Abstract
Background
Inherited mutations in the BRCA2 gene greatly increase the risk of developing breast cancer. Consistent with an important role for BRCA2 in error-free DNA repair, complex genomic changes are frequently observed in tumors derived from BRCA2 mutation carriers. Here, we explore the impact of DNA copy-number changes in BRCA2 tumors with respect to phenotype and clinical staging of the disease.
Methods
Breast tumors (n = 33) derived from BRCA2 999del5 mutation carriers were examined in terms of copy-number changes with high-resolution aCGH (array comparative genomic hybridization) containing 385 thousand probes (about one for each 7 kbp) and expression of phenotypic markers on TMAs (tissue microarrays). The data were examined with respect to clinical parameters including TNM staging, histologic grade, S phase, and ploidy.
Results
Tumors from BRCA2 carriers of luminal and basal/triple-negative phenotypes (TNPs) differ with respect to patterns of DNA copy-number changes. The basal/TNP subtype was characterized by lack of pRb (RB1) coupled with high/intense expression of p16 (CDKN2A) gene products. We found increased proportions of Ki-67-positive cells to be significantly associated with loss of the wild-type (wt) BRCA2 allele in luminal types, whereas BRCA2wt loss was less frequent in BRCA2 tumors displaying basal/TNP phenotypes. Furthermore, we show that deletions at 13q13.1, involving the BRCA2wt allele, represents a part of a larger network of co-occurring genetic changes, including deletions at 6q22.32-q22.33, 11q14.2-q24.1, and gains at 17q24.1. Importantly, copy-number changes at these BRCA2-linked networking regions coincide with those associated with advanced progression, involving the capacity to metastasize to the nodes or more-distant sites at diagnosis.
Conclusions
The results presented here demonstrate divergent paths of tumor evolution in BRCA2 carriers and that deletion of the wild-type BRCA2 allele, together with co-occurring changes at 6 q, 11 q, and 17 q, are important events in progression toward advanced disease.
Introduction
Germline mutations in one allele of the BRCA2 tumor-suppressor gene confer greatly increased risk of developing breast cancer [1]. The BRCA2 gene is known to be involved in error-free DNA repair of double-strand breaks (DSBs) through homologous recombination (HR) [2]. Defects in this mechanism lead to repair of DSBs by error-prone nonhomologous end joining (NHEJ) or single-strand annealing (SSA). The use of error-prone mechanisms for DSB repair can result in genetic changes involving rearrangements and sequence modification at the damaged sites [3]. In this relation, it has been shown that chromosomal instability arises after inactivation of murine Brca2 gene, which is in agreement with complex chromosomal changes observed in human breast tumors derived from BRCA2 carriers [4,5]. Consistent with Knudson's two-hit hypothesis for tumor suppression, loss of the wild-type (wt) BRCA2 allele has been described as an essential event in tumor initiation in BRCA2 carriers [6]. This notion has, however, recently been questioned with data demonstrating that loss of Brca2 heterozygosity is not a required event in murine models of familial pancreatic cancer and, for human carriers of the BRCA2 999del5 mutation, that loss of heterozygosity (LOH) might not be needed to promote carcinogenesis in the pancreas [7]. In addition, breast cancers arising in BRCA1 and BRCA2 mutation carriers are heterogenic for LOH, suggesting haploinsufficient effects in pathogenesis [8].
Genome-wide expression analyses of breast cancers have led to the identification of biologically and clinically relevant subtypes (that is, luminal-A (LumA), luminal-B (LumB), basal-like, HER2, and normal-like) [9]. The identified subtypes have shown substantial differences with respect to copy-number changes, suggesting that their development adheres to changes in distinct sets of genes, leading to diverging tumor-evolution pathways [10,11]. Of these, the luminal subtypes comprise the majority of all cases (60% to 70%), both characterized by expression of GATA3-regulated genes. The LumB subtype differs from LumA by high expression of proliferation-associated genes with increased levels of MKI67 (Ki-67) as a prominent feature, and are associated with less-favorable disease outcome [12]. Basal-like breast cancers are characterized by expression of genes that pertain to the basal layer of cells in normal mammary glands, such as CK5/6 and CK14. The basal-like and HER2 subtypes are both highly proliferative, poorly differentiated, and, in concordance, show the least-favorable disease outcome [13]. Tumors arising in BRCA2 germline mutation carriers mostly display luminal phenotypes of high histologic grade and have prominent expression of proliferation markers, suggesting similarities to the LumB subtype [14]. Relatively few BRCA2 tumors have been profiled for copy-number changes, and have mostly been studied by using low-resolution array design, resulting in relatively crude patterns and lack of clarity in identification of key drivers. In our previous report, we found that BRCA2 tumors often show highly complex genomes coupled with a unique pattern of copy-number changes [15]. To follow up on these results, we have studied copy-number changes in a larger number of BRCA2 tumors by high-resolution array-comparative hybridization (aCGH) microarrays containing 385 thousand probes (about one for each 7 kbp) in breast tumors derived from BRCA2 germline mutation carriers (n = 33) combined with analysis of phenotypic markers by immunohistochemistry (IHC) on tissue microarrays (TMAs). The purpose of the study was to characterize the spectrum of copy-number changes in tumors obtained from BRCA2 carriers with respect to phenotype and clinical parameters.
Materials and methods
Study group
Tumor DNA samples (n = 33) were derived from patients carrying the 999del5 BRCA2 germline mutation [16]. The tumors were macroscopically examined by a pathologist to identify portions with invasive tumor cells. DNA extraction was performed on freshly frozen tumor portions by following protocols based on standard procedures by using phenol-chloroform and proteinase K. Clinical parameters, including TNM staging and histologic grade, were obtained from the Department of Pathology, Landspitali University, and are given in Additional File 1. Data on S phase and ploidy, obtained with flow cytometry, were available for 30 of the 33 breast tumors analyzed in this study. Data on patient relapse, that is, events of recurrences or metastases after diagnosis and treatment, were obtained from Landspitali University Hospital. This work was carried out according to permits from the Icelandic Data Protection Commission (2006050307) and Bioethics Committee (VSNb2006050001/03-16). Informed consent was obtained from all patients.
Array-comparative genomic hybridization
Copy-number changes were studied with high-resolution 385 K oligonucleotide CGH microarrays (Roche NimbleGen, Inc., Reykjavik, Iceland) containing 385 thousand probes covering the human genome with about one probe for each 7 kbp. DNA derived from tumor- and normal blood samples from the same individual were Cy3 and Cy5 labeled, respectively. Sample labeling and hybridizations were carried out according to protocols developed by the manufacturer (NimbleGen Arrays User's Guide-CGH Analysis; Roche NimbleGen, Inc., Reykjavik, Iceland). The aCGH data are available online from ArrayExpress [17] under the accession number E-TABM-1199.
Fine-tiling aCGH analysis was performed by using the 385 K oligonucleotide CGH microarrays covering chromosome 6 with about one probe for each 400 bp (Roche NimbleGen, Inc.), following the manufacturers protocols for sample labeling and hybridization (Roche NimbleGen).
