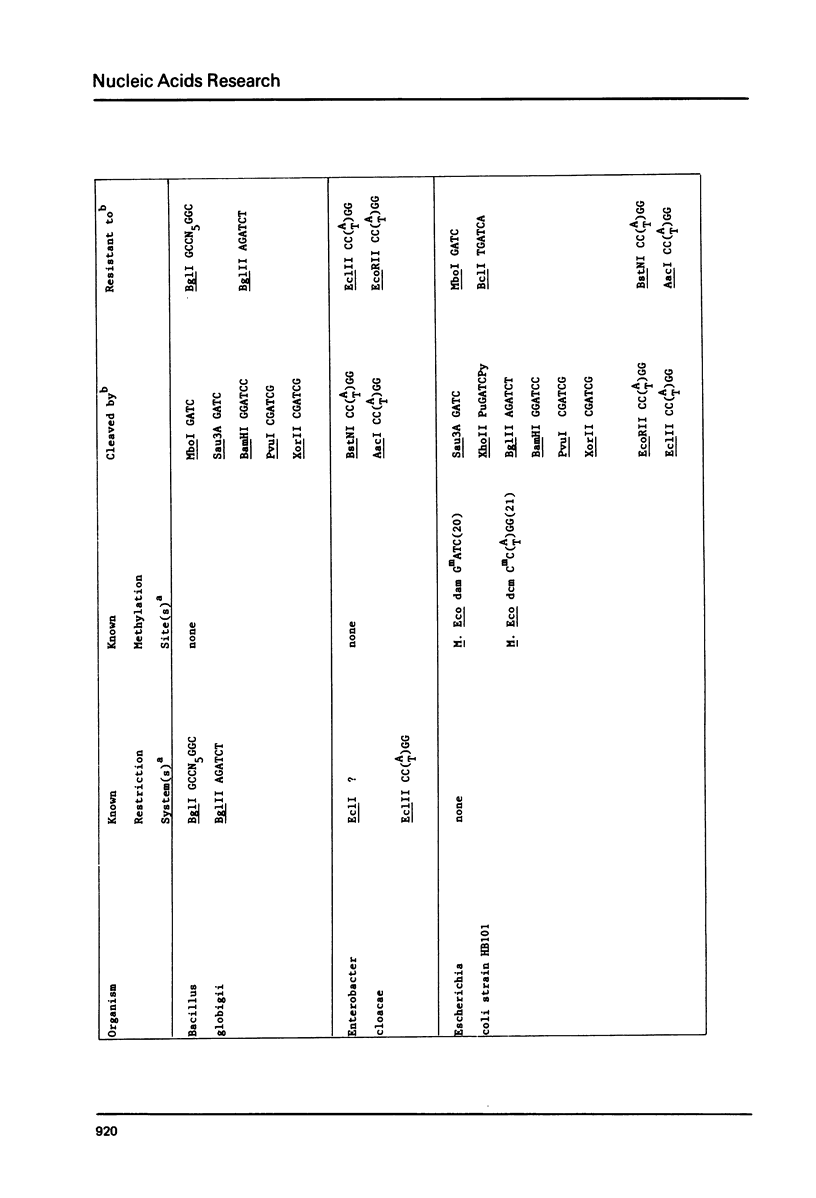
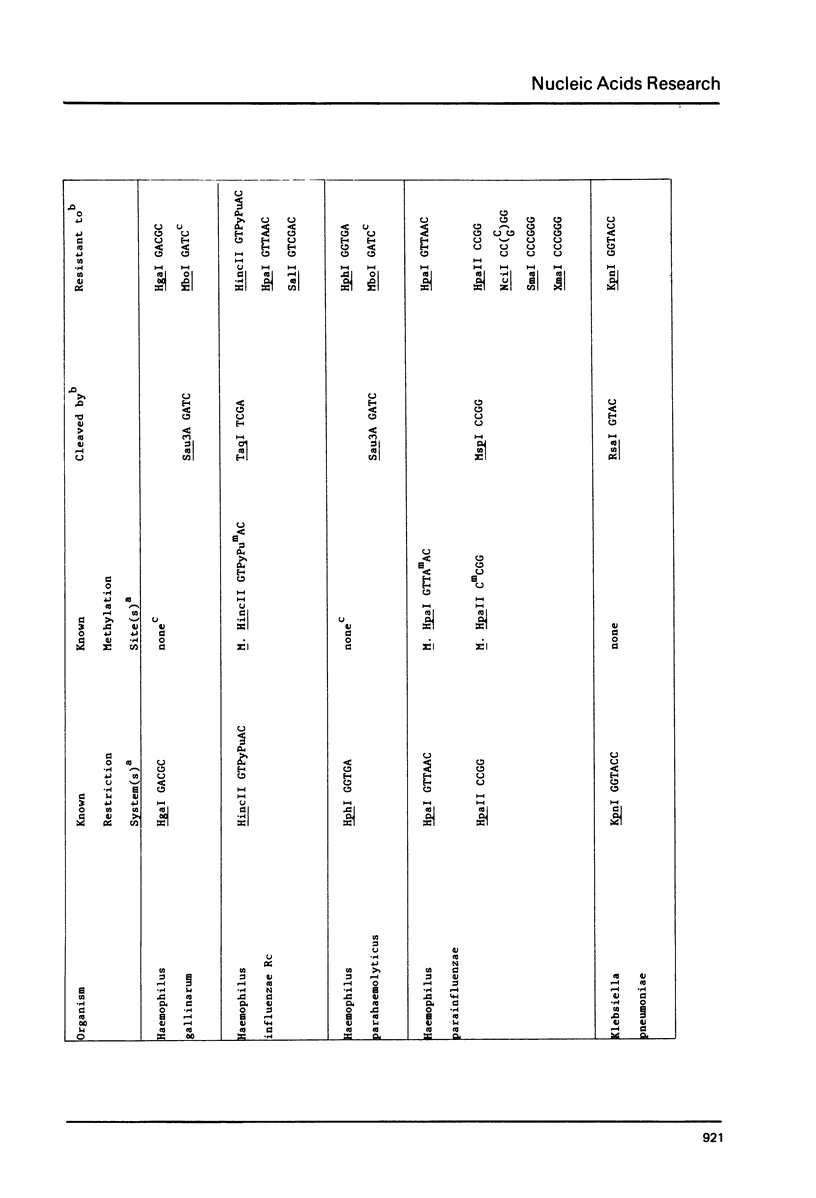
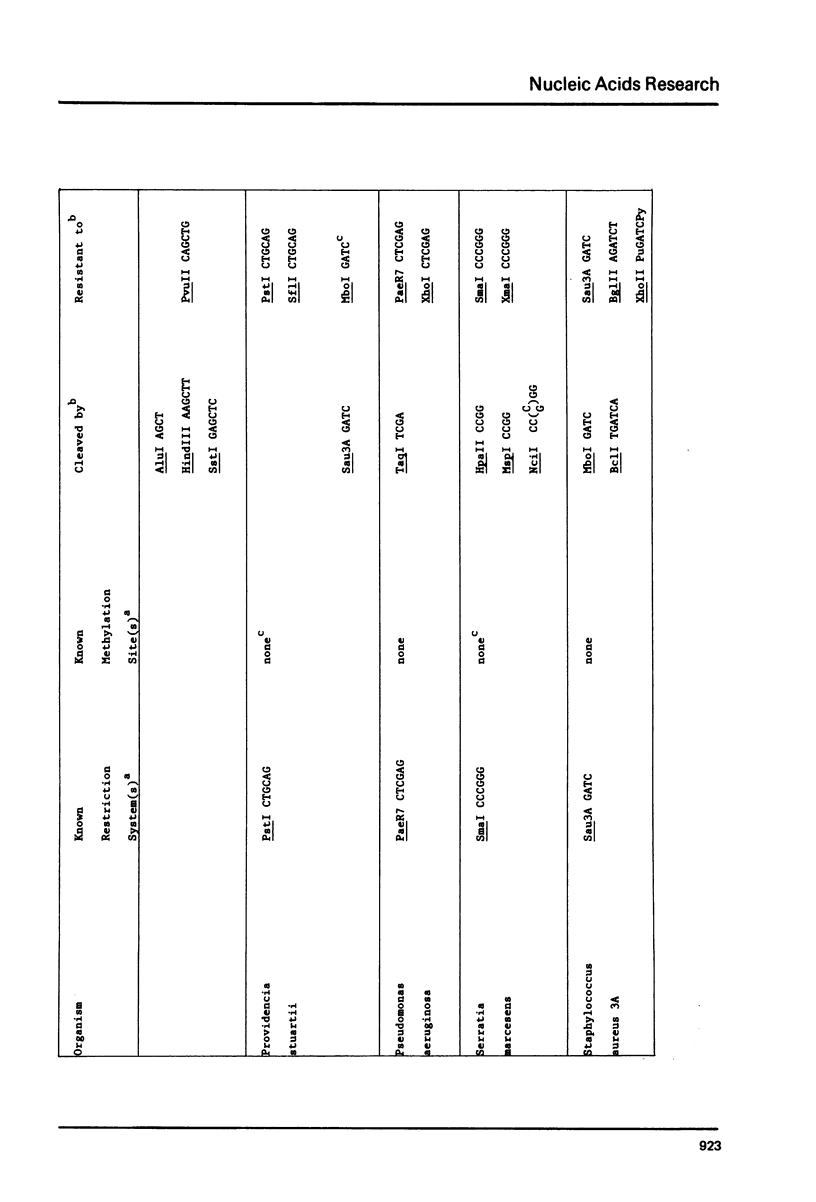
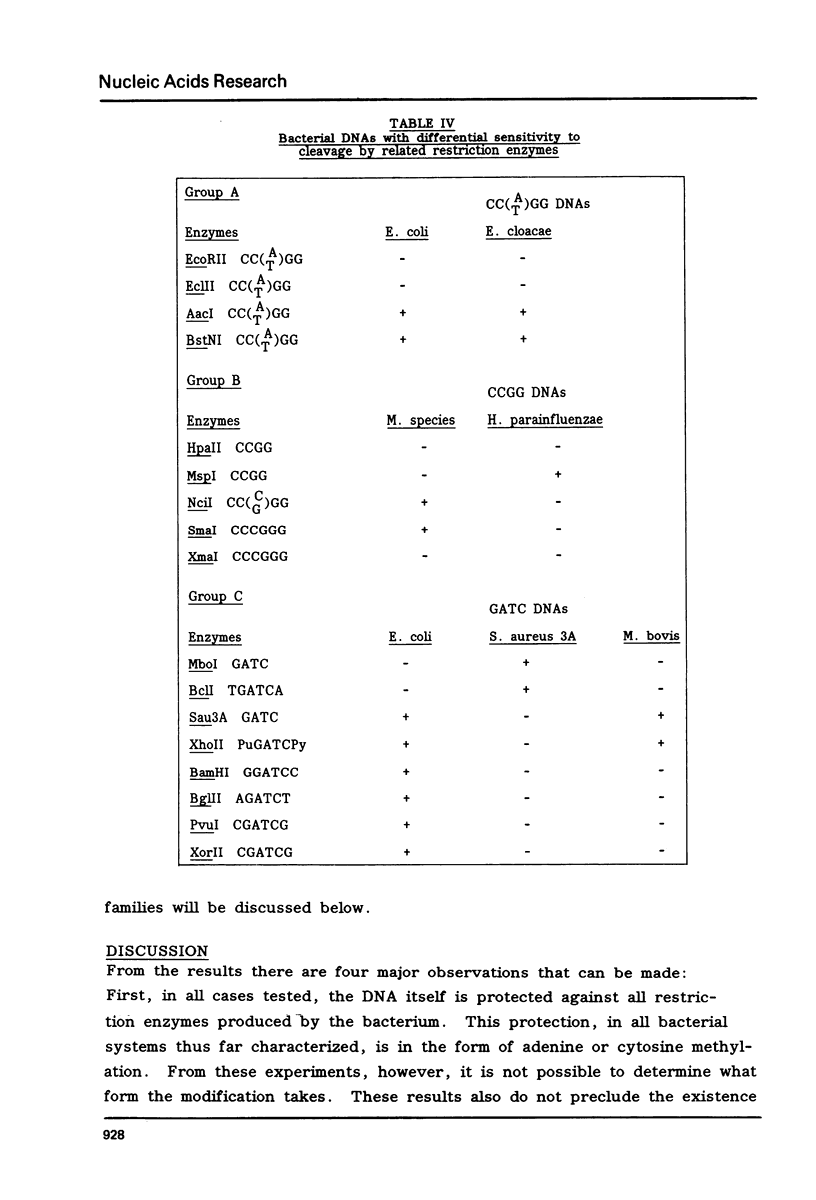
DNAs were prepared from twenty-six bacterial species and digested with a variety of restriction endonucleases to determine what modifications the DNAs carry. Several general conclusions could be made: 1) First, in no instance was the DNA of a restriction enzyme. 2) The specificity of the DNA modification was the same as that of its restriction counterpart; there were no cases of the DNAs being modified against a less specific class of restriction enzymes. 3) In most (but not all) cases, the resistance of a bacterium's DNA to its own restriction enzyme could be generalized to include resistance to all other restriction enzymes with the same specificity (isoschizomers). 4) DNA modified within the central tetramer of a recognition sequence is usually protected against cleavage by all related hexameric enzymes possessing that central tetramer. Only three families of DNA presented in this study disobey this rule. 5) Finally, a significant number of cases emerge where bacterial DNA carries a modification but no corresponding restriction endonuclease activity.