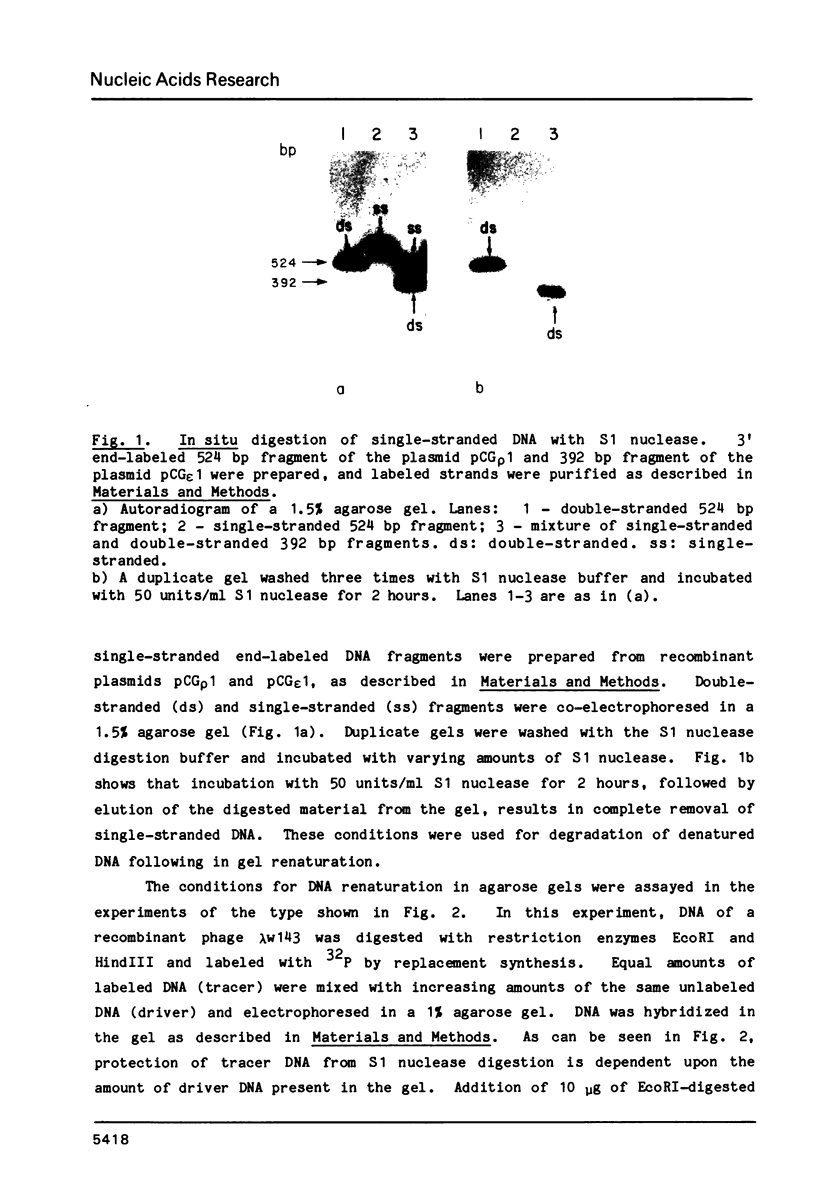
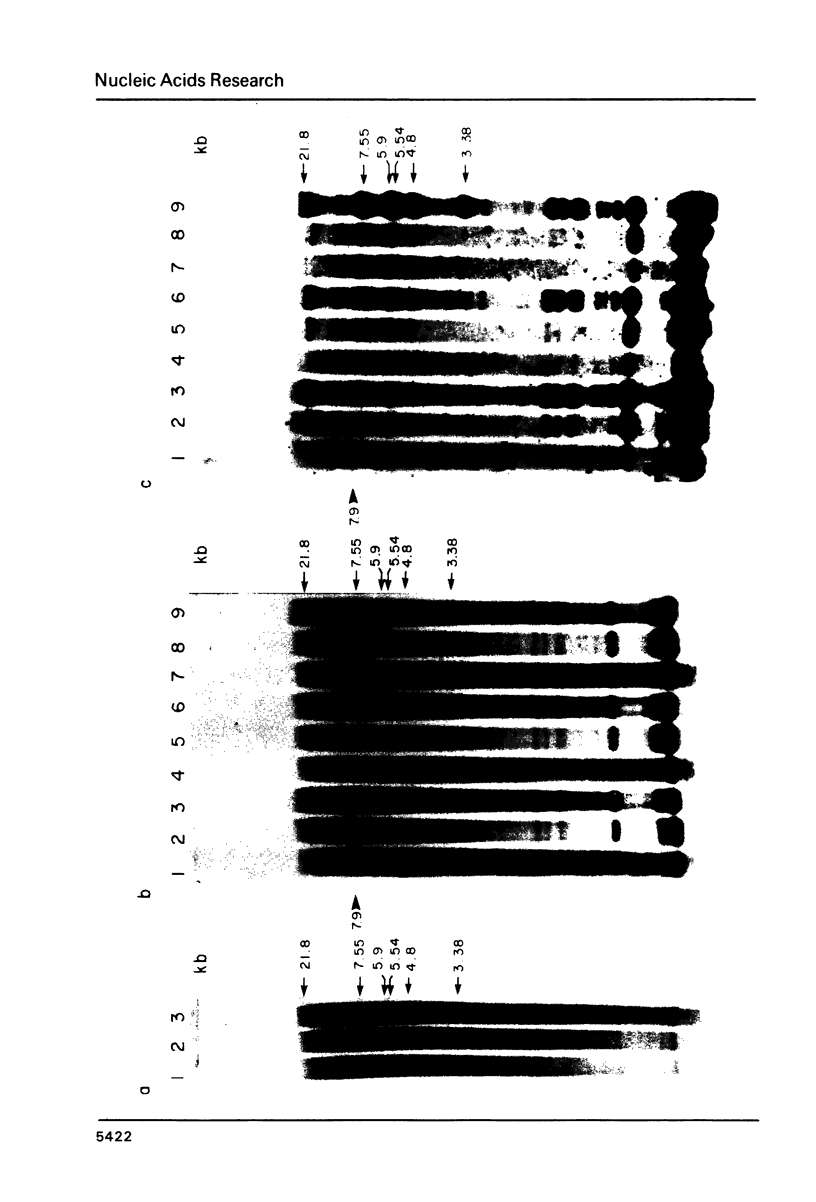
Abstract
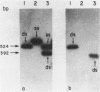
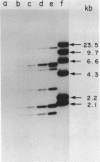
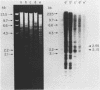
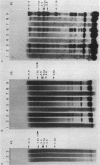
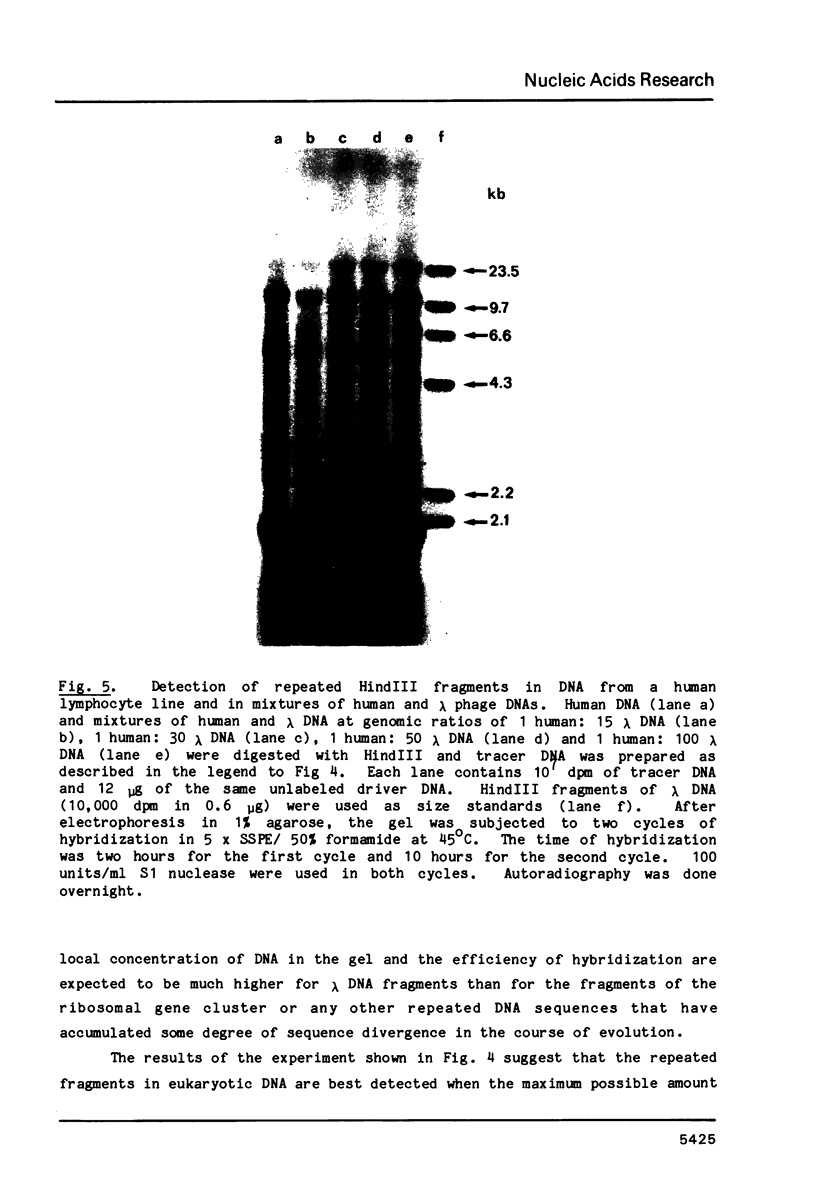
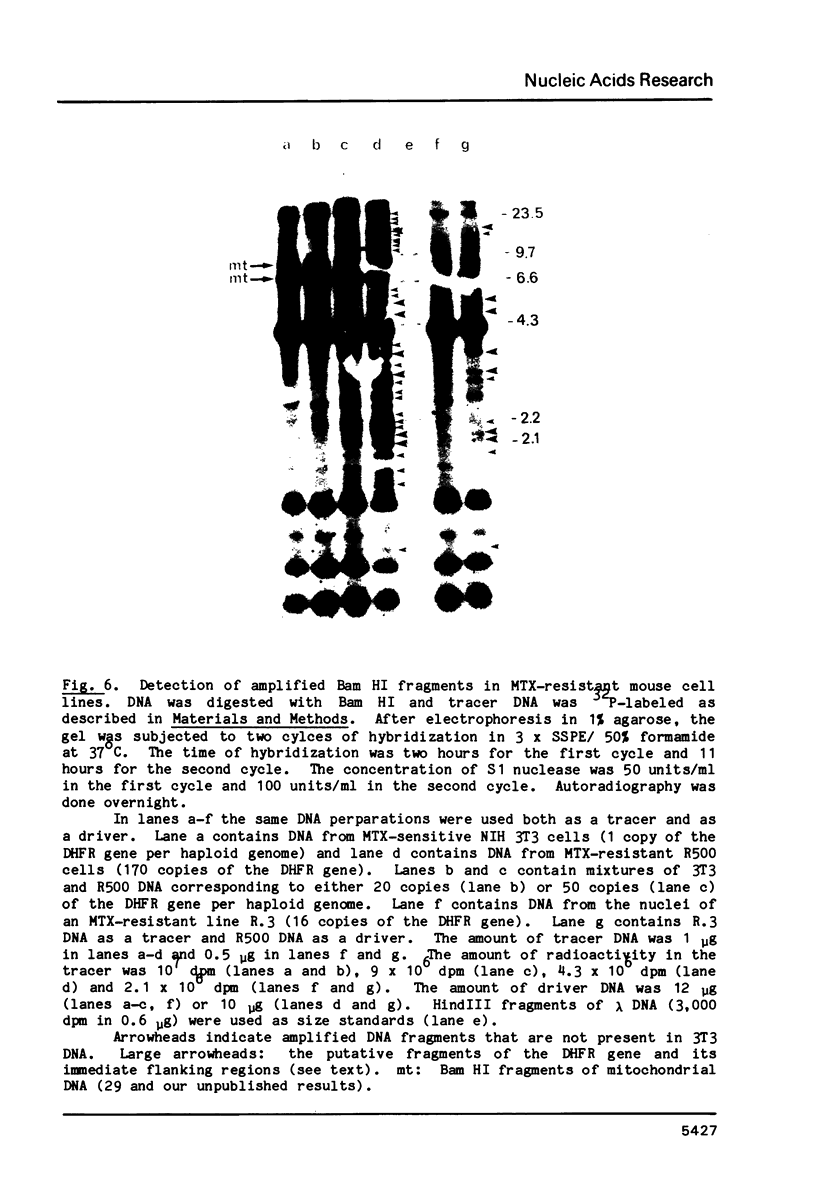
A new molecular hybridization approach to the analysis of complex genomes has been developed. Tracer and driver DNAs were digested with the same restriction enzyme(s), and tracer DNA was labeled with 32P using T4 DNA polymerase. Tracer DNA was mixed with an excess amount of driver, and the mixture was electrophoresed in an agarose gel. Following electrophoresis, DNA was alkali-denatured in situ and allowed to reanneal in the gel, so that tracer DNA fragments could hybridize to the driver only when homologous driver DNA sequences were present at the same place in the gel, i.e. within a restriction fragment of the same size. After reannealing, unhybridized single-stranded DNA was digested in situ with S1 nuclease. The hybridized tracer DNA was detected by autoradiography. The general applicability of this technique was demonstrated in the following experiments. The common EcoRI restriction fragments were identified in the genomes of E. coli and four other species of bacteria. Two of these fragments are conserved in all Enterobacteriaceae. In other experiments, repeated EcoRI fragments of eukaryotic DNA were visualized as bands of various intensity after reassociation of a total genomic restriction digest in the gel. The situation of gene amplification