Tissue microarrays and expression analysis by immunohistochemistry
TMAs construction and their application in expression analysis by immunohistochemistry (IHC) have been described previously [15]. Tumors were assigned to molecular subtypes by using the five-biomarker scheme described in Cheang et al. [13] by expression of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER-2), epidermal growth factor receptor (EGFR), and cytokeratin 5/6 (CK5/6). Expression of ER and PR were scored positive (IHC score ≥ 1+) when IHC staining was observed in more than 1% of tumor cell nuclei. HER-2 positivity was scored for strong membranous staining (IHC score 3+), whereas EGFR was scored positive for any, weak or strong, membranous staining (IHC score ≥ 1+). Expression of CK5/6 was defined as positive when cytoplasmic and/or membranous staining was observed and otherwise considered negative. Hormone-receptor-positive tumors, those positive for either ER or PR, were assigned to the luminal subtype. Tumors negative for ER, PR, and HER-2 were categorized as triple-negative phenotype (TNP), whereas ER- and PR-negative tumors displaying HER-2 positivity (IHC 3+) were assigned to the HER2 subtype. Basal-like phenotype was assigned to triple-negative tumors displaying expression of either EGFR or CK5/6. Estimates for the expression of Ki-67 (MKI67), cyclin-D1 (CCND1), and topoisomerase II alpha (TOP2A) gene products represent counts for the proportion of positive tumor cells, based on nuclear expression, reported on a scale from 0.0 to 1.0 within intervals of 0.05. Luminal tumors showing Ki-67 expression ≥ 0.14 were assigned to the luminal B subtype, whereas the remaining part was considered luminal A [12]. Expression of p16 (CDKN2A) and pRb (RB1) was scored on a discontinuous scale based on counts for the proportion of tumor cells showing nuclear expression (that is, 0 (< 10%), 1+ (10% to 25%), 2+ (25% to 50%), and 3+ (> 50%). The antibodies and dilutions used in IHC expression analysis are listed in Additional File 2.
TaqMan BRCA2 allele-specific quantitative PCR
The proportion of BRCA2 wild-type (wt) alleles was quantitatively measured relative to 999del5 BRCA2 mutated alleles in tumor DNA samples by quantitative polymerase chain reaction (PCR) (7500 Realtime PCR system; Applied Biosystems) with a TaqMan method by using a single BRCA2-specific, minor groove-binding probe (MGB-probe) 5'-end-labeled with FAM and a nonfluorescent quencher (NFQ) at the 3' end, a single BRCA2-specific forward primer, and two allele-specific reverse primers. Therefore, the PCR for wt and mutant alleles was performed in separate wells. The qPCR assay primers and TaqMan-MGB probe were as follows: Forward primer: 5'-CATGATGAAAGTCGTAAGAAA-3', Reverse primer (mut): 5'-CATGACTTGCAGCTTCTCTTTGTG-3', Reverse primer (wt): 5'-CATGACTTGCAGCTTCTCTTTGAT-3', TaqMan-MGB probe: 5'-TTTATCGCTTCTGTGACA-3'. Reactions were at a 12-μl final volume per well, each containing: TaqMan-MGB at 50 nM, forward and reverse primers at 0.5 nM each, 6 μl TaqMan Universal PCR Master Mix (Applied Biosystems, Pn. 4304437), and 0.25 μg of sample DNA, with H2O to final volume. The PCR: Step(S)1: 50°C for 2 minutes, S2: 95°C for 10 minutes, S3: 95°C for 15 seconds, S4: 60°C for 1 minute, with S3 and S4 were repeated for 40 cycles. The BRCA2 wt- to mutant-allele ratios were quantified by measuring differences in fluorescence intensity of FAM performed in duplicate, and the Ct values (number of cycles to reach intensity threshold) were averaged. Ct measures with standard deviation higher than 0.5 (SD > 0.5) were rejected and remeasured. The wild-type to mutant-allele ratios were calculated to wt/allele frequencies by the following equation:
where ΔCt = (Ct of allele1 - Ct of allele2) [18].
Statistical analysis and data mining of aCGH data
Preprocessing and segmentation of the aCGH data were carried out as described previously [15]. Normalized log2 ratios were replaced with corresponding segment means for each probe represented on the arrays to derive a segmented profile for each tumor. These data were used as input for SAMr (significance analysis of microarrays for R) in two-class or quantitative analysis, in which repeated permutations are used to address multiple testing [19].
Genetic networks represent a set of associated copy-number changes (links) between distinct regions (nodes) of the genome. Here, the average of segmented aCGH log2 ratios was computed for each of 811 cytobands with the nucleotide cytoband-locations downloaded by using the UCSC table browser [20]. The pair-wise cytoband-cytoband associations were then determined by hypothesis testing for Spearman's rho (Stats package in R). The P values for each of these comparisons were then adjusted for multiple testing by the Holm procedure (Multitest package in Bioconductor for R). Links were then established for associations with Padj < 0.001, while also having minimum event frequency of 25%. The associations, or "genetic links," represent nonrandom patterns of co-occurring copy-number changes. We then split the links into sets of inter- and intrachromosomal links involving associations between distinct chromosomes (inter), or associations between distinct arms on the same chromosome (intra), respectively. The resulting networks of co-occurring genetic changes were visualized as undirected node-and-edge graphs by making use of the Rgraphviz package in Bioconductor for R.
Results
Breast cancer subtypes in BRCA2 mutated tumors
Thirty-three female breast cancer patients carrying the same 999del5 BRCA2 germline mutation constituted the study group analyzed with array CGH (comparative genomic hybridization). The year of diagnosis was between 1989 and 1999, with the mean age of 50 years (standard deviation (SD) = 12.2) (Additional File 1). Of the 33 tumors analyzed with aCGH, 32 were available on tissue microarrays (TMAs). Expression analysis of subtype-specific biomarkers, that is, ER, PR, HER-2, Ki-67, CK5/6, and EGFR, was performed on TMAs with IHC. The results show that most BRCA2 tumors express luminal markers (24 of 32; 75.0%), of which the majority subcategorize as luminal B (16 of 24; 67%) rather than luminal A (eight of 24; 33.3%) (Table 1). Notably, none of the 32 BRCA2 tumors displayed overexpression or high-level amplification of the human epidermal growth factor receptor 2 (HER2) oncogene (Table 1). Of the eight hormone-receptor negative BRCA2 tumors, all but one was subcategorized as basal-like (seven of eight, 87.5%), of which four displayed EGFR expression, and five were positive for CK5/6 (Table 1).
Table 1.