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clewell D. B., Helinski D. R. Effect of growth conditions on the formation of the relaxation complex of supercoiled ColE1 deoxyribonucleic acid and protein in Escherichia coli. J Bacteriol. 1972 Jun;110(3):1135–1146. doi: 10.1128/jb.110.3.1135-1146.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S., Keister T., Gottehrer A. Sequence specificity of DNA methylases from Bacillus amyloliquefaciens and Bacillus brevis. J Mol Biol. 1978 Oct 5;124(4):701–711. doi: 10.1016/0022-2836(78)90178-x. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. 1975 Jun 10;250(11):4060–4066. [PubMed] [Google Scholar]
- Makino O., Saito H., Ando T. Bacillus subtilis-phage phi 1 overcomes host-controlled restriction by producing BamNx inhibitor protein. Mol Gen Genet. 1980;179(3):463–468. doi: 10.1007/BF00271735. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Rao R. N., Smith H. O. Cloning of restriction and modification genes in E. coli: the HbaII system from Haemophilus haemolyticus. Gene. 1978 Apr;3(2):97–112. doi: 10.1016/0378-1119(78)90054-9. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. S., Hattman S. Analysis of bacteriophage deoxyribonucleic acid sequences methylated by host- and R-factor-controlled enzymes. J Bacteriol. 