was modeled by the addition of varying amounts of lambda phage DNA to eukaryotic DNA prior to restriction enzyme digestion. Restriction fragments of lambda DNA were detectable at a ratio of 15 copies per chicken genome and 30 copies per human genome. This approach was used to detect amplified DNA fragments in methotrexate (MTX)-resistant mouse cells and to identify commonly amplified fragments in two independently derived MTX-resistant lines.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Johnson K. E., Citarella R. V., Falkow S. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol. 1969 May;98(2):637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brison O., Ardeshir F., Stark G. R. General method for cloning amplified DNA by differential screening with genomic probes. Mol Cell Biol. 1982 May;2(5):578–587. doi: 10.1128/mcb.2.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. C., Beverley S. M., Schimke R. T. Relationship of amplified dihydrofolate reductase genes to double minute chromosomes in unstably resistant mouse fibroblast cell lines. Mol Cell Biol. 1981 Dec;1(12):1077–1083. doi: 10.1128/mcb.1.12.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Englund P. T. Specific labeling of 3' termini with T4 DNA polymerase. Methods Enzymol. 1980;65(1):39–43. doi: 10.1016/s0076-6879(80)65008-3. [DOI] [PubMed] [Google Scholar]
- Cowell J. K. Double minutes and homogeneously staining regions: gene amplification in mammalian cells. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- George D. L., Powers V. E. Cloning of DNA from double minutes of Y1 mouse adrenocortical tumor cells: evidence for gene amplification. Cell. 1981 Apr;24(1):117–123. doi: 10.1016/0092-8674(81)90507-9. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Knöchel W., Vogelsberg C., Brost E. F. Cleavage sites of restriction endonucleases EcoRI, BamHI and HindIII on chicken embryo rDNA. FEBS Lett. 1979 Jun 15;102(2):287–290. doi: 10.1016/0014-5793(79)80020-4. [DOI] [PubMed] [Google Scholar]
- Kretschmer P. J., Kaufman R. E., Coon H. C., Chen M. J., Geist C. E., Nienhuis A. W. Hemoglobin switching in sheep. Cloning and characterization of the beta A and beta-like embryonic globin genes from genomic DNA. J Biol Chem. 1980 Apr 10;255(7):3204–3211. [PubMed] [Google Scholar]
- Lehmann C., Warnhoff M., Knochel W., Lange D., Born J., Tiedemann H. Chicken ribosomal DNA: gene frequency and purification by R-loop hybridization. Mol Biol Rep. 1979 Feb 15;4(4):217–221. doi: 10.1007/BF00777557. [DOI] [PubMed] [Google Scholar]
- MASTER R. W. POSSIBLE SYNTHESIS OF POLYRIBONUCLEOTIDES OF KNOWN BASE-TRIPLET SEQUENCES. Nature. 1965 Apr 3;206:93–93. doi: 10.1038/206093b0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moore K. H., Johnson P. H., Chandler S. E., Grossman L. I. A restriction endonuclease cleavage map of mouse mitochondrial DNA. Nucleic Acids Res. 1977;4(5):1273–1289. doi: 10.1093/nar/4.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall M. L. Gene-amplification model of carcinogenesis. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2465–2468. doi: 10.1073/pnas.78.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson I. B., Ingram V. M. Gene evolution in the chicken beta-globin cluster. Cell. 1982 Mar;28(3):515–521. doi: 10.1016/0092-8674(82)90206-9. [DOI] [PubMed] [Google Scholar]