Phenotypic characterization of breast tumors derived from BRCA2 germline mutation carriers
| Biomarkers for definition of phenotypes | 0 | I-II | III |
|---|---|---|---|
| ER | 8 (25%) | 7 (22%) | 17 (53%) |
| PR | 12 (37%) | 14 (44%) | 6 (19%) |
| HER2 | 22 (69%) | 10 (31%) | 0 (0) |
| Cellular proliferation; The proportion of cells positive for Ki-67 | |||
| Ki-67 | 0-14% (1st Qu.) | > 14%-35% (IQR) | > 35% (3rd Qu.) |
| Luminal | 8 (33%) | 14 (58%) | 2 (8%) |
| Triple-negative | 0 (0) | 3 (37%) | 5 (63%) |
| Χ2 test; P = 0.004 | |||
| Expression of basal features | |||
| EGFR | 0 | I | ≥ II |
| Luminal | 22 (96%) | 1 (4%) | 0 (0) |
| Triple-negative | 4 (50%) | 1 (13%) | 3 (37%) |
| Χ2 test; P = 0.004 | |||
| CK5/6 | Negative | Positive | |
| Luminal | 24 (100%) | 0 | |
| Triple-negative | 3 (37%) | 5 (63%) | |
| Χ2 test; P = 0.0003 | |||
| pRb/p16 pathway | |||
| p16 (CDKN2A) | 0 | I-II | III |
| Luminal A | 3 (38%) | 4 (50%) | 1 (12%) |
| Luminal B | 9 (60%) | 5 (33%) | 1 (7%) |
| Triple-negative | 0 | 1 (12%) | 7 (88%) |
| Χ2 test; P = 0.0007 | |||
| Cyclin-D1 (CCND1) | 0-15% (1st Qu.) | > 15%-75% (IQR) | > 75% (3rd Qu.) |
| Luminal A | 1 (12%) | 4 (50%) | 3 (38%) |
| Luminal B | 1 (6%) | 8 (50%) | 7 (44%) |
| Triple-negative | 6 (75%) | 2 (25%) | 0 |
| Χ2 test; P = 0.005 | |||
| pRb (RB1) | 0 (Negative) | I (Positive) | |
| Luminal A | 1 (13%) | 7 (87%) | |
| Luminal B | 2 (13%) | 14 (87%) | |
| Triple-negative | 4 (50%) | 4 (50%) | |
| Χ2 test; P = 0.085 | |||
| Morphologic features | |||
| Histologic grade | I | II | III |
| Luminal | 2 (8%) | 11 (44%) | 12 (48%) |
| Triple-negative | 1 (12%) | 1 (12%) | 6 (75%) |
| Χ2 test, P = 0.27 | |||
MKI67 (Ki-67) gene products tightly associate with cellular proliferation and are often used to estimate tumor growth rates [21]. Counting the number of tumor cells positive for Ki-67 against the total number of tumor cells reveals generally higher proliferation rates among triple-negative BRCA2 tumors compared with luminal BRCA2 tumors (χ2 test; P = 0.004; Table 1; Additional File 3). These differences coincide with high p16 (CDKN2A) (χ2 test; P = 0.0007), and low pRb (RB1) in the triple-negative subtype of BRCA2 mutated breast tumors (χ2 test; P = 0.085) (Table 1). Increased p16 expression levels were highly correlated with decreased pRb levels (Spearman correlation rho = -0.63; P = 0.0001). Accordingly, low pRb levels coupled with high p16, that is pRblow/p16high, was commonly found in triple-negative BRCA2 tumors (four of eight; 50%), but rarely in luminal BRCA2 tumors (two of 23; 8.7%) (χ2 test; P = 0.043) (Additional File 3). In contrast, the luminal subtypes of BRCA2 mutated breast tumors were associated with high expression of cyclin-D1 gene products (χ2; P = 0.005) (Table 1).
DNA copy-number changes with respect to phenotype in BRCA2-associated tumors
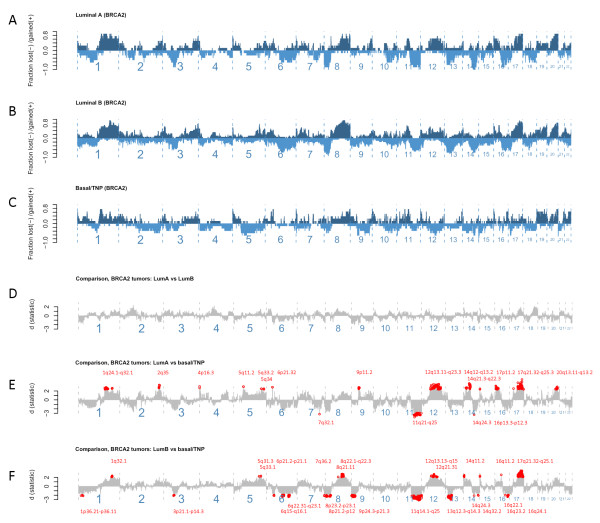
Event-frequency plots for copy-number changes in BRCA2 tumors displaying luminal A, luminal B, and basal/TNP describe the fraction of tumors within each class, showing gains or losses in order of genomic position (Figure 1a-c). Clearly, certain events occur more frequently in the luminal types of BRCA2 tumors compared with those of basal/TNP phenotypes and vice versa. An important observation in this respect is that some changes are inversely altered between the two phenotypic subgroups (that is, luminal and TNP/basal). To illustrate this point, it can be seen that changes over 12 q predominantly occur as gains in the luminal subtypes, whereas they mostly occur as deletions in the basal/TNP subtypes (Figure 1a-c). Detailed comparison between each phenotypic subclass of BRCA2 tumors shows that luminal A and B do not differ (Figure 1d). However, substantial differences were revealed between either luminal A or B when compared with TNP/basal types of BRCA2 tumors (Figure 1e, f). Some of these differences probably reflect phenotype-related effects independent of BRCA2 abnormalities (for example, the events at 5 q and 8 q have been described with respect to phenotype in sporadic tumors elsewhere [10,22]). However, other differences may relate to BRCA2 defects; for example, deletions at 13q13.1, covering the BRCA2 gene locus, were highly prominent in BRCA2 tumors of luminal B types compared with those displaying basal/TNP features (Figure 1f).
Figure 1.
Event-frequency plots for copy-number alterations and comparison between phenotypic subgroups of BRCA2 tumors. (a) The proportion of gains (positive, dark blue) and deletions (negative, light blue) plotted on y-axis for each genomic location ordered from p- to q-arm on x-axis left to right in BRCA2 tumors displaying luminal A (n = 8), and (b) luminal B (n = 16), and (c) basal/TNP phenotypes (n = 8). (d) Comparison in copy-number alterations with significance levels (d) shown on y-axis and differences below a false-discovery rate of 0.05 indicated in red (cytoband locations) derived from comparisons between BRCA2 tumors displaying luminal A and luminal B phenotypes, (e) between BRCA2 tumors displaying luminal A and basal/TNP, and (f) between BRCA2 tumors displaying luminal B and basal/TNP.
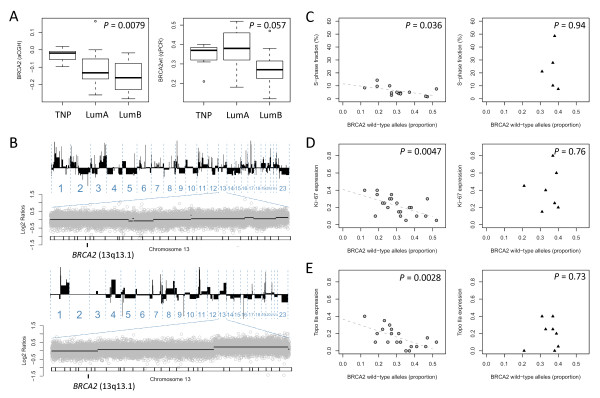
Given the importance of the BRCA2 locus in this study, we specifically explored the segmented aCGH log2 ratios covering the BRCA2 gene along with analysis of allele-specific BRCA2 loss with qPCR analysis. These methods showed strong concordance (that is, changes in aCGH log2 ratios at the BRCA2 locus significantly correlate with estimates of BRCA2 wild-type allele proportions by qPCR; Pearson correlation r = 0.69; P = 0.000025). Looking separately at each of the two BRCA2 phenotypic subgroups (luminal and basal/TNP), a clear trend toward deletion at the BRCA2 locus emerged for the luminal types of BRCA2 tumors, in particular, in the more-proliferative luminal B type (Figure 2a). Two individual cancer genomes analyzed by aCGH are shown to illustrate lack of deletion events affecting the BRCA2 gene locus at 13q13.1 in the TNP subtype of BRCA2 tumors (Figure 2b). To examine this further, we studied the relation on a continuous scale with markers of cellular proliferation that are S phase by flow cytometry, along with expression of proliferation-associated genes measured on a semicontinuous scale that is, Ki-67, and Topo IIα. Estimates for S-phase fractions, indicative of cells undergoing DNA replication, were found to be associated with decreased proportions of BRCA2 wild-type alleles in BRCA2 tumors of the luminal phenotype (Pearson correlation, r = -0.56; P = 0.036) (Figure 2c). This relation was not found in BRCA2 tumors displaying basal/TNP features, wherein very high proliferation rates are attained independent of deletion events over the BRCA2 locus at 13q13.1. Expression of Ki-67, as a measure of cellular proliferation, has the benefit of enabling stromal cells to be ignored, which is more difficult in analysis of S phase with flow cytometry. Estimates for the proportion of Ki-67-positive tumors cells were found to be correlated with decreased proportions of wild-type BRCA2 alleles in BRCA2 tumors displaying luminal characteristics (Pearson correlation, r = -0.59; P = 0.0047), whereas the same was not found in BRCA2 tumors displaying basal/TNP phenotype (Pearson correlation, r = 0.13, P = 0.76) (Figure 2d). In support for the preceding, the same associations, with respect to BRCA2 wild-type allele loss and phenotype, were identified for estimates of the cell-cycle associated gene product Topo IIα (Figure 2e).