1975 Aug;123(2):768–770. doi: 10.1128/jb.123.2.768-770.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. Structures and mechanisms of DNA restriction and modification enzymes. Q Rev Biophys. 1979 Aug;12(3):315–369. doi: 10.1017/s0033583500005461. [DOI] [PubMed] [Google Scholar]
- Nathans D., Smith H. O. Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Norlander L., Davies J. K., Hagblom P., Normark S. Deoxyribonucleic acid modifications and restriction endonuclease production in Neisseria gonorrhoeae. J Bacteriol. 1981 Feb;145(2):788–795. doi: 10.1128/jb.145.2.788-795.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1981 Jan 10;9(1):r75–r96. doi: 10.1093/nar/9.1.213-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. I. Purification and properties. J Mol Biol. 1973 Dec 25;81(4):427–444. doi: 10.1016/0022-2836(73)90515-9. [DOI] [PubMed] [Google Scholar]
- Sato S., Nakazawa K., Shinomiya T. A DNA methylase from Thermus thermophilus HB8. J Biochem. 1980 Sep;88(3):737–747. doi: 10.1093/oxfordjournals.jbchem.a133026. [DOI] [PubMed] [Google Scholar]
- Schrenk W. J., Weisberg R. A. A simple method for making new transducing lines of coliphage lambda. Mol Gen Genet. 1975;137(2):101–107. doi: 10.1007/BF00341676. [DOI] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Sneider T. W. The 5'-cytosine in CCGG1 is methylated in two eukaryotic DNAs and Msp I is sensitive to methylation at this site. Nucleic Acids Res. 1980 Sep 11;8(17):3829–3840. doi: 10.1093/nar/8.17.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J Mol Biol. 1975 May 15;94(2):283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- Stuy J. H. Restriction enzymes do not play a significant role in Haemophilus homospecific or heterospecific transformation. J Bacteriol. 1976 Oct;128(1):212–220. doi: 10.1128/jb.128.1.212-220.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Walder R. Y., Hartley J. L., Donelson J. E., Walder J. A. Cloning and expression of the Pst I restriction-modification system in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1503–1507. doi: 10.1073/pnas.78.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori R., Roulland-Dussoix D., Boyer H. W. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J Bacteriol. 1972 Dec;112(3):1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Annu Rev Biochem. 1981;50:285–319. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]