Abstract
A conjugate of lysozyme with avarone, a bioactive sesquiterpene quinone of marine origin, and its three derivatives, was synthesized. MALDI TOF mass spectral analysis and tryptic digestion showed that the only residue in lysozyme that was modified by all derivatives was lysine 97. The identity of the residue was in full correlation with the prediction obtained by molecular modelling. All bioconjugates preserved most of the enzymatic activity of lysozyme. The melting point of the conjugates was slightly increased in comparison to lysozyme, indicating a slight stabilization of structure. The antibacterial activity of all the conjugates to both Gram positive and Gram negative bacteria was stronger than the activity of either lysozyme or the quinones, the MIC values being in low micromolar range for some conjugates.
INTRODUCTION
Lysozyme is a globular basic protein, present in almost all secretion body fluids and tissues of human and animal organisms.1 It has also been isolated from some plants, bacteria and bacteriophages.2 Lysozyme monomer exhibits strong antibacterial activity against Gram-positive organisms.3 This enzyme is widely used in food industry4 as a preservative for meat, fish and their products, as well as for milk and dairy products and fruits and vegetables.5 In pharmacy and medicine it is used in the manufacture of adjuvant drugs for antibiotics in viral and bacterial infections, in treatment of leukemia and neoplastic diseases6 and also as a diagnostic agent, being an indicator of the occurence and the progression of pathological changes in humans and animals.7 Various modifications of lysozyme caused changes of enzyme properties. Thermal modification induced formation of a dimeric form of lysozyme,8,9 which exhibits antiviral and anti-inflammatory properties.10 Many techiques have been used to modify enzyme surface and simultaneously modify enzyme properties. The most frequent is the covalent binding of appropriate molecules to the enzyme’s surface aminoacids in order to improve lysozyme antibacterial activity.11-13
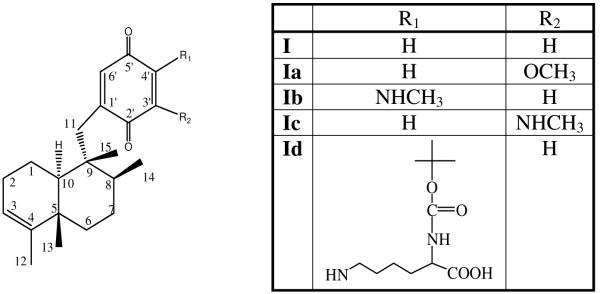
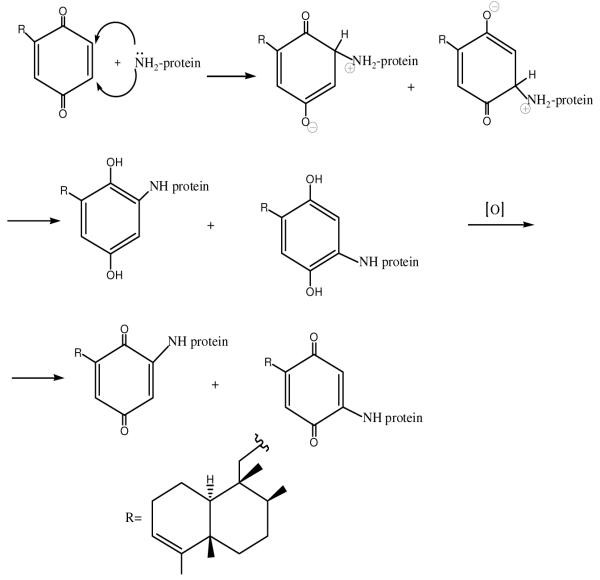
In this work, a modification of lysozyme with avarone14 (I), a sesquiterpene quinone isolated from the marine sponge Dysidea avara,15,16 and its derivatives (Ia-c) (Chart 1) was performed. This quinone was selected because of its antibacterial activity, availability, and previous knowledge that it modifies predominantly lysine residues in proteins by nucleophilic addition (Scheme 1).17,18 Such modifications are expected not to interfere much with the enzyme activity, since the mechanism of lysozyme is based on participation of carboxylic groups from aspartic and glutamic acid residues.
Chart 1.
Structures of quinones
Scheme 1.
Mechanism of protein amino groups modification by avarone
EXPERIMENTAL PROCEDURES
General information
All reagents used were commercial products purchased from Merck and Sigma-Aldrich. Column chromatography was carried out on silica gel (0.063-0.200 mm, Merck). Analytical thin-layer chromatography (TLC) was performed on precoated aluminium-backed plates (silica gel 60 F254, Merck). 1H and 13CNMR spectra were recorded at 200 MHz (Oxford NMR YH) in deuterated methanol (CD3OD). Chemical shifts are reported in parts per milion (ppm), relative to tetramethylsilane. Coupling constants are reported in hertz (Hz). Mass spectra were recorded on mass spectrometer 6210 Time-of-Flight LC-MS system (Agilent Technologies). [