Figure 2.
The effects of BRCA2 deletion in phenotypic context. (a) Deletion at the BRCA2 locus measured by segmented aCGH log2 ratios (left) and TaqMan qPCR (right) in relation to phenotype. (b) Example profiles derived from two BRCA2 tumors (a19861102, a128049) of the triple-negative subtype analyzed by using 385 K aCGH (about one probe for each 7 kb). DNA gains or deletions are represented on the y-axis as black bars toward positive or negative values, respectively, in order of the chromosomal location from p- to q-arm (left to right). Focused views of chromosome 13 are displayed below each of the two profiles showing log2 ratios (gray dots) on y-axis by nucleotide position on x-axis from p-arm to q-arm (left to right) with cytobands and the BRCA2 gene locus at 13q13.1 indicated as shown at the bottom of each plot. (c) S phase, (d) Ki-67, and (e) Topo IIa in relation to changes at the BRCA2 gene locus, affecting the wild-type allele, in luminal- (left panel, gray circles) and triple-negative phenotypes (right panel, black triangles).
Genetic networks
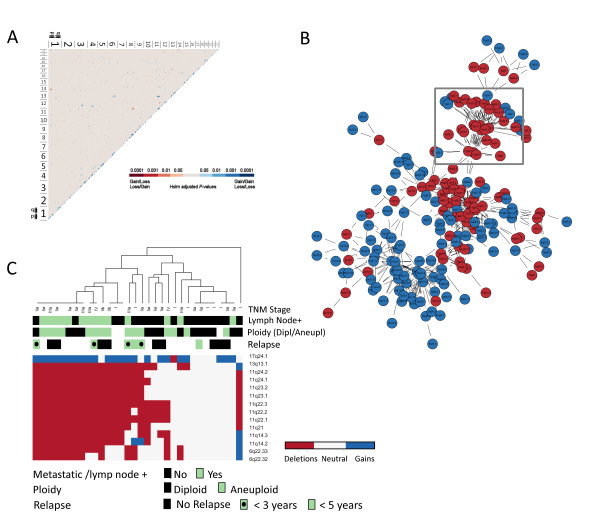
DNA copy-number changes showing significant associations between distinct chromosomal regions in the set of 33 BRCA2 tumors were identified, and visualized as an undirected node-and-edge graph in which significant correlations are represented as edges, with genomic regions as nodes (Figure 3a, b; Additional File 4). This analysis identified a set of 14 cytobands involving three distinct interchromosomal links in association with the BRCA2 locus at 13q13.1, that is, deletions at 6q22.32-q22.33 and 11q14.2-q24.1, along with copy-number gains at 17q24.1 (Figure 3b, Additional File 4). It is important to note here that the data on BRCA2wt-specific loss confirmed the links identified with respect to 6q22.32-q22.33 and 11q14.2-q24.1 (Additional File 5). Based on the BRCA2-linked networking regions (6q22.32-q22.33, 11q14.2-q24.1, 13q13.1, and 17q24.1), two subgroups were identified through cluster analysis (Figure 3c). Evidently, these subgroups differ by the degree of change over the regions identified in association with the BRCA2 gene locus. Here, measures of higher clinical staging at the time of diagnosis, involving metastasis to the lymph nodes, were more prominent in the subgroup showing changes over the BRCA2-linked networking regions (Figure 3c). Data on patient relapse were available in some of these cases, defined as an instance of subsequent metastasis or recurrence, which provided support for the hypothesis generated here that DNA copy-number changes over the BRCA2-linked networking regions promote progression toward more-advanced forms of the disease (see Figure 3c). In all cases, metastasis to more-distant sites of the body was involved, of which all but one occurred as bone metastases. To determine whether the network of genetic changes identified in relation to the BRCA2 locus at 13q13.1 is specific for BRCA2 mutated breast cancers, we looked at our previously published data on sporadic breast cancers [15]. As expected, the BRCA2 locus at 13q13.1 was not represented within the genetic networks constructed by using the same approach for sporadic breast cancers (Additional File 4).
Figure 3.
Genetic networks in BRCA2 tumors. (a) Matrix of associations with blue and red reflecting P values (Holm adjustment for multiple testing) for distinct chromosomal regions where copy-number changes co-occur in patterns of loss-loss or gain-gain relations, represented in blue, or gain-loss relations represented in red. (b) Genetic networks; interchromosomal links constructed for relations below Holm-adjusted P values of 0.001 (edges) between distinct genomic regions (nodes), with deletions in red and gains in blue, showing multiple links adjacent to the BRCA2 locus at 13q13.1 (box). (c) Hierarchic cluster analysis of copy-number changes, deletions in red and gains in blue, over the subnet of chromosomal regions linked to 13q13.1 (BRCA2) in relation to the occurrence of lymph node metastasis at diagnosis, and later relapse involving distant metastases represented in colors of green (positive) or black (negative), ploidy as green for diploid and black for aneuploid (see color codes at bottom of heatmap), along with TNM stage.
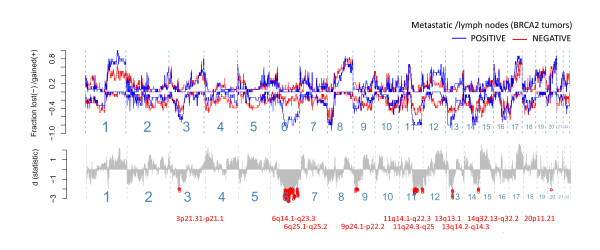
In a two-class comparison for BRCA2 tumors with or without evidence of metastasis to the lymph nodes, or other more distant sites, loss over the BRCA2 locus at 13q13.1 was identified in this analysis as significantly associated with metastasis to the lymph nodes, along with the BRCA2-linked networking regions at 6 q and 11 q (Figure 4). Of these, deletions at chromosome 6 q were the most predictive for lymph node positivity, or "metastatic capacity," with the strongest associations (significance peaks) found at 6q16.3 and 6q22.33. Looking at the frequency of copy-number changes at 6q22.33, we identified a shift toward increased frequency of deletions at a location close to the PTPRK gene, which affected four other genes, that is EEF1DP5, LAMA2, ARHGAP18, and C6orf191 (Additional File 6). Fine-tiling aCGH analysis at exon-level resolution (about one probe for each 400 bp) covering chromosome 6 confirmed deletions at 6q22.33 (Additional File 7). This analysis shows that the events at 6q-arm occur as complex deletions confined within the 6 q chromosomal region. Here, one of the tumors had acquired a breakpoint event located within the 6q22.33 cytoband proximal to the PTPRK gene affecting upstream regions (Additional File 7). This subregion within the 6q22.33 cytoband was not affected in the lymph-node-negative tumor, but rather involved a region downstream of the PTPRK gene (Additional File 7).