- Roninson I. B., Ingram V. M. cDNA sequence of a new chicken embryonic rho-globin. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4782–4785. doi: 10.1073/pnas.78.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lund E., Smithies O., Blattner F. R. Hybridization of labeled RNA to DNA in agarose gels. Nucleic Acids Res. 1975 Oct;2(10):1911–1929. doi: 10.1093/nar/2.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis R. J., Goldstein S. Loss of reiterated DNA sequences during serial passage of human diploid fibroblasts. Cell. 1980 Oct;21(3):739–749. doi: 10.1016/0092-8674(80)90437-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stumph W. E., Kristo P., Tsai M. J., O'Malley B. W. A chicken middle-repetitive DNA sequence which shares homology with mammalian ubiquitous repeats. Nucleic Acids Res. 1981 Oct 24;9(20):5383–5397. doi: 10.1093/nar/9.20.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C., Alderson T. Gene amplification in methotrexate-resistant mouse cells. I. DNA rearrangement accompanies dihydrofolate reductase gene amplification in a T-cell lymphoma. J Mol Biol. 1981 Dec 5;153(2):203–218. doi: 10.1016/0022-2836(81)90274-6. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Phorbol ester dramatically increases incidence of methotrexate-resistant mouse cells: possible mechanisms and relevance to tumor promotion. Cell. 1981 Aug;25(2):561–572. doi: 10.1016/0092-8674(81)90074-x. [DOI] [PubMed] [Google Scholar]