Figure 4.
Two-class comparative analysis displaying event-frequency plots for BRCA2 tumors with (blue) and without (red) evidence for lymph node metastases at diagnosis. The fraction of tumors with acquired changes are represented on the y-axis with positive values for gains, and negative for deletions, ordered by chromosomal location on x-axis from p- to q-arm, from left to right. The significance scores (d), derived from a permutation-based procedure (SAM two-class unpaired), representing the strength of association, are shown in the lower panel with relations below a false-discovery rate of 0.05 indicated in red (cytoband locations).
The analysis of ploidy in BRCA2 mutated breast cancers with respect to DNA copy-number changes by aCGH
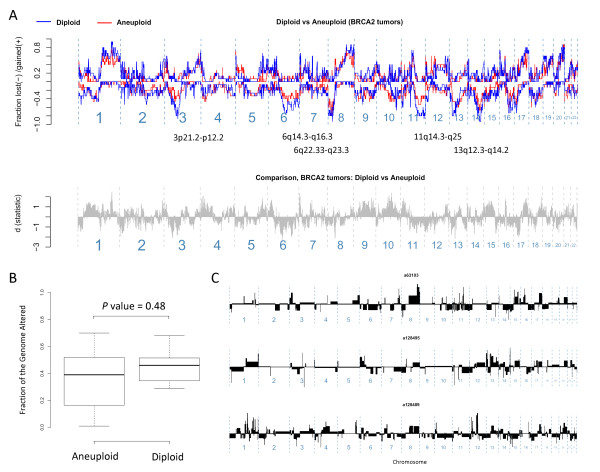
Data available on ploidy were analyzed with respect to DNA copy-number changes to follow-up on recent results demonstrating poor disease outcome for BRCA2 carriers diagnosed with breast tumors of the diploid type (Tryggvadottir et al., 2011; unpublished data). In line with the less-favorable prognosis associated with the diploid type of BRCA2 tumors, we found that BRCA2 mutated tumors, having acquired changes over the BRCA2-networking regions, were often identified as diploid with flow cytometry (Figure 3c).
To study this further, we compared event-frequency plots and the altered genome fractions for BRCA2 tumors of the diploid and aneuploid types. This analysis shows that the amounts of genomic changes are not fewer in diploids (Figure 5a) and, although not statistically significant, that the regions affected more frequently by DNA copy-number changes in the diploid types overlap with those referred to here as the BRCA2-networking regions (Figure 5b). It is clear that the genomes of BRCA2 mutated breast cancers classified as diploid are extensively altered, and individual examples are presented here to demonstrate complex patterns of genomic changes (Figure 5c).
Figure 5.
The effects of ploidy on DNA copy-number changes in BRCA2 breast tumors. (a) Event-frequency plots to compare the genomic regions affected in diploid (blue) and aneuploid (red) types of BRCA2 tumors. (b) The proportion of the genome altered in BRCA2 tumors with respect to ploidy. (c) Three example genomic profiles analyzed by aCGH to illustrate complex changes in BRCA2 tumors classified as diploid by flow-cytometry. The genomic profiles represent DNA copy-changes number toward gains (above) or losses (below) on the y-axis by chromosomal position on the x-axis from p- to q-arms, left to right, respectively.
Discussion
In this study, we show that breast tumors from BRCA2 999del5 mutation carriers, displaying luminal- or basal/triple-negative phenotypes (TNPs), differ with respect to patterns of copy-number changes and markers of pRb/p16 pathway dysfunction, suggesting divergent paths of tumor evolution. This finding led to important observations regarding loss of the wild-type (wt) BRCA2 allele in luminal types of BRCA2 tumors, where it associates with increased tumor growth rates. Furthermore, we find that deletion of the BRCA2wt allele represents a part of a larger network of co-occurring genetic changes that relate to advanced progression, involving deletions at 6q22.32-q22.33, 11q14.2-q24.1, as well as gains at 17q24.1. These results suggest that loss of the BRCA2wt allele is not required in tumor initiation, but rather plays an important role later in disease progression. Recently, it was demonstrated that heterozygosity for Brca2 is sufficient to promote carcinogenesis in a murine model of familial pancreatic cancer and also that LOH might not be essential for human pancreatic ductal carcinogenesis in BRCA2 999del5 mutation carriers [7]. In addition, an earlier study on breast cancer samples from carriers of different BRCA2 mutations showed retention of the BRCA2wt allele in some cases, providing support for our conclusion [8]. We conclude that loss of the BRCA2wt allele, although not required in tumor initiation, is not a randomly occurring passenger but rather an important driver event in leading to selective growth advantages and higher clinical staging of the disease. These effects are likely indirect, resulting from accelerated mutation rates, from which advantageous mutations can arise and lead to a more aggressive phenotype.
The majority of the BRCA2-associated tumors under study displayed luminal characteristics, whereas the remaining part was of basal/triple-negative phenotype (TNP). Notably, none of the BRCA2 tumors showed overexpression or high-level amplification of the HER-2 oncogene. This subdivision of BRCA2 tumors into phenotypic subgroups is in agreement with our previous study and those of others [14,15]. The loss of pRb observed in the basal/TNP subtype of BRCA2 mutated breast tumors likely reflects acquired defects in the RB1 gene. Indeed, high expression of CDKN2A (p16) gene products, indicative of functional defects in RB1, has been described as a defining feature of basal-like breast cancers, and it has been suggested that this may represent an important early event in bypassing cellular stress and replicative senescence [23,24]. The negative-feedback loop between high p16 and low pRb has been described and is known to be the result of pRb binding to the CDKN2A (p16) gene promoter, leading to transcriptional repression of CDKN2A [25]. Increased expression of CDKN2A (p16) gene products arises under normal circumstances as a response to stress signals, such as telomere crisis or DNA damage. Thus, unimpeded cellular proliferation, indicated by expression of cell-cycle markers such as Ki-67, in combination with the presence of high p16 expression levels, could be indicative of abrogated p16/RB checkpoint control [23]. We found high expression of MKI67 (Ki-67) gene products, a marker for cellular proliferation, in basal/TNP BRCA2 tumors, together with low pRb and high p16 levels. In contrast, the luminal subgroup of BRCA2 tumors rarely showed high p16 expression levels and almost invariably showed pRb expression. Given the association for loss of BRCA2wt alleles with respect to the luminal phenotype, this finding would have been unexpected as the RB1 (pRb) gene resides close to BRCA2 on chromosome 13q. Nevertheless, the association of p16/pRb dysfunction to the basal/TNP subtype, identified here in BRCA2 mutated breast tumors, is similar to that reported in sporadic tumors suggesting that the events leading to the basal/TNP phenotype in carriers and noncarriers could be the same. In this respect, it has been demonstrated that BRCA1 inactivation in sporadic disease leads to development of basal/TNP breast cancers [26,27]. In our recent report, we described the loss of pRb, coupled with high p16, in BRCA1-defective breast cancers [27]. We therefore postulate that the event of acquired defects in the BRCA1 gene could have the same effect in BRCA2 mutation carriers, as seen in sporadic breast cancers. Presumably, this would alleviate selection pressure for loss of the BRCA2wt allele in BRCA2 mutation carriers. This is consistent with the data presented here, demonstrating lack of definitive BRCA2wt loss in the basal/TNP subclass of BRCA2 tumors.
Whole-genome copy-number changes in this same set of BRCA2 tumors analyzed with high-resolution (~7 kbp) aCGH demonstrated substantial differences between luminal- and basal/TNP subtypes of BRCA2 tumors. These differences involved frequent alterations at multiple chromosome locations in luminal BRCA2 tumors that were infrequently or inversely altered in basal/TNP BRCA2 tumors. Their unique patterns of copy-number changes suggest that the two phenotypic subclasses of BRCA2 tumors develop in different ways. Of particular interest here are the luminal subtypes of BRCA2 tumors showing more-frequent deletions over the BRCA2 gene locus. Here, gradual decreases in the proportion of BRCA2wt alleles present within the tumor mass were correlated with increased growth rates, suggesting selective advantages for the event of BRCA2wt loss. It can be hypothesized that this relation reflects a progression pattern involving clonal expansion of tumor cells that have acquired deletion of the BRCA2wt allele. This appears contradictory to previous studies demonstrating that targeted disruption of the Brca2 gene negatively affects cell-cycle progression [28]. Other changes could therefore be required to overcome the deleterious effects of BRCA2 deficiency. In this respect, the network of genetic changes affecting 6q22.32-q22.33, 11q14.2-q24.1, and 17q24.1 identified in association with BRCA2wt loss could be of critical importance in promoting survival of BRCA2-defective tumor cells. This would give rise to cancer cells with BRCA2 deficiency, thereby leading to accelerated mutation rates and, through the forces of natural selection, advantageous changes could accumulate and eventually lead to disease progression. Indeed, we find that the set of co-occurring changes linked to the BRCA2 region at 13q.13.1 coincide with those that associate with advanced disease progression involving metastasis to the nodes, and patient relapse involving distant metastases. Here, deletion events at chromosome 6 q were the strongest predictors of metastatic capacity. To gain further insight into the genetic events occurring at this location, we performed fine-tiling aCGH analysis (exon-level resolution). Detailed views of changes at 6q22.33 (that is, the highest ranking of the BRCA2-linked genomic regions) suggested the involvement of genes located at, or downstream, of a breakpoint event disrupting protein tyrosine phosphatase, receptor type, K (PTPRK), a putative tumor suppressor gene known to be involved in TGF-β signaling [29]. Interestingly, the 6q22.33 region has been identified as a breast cancer susceptibility locus where two candidate genes are located, that is, RING finger protein 146 (RNF146) and enoyl coenzyme A hydratase domain-containing 1 (ECHDC1) [30]. Looking at breakpoint events at chromosome 6 q by using the fine-tiling aCGH analysis, we did not find strong evidence for gross genetic changes indicative of error-prone DNA repair through NHEJ, similar to that reported for the PTEN gene at chromosome 10 q in BRCA1 mutated breast tumors [31]. However, it is clear that, in most cases, these events involve complex changes as they are not characterized by loss of the entire 6 q arm. Deletions at 11 q have been described previously in association with BRCA2 breast tumors wherein aberrant expression of ATM has been demonstrated [32]. However, the region at 11q14.2-q22.2, including MRE11, has not been studied previously in this respect. The subnet of genetic links associated with the BRCA2 gene locus warrants further investigation in terms of which genes are involved and whether they explain disease outcome in BRCA2 carriers and other patients with or without a family history of the disease. This could be of potential significance in defining a set of biomarkers for use in clinical settings to identify HR defects (that is, markers of BRCAness) [33].
In looking at data from flow-cytometry, we observed diploidy as a common feature of those BRCA2 tumors displaying changes over the BRCA2-linked regions. Clearly, this was unexpected, as diploid tumors are usually associated with genomes characterized by few or no genomic changes (that is, simple genomes) [34]. This finding has relevance to a recent study on survival in BRCA2 mutation carriers, wherein tumors classified as diploid were found to be associated with fast disease progression independent of hormone receptors, histologic grade, and TNM stage (Tryggvadottir et al., 2011, unpublished data). Here, we add important insight into this, with data demonstrating complex genetic changes in genomes of BRCA2 tumors identified as diploid with flow-cytometry. The presence of genomic complexity in BRCA2 tumors of the diploid class demonstrates that they are pseudo-diploids. Nevertheless, the differences in disease outcome with respect to ploidy in BRCA2 carriers are compelling and suggestive of divergent tumor-progression paths. In this respect, it has been shown that approximately 30% of all breast tumors classified as diploid by flow-cytometry have acquired complex patterns of genetic changes, as identified through aCGH-based method (ROMA) [34]. Importantly, the two different instability-types described (that is, firestorm and sawtooth), were associated with poor disease outcome. Thus, it seems likely that at least a certain proportion of tumors classified as diploid by conventional flow-cytometry could be promoted by acquired defects in components of the HR machinery, possibly involving the BRCA2 gene.
Inhibitors of PARP1 gene products have been shown to kill BRCA1- or BRCA2-defective cells selectively, with no effects on heterozygous cells, and such inhibitors are currently being tested in clinical trials [35]. Our results could have clinical implications regarding the use of PARP inhibitors, with observations demonstrating late involvement for loss of the BRCA2wt allele. In this respect, PARP inhibitors will probably not be sufficient alone (that is, as monotherapy), for treatment of BRCA2 carriers. Nevertheless, the risk of progression toward advanced disease in the event of BRCA2wt allele loss is of considerable concern, and our results therefore strongly support adjuvant use of PARP inhibitors in standard treatment of all BRCA2 mutation carriers.
Conclusions
In conclusion, our results demonstrate phenotypic heterogeneity of breast cancers arising in carriers of the same 999del5 BRCA2 mutation. We show that the luminal- and basal/TN phenotypes develop through divergent tumor evolutionary paths. Here, loss of the BRCA2 wild-type allele occurs more frequently in the luminal subtype, where it correlates with increased tumor growth rates. Furthermore, the event of BRCA2wt allele loss occurs in the context of other genetic changes, involving deletions at 6 q, 11 q, and copy-number gains at 17 q. These changes, including deletions over the BRCA2 gene locus, were found to be associated with higher clinical staging at diagnosis involving metastasis to the lymph nodes, and correlated with patient relapse. These observations could explain why BRCA2 carriers have predilection for developing breast cancers of the luminal B phenotype. The findings presented here, suggesting that loss of the BRCA2wt allele is a late, rather than early, event in progression toward metastatic disease, have clinical relevance supporting adjuvant, and not single-agent, use of PARP inhibitors in treatment of BRCA2 carriers.
Abbreviations
CGH: comparative genomic hybridization; DSB: double-stranded break; HR: homologous recombination; IHC: immunohistochemistry; LumA: Luminal-A; LumB: Luminal-B; NHEJ: nonhomologous end joining; SSA: single-strand annealing; TMA: tissue microarrays; TNP: triple-negative phenotype.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
OAS contributed to the study design and performed the aCGH analysis along with statistical analysis, data mining, and writing of the manuscript. OAS and JGJ selected tumor tissue for TMAs. KO constructed the TMAs and performed IHC analysis. OAS and JGJ scored the IHC results. JGJ scored histologic grade and TNM staging. OTJ contributed clinical data, and SV contributed to the aCGH analysis. HB and SKB performed the TaqMan BRCA2 allele-specific quantitative PCR (qPCR). JEE conceived of the study and was in charge of its design and the coordination and writing of the manuscript. All authors read and approved of the manuscript.
Supplementary Material
Phenotype and clinical parameters. Table listing tumor phenotype and clinical parameters including breast cancer subtype, histologic grade, and TNM stage.
Antibodies for expression analysis on TMAs. Table listing the antibodies used for immunohistochemistry on tissue microarrays (TMAs).
The expression of Ki-67, p16, and pRb in BRCA2 mutated breast cancers by phenotype. Expression of Ki-67, p16, and pRb by IHC on tissue microarrays (TMAs) in subtypes of BRCA2 breast tumors. (a) The distribution of Ki-67 expression levels, reflecting cellular proliferation, compared between BRCA2 tumors of basal/triple-negative and luminal phenotypes. Example IHC analysis for Ki-67 shows higher proportion of positive cells in BRCA2 tumors of triple-negative (lower panel) compared with luminal phenotypes (upper panel). (b) Expression of p16 (CDKN2A) in BRCA2 tumors by phenotype with low expression of pRb (IHC null) indicated, see Table 1 for details. Examples show representative BRCA2 tumors of luminal- (upper panel) and triple-negative (lower panel) phenotypes.
The complete set of links identified in breast cancers graphically represented as node-and-edge graphs, shown here separately for those derived from BRCA2 carriers and sporadic cases. Genetic networks constructed for relations below Holm-adjusted P values of 0.001 (edges) between distinct genomic regions (nodes), with deletions in red and gains in blue. The links identified, reflecting co-occurring genetic changes, were split into sets of inter- and intrachromosomal links characterized as those involving distinct chromosomes (inter, left on figure) or distinct arms on the same chromosome (intra, right on figure), respectively. (a) The genetic networks constructed for BRCA2 mutated breast cancers, and (b) those arising in the sporadic setting.
BRCA2 wild-type allele specific qPCR examined with respect to DNA copy-number changes. Deletion of wild-type BRCA2 alleles by TaqMan qPCR analysis were examined on a continuous scale in relation with the genome-wide segmented aCGH log2 ratios by using a regression model with repeated permutations to address the problem of multiple testing, as implemented in SAM (quantitative analysis). The significance scores (r) derived from this analysis are plotted on the y-axis for each genomic location ordered from p- to q-arm on the x-axis, from left to right, with relations below a false-discovery rate of 0.0001 indicated by red dots.
Detailed analysis of event frequencies at chromosome 6 q in BRCA2 mutated breast cancers. Analysis of DNA copy-number changes occurring at chromosome 6 in BRCA2 tumors. (a) Top: Comparison for the frequency of events in BRCA2 tumors with and without evidence for metastatic spread to the lymph nodes or other sites at diagnosis, a trait reflecting metastatic capacity, represented in red and orange, respectively (see legend at the top right corner). Bottom: Zooming in on the frequency of events at 6q22.33, selected for further analysis, as this region strongly associates with metastatic capacity in BRCA2 tumors while also being linked to deletions over the BRCA2 gene locus at 13q13.1. (b) Top: Example of fine-tiling aCGH analysis (exon-level resolution) for chromosome 6 in one BRCA2 breast tumor where aCGH log2 ratios are plotted on the y-axis in order of nucleotide position on the x-axis. Dashed vertical lines represent locations of cytoband boundaries, as indicated below, and solid horizontal red lines represent output derived from CBS analysis for detecting copy-number changes with negative values of segmented aCGH log2 ratios reflecting change toward deletions, and positive values as change toward copy-number gains. Bottom: Enlarged, detailed view of the entire 6q22.33 region in this tumor shows a breakpoint event occurring nearby the PTPRK gene, a frequent site of deletion events in metastatic BRCA2 tumors (see data in Figure 5a, bottom panel) affecting four other genes within this cytoband (EEF1DP5, LAMA2, ARHGAP18, and C6orf191).
aCGH analysis using fine-tiling arrays (400 bp) for chromosome 6 in BRCA2 mutated breast cancers. Fine-tiling aCGH analysis (exon-level resolution) covering chromosome 6, with about one probe for each 400 bp, was performed to reanalyze and confirm events at this chromosome with greater clarity in three of the BRCA2 tumors. The aCGH log2 ratios are plotted on the y-axis in order of nucleotide position on the x-axis, with dashed vertical lines representing locations of cytoband boundaries, as indicated below. Solid horizontal red lines represent output derived from CBS analysis for detecting copy-number changes with negative values of segmented aCGH log2 ratios reflecting change toward deletions, and positive values as change toward copy-number gains.
Contributor Information
Olafur A Stefansson, Email: oas@hi.is.
Jon G Jonasson, Email: jongj@landspitali.is.
Kristrun Olafsdottir, Email: kristrol@landspitali.is.
Hordur Bjarnason, Email: hob3@hi.is.
Oskar Th Johannsson, Email: oskarjoh@landspitali.is.
Sigridur K Bodvarsdottir, Email: skb@hi.is.
Sigridur Valgeirsdottir, Email: sigridur.valgeirsdottir@roche.com.
Jorunn E Eyfjord, Email: jorunne@hi.is.
Acknowledgements
The authors thank Holmfridur Hilmarsdottir for BRCA2 germline mutation screening and technical assistance. We also thank the staff at Roche NimbleGen Systems in Iceland for assistance with the aCGH analysis. This work was funded by Eimskipafelag University Fund (OAS), Minningarsjodur Bergthoru Magnusdottur and Jakobs J Bjarnasonar (OAS), Gongum Saman (OAS), Icelandic Cancer Research Fund SKI (OAS), Icelandic Centre for Research RANNIS (OAS, JGJ, OTJ, JEE), and The University of Iceland Research Fund (OAS, JEE).
References
- Oldenburg RA, Meijers-Heijboer H, Cornelisse CJ, Devilee P. Genetic susceptibility for breast cancer: how many more genes to be found? Crit Rev Oncol Hematol. 2007;63:125–149. doi: 10.1016/j.critrevonc.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Murphy CG, Moynahan ME. BRCA gene structure and function in tumor suppression: a repair-centric perspective. Cancer J. 2010;16:39–47. doi: 10.1097/PPO.0b013e3181cf0204. [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S, Thorlacius S, Valgardsdottir R, Gudlaugsdottir S, Sigurdsson S, Steinarsdottir M, Jonasson JG, Anamthawat-Jonsson K, Eyfjord JE. BRCA2 and p53 mutations in primary breast cancer in relation to genetic instability. Cancer Res. 1998;58:859–862. [PubMed] [Google Scholar]
- Collins N, McManus R, Wooster R, Mangion J, Seal S, Lakhani SR, Ormiston W, Daly PA, Ford D, Easton DF. et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- Skoulidis F, Cassidy LD, Pisupati V, Jonasson JG, Bjarnason H, Eyfjord JE, Karreth FA, Lim M, Barber LM, Clatworthy SA, Davies SE, Olive KP, Tuveson DA, Venkitaraman AR. Germline Brca2 heterozygosity promotes Kras(G12D)-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- King TA, Li W, Brogi E, Yee CJ, Gemignani ML, Olvera N, Levine DA, Norton L, Robson ME, Offit K, Borgen PI, Boyd J. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann Surg Oncol. 2007;14:2510–2518. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- Melchor L, Honrado E, Garcia MJ, Alvarez S, Palacios J, Osorio A, Nathanson KL, Benitez J. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2008;27:3165–3175. doi: 10.1038/sj.onc.1210975. [DOI] [PubMed] [Google Scholar]
- Stefansson OA, Jonasson JG, Johannsson OT, Olafsdottir K, Steinarsdottir M, Valgeirsdottir S, Eyfjord JE. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11:R47. doi: 10.1186/bcr2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson JG, Tavtigian SV, Tulinius H, Ogmundsdottir HM, Eyfjord JE. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, Kurbatova N, Lukk M, Malone J, Mani R, Pilicheva E, Rustici G, Sharma A, Williams E, Adamusiak T, Brandizi M, Sklyar N, Brazma A. ArrayExpress update: an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. pp. D1002–1004. [DOI] [PMC free article] [PubMed]
- Germer S, Higuchi R. Homogeneous allele-specific PCR in SNP genotyping. Methods Mol Biol. 2003;212:197–214. doi: 10.1385/1-59259-327-5:197. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Kent WJ. The UCSC genome browser. Curr Protoc Bioinformatics. 2009;Chapter 1:Unit1–4. doi: 10.1002/0471250953.bi0104s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast Cancer Res. 2006;8:216. doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R, Lambros MB, Rodríguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchió C, Vatcheva R, Rayter S, Mahler-Araujo B, Fulford LG, Hungermann D, Mackay A, Grigoriadis A, Fenwick K, Tamber N, Hardisson D, Tutt A, Palacios J, Lord CJ, Buerger H, Ashworth A, Reis-Filho JS. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–2722. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhawong AP, Subhawong T, Nassar H, Kouprina N, Begum S, Vang R, Westra WH, Argani P. Most basal-like breast carcinomas demonstrate the same Rb-/p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33:163–175. doi: 10.1097/PAS.0b013e31817f9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J, Raffeld M, Yakhini Z, Ben-Dor A, Dougherty E. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT, Eyfjord JE. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6:638–649. doi: 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/S1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- Wang SE, Wu FY, Shin I, Qu S, Arteaga CL. Transforming growth factor {beta} (TGF-{beta})-Smad target gene protein tyrosine phosphatase receptor type kappa is required for TGF-{beta} function. Mol Cell Biol. 2005;25:4703–4715. doi: 10.1128/MCB.25.11.4703-4715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, Friedman E, Narod S, Olshen AB, Gregersen P, Kosarin K, Olsh A, Bergeron J, Ellis NA, Klein RJ, Clark AG, Norton L. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Gruvberger-Saal SK, Persson C, Lovgren K, Jumppanen M, Staaf J, Jonsson G, Pires MM, Maurer M, Holm K, Koujak S, Subramaniyam S, Vallon-Christersson J, Olsson H, Su T, Memeo L. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, Aittomaki K, Hofstetter B, Lukas J, von Smitten K, Blomqvist C, Ristimäki A, Heikkilä P, Bartek J, Nevanlinna H. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–2506. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. Hallmarks of "BRCAness" in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, Yoon S, Wigler M, Ye K, Børresen-Dale AL, Naume B, Schlicting E, Norton L, Hägerström T. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotype and clinical parameters. Table listing tumor phenotype and clinical parameters including breast cancer subtype, histologic grade, and TNM stage.
Antibodies for expression analysis on TMAs. Table listing the antibodies used for immunohistochemistry on tissue microarrays (TMAs).
The expression of Ki-67, p16, and pRb in BRCA2 mutated breast cancers by phenotype. Expression of Ki-67, p16, and pRb by IHC on tissue microarrays (TMAs) in subtypes of BRCA2 breast tumors. (a) The distribution of Ki-67 expression levels, reflecting cellular proliferation, compared between BRCA2 tumors of basal/triple-negative and luminal phenotypes. Example IHC analysis for Ki-67 shows higher proportion of positive cells in BRCA2 tumors of triple-negative (lower panel) compared with luminal phenotypes (upper panel). (b) Expression of p16 (CDKN2A) in BRCA2 tumors by phenotype with low expression of pRb (IHC null) indicated, see Table 1 for details. Examples show representative BRCA2 tumors of luminal- (upper panel) and triple-negative (lower panel) phenotypes.
The complete set of links identified in breast cancers graphically represented as node-and-edge graphs, shown here separately for those derived from BRCA2 carriers and sporadic cases. Genetic networks constructed for relations below Holm-adjusted P values of 0.001 (edges) between distinct genomic regions (nodes), with deletions in red and gains in blue. The links identified, reflecting co-occurring genetic changes, were split into sets of inter- and intrachromosomal links characterized as those involving distinct chromosomes (inter, left on figure) or distinct arms on the same chromosome (intra, right on figure), respectively. (a) The genetic networks constructed for BRCA2 mutated breast cancers, and (b) those arising in the sporadic setting.
BRCA2 wild-type allele specific qPCR examined with respect to DNA copy-number changes. Deletion of wild-type BRCA2 alleles by TaqMan qPCR analysis were examined on a continuous scale in relation with the genome-wide segmented aCGH log2 ratios by using a regression model with repeated permutations to address the problem of multiple testing, as implemented in SAM (quantitative analysis). The significance scores (r) derived from this analysis are plotted on the y-axis for each genomic location ordered from p- to q-arm on the x-axis, from left to right, with relations below a false-discovery rate of 0.0001 indicated by red dots.
Detailed analysis of event frequencies at chromosome 6 q in BRCA2 mutated breast cancers. Analysis of DNA copy-number changes occurring at chromosome 6 in BRCA2 tumors. (a) Top: Comparison for the frequency of events in BRCA2 tumors with and without evidence for metastatic spread to the lymph nodes or other sites at diagnosis, a trait reflecting metastatic capacity, represented in red and orange, respectively (see legend at the top right corner). Bottom: Zooming in on the frequency of events at 6q22.33, selected for further analysis, as this region strongly associates with metastatic capacity in BRCA2 tumors while also being linked to deletions over the BRCA2 gene locus at 13q13.1. (b) Top: Example of fine-tiling aCGH analysis (exon-level resolution) for chromosome 6 in one BRCA2 breast tumor where aCGH log2 ratios are plotted on the y-axis in order of nucleotide position on the x-axis. Dashed vertical lines represent locations of cytoband boundaries, as indicated below, and solid horizontal red lines represent output derived from CBS analysis for detecting copy-number changes with negative values of segmented aCGH log2 ratios reflecting change toward deletions, and positive values as change toward copy-number gains. Bottom: Enlarged, detailed view of the entire 6q22.33 region in this tumor shows a breakpoint event occurring nearby the PTPRK gene, a frequent site of deletion events in metastatic BRCA2 tumors (see data in Figure 5a, bottom panel) affecting four other genes within this cytoband (EEF1DP5, LAMA2, ARHGAP18, and C6orf191).
aCGH analysis using fine-tiling arrays (400 bp) for chromosome 6 in BRCA2 mutated breast cancers. Fine-tiling aCGH analysis (exon-level resolution) covering chromosome 6, with about one probe for each 400 bp, was performed to reanalyze and confirm events at this chromosome with greater clarity in three of the BRCA2 tumors. The aCGH log2 ratios are plotted on the y-axis in order of nucleotide position on the x-axis, with dashed vertical lines representing locations of cytoband boundaries, as indicated below. Solid horizontal red lines represent output derived from CBS analysis for detecting copy-number changes with negative values of segmented aCGH log2 ratios reflecting change toward deletions, and positive values as change toward copy-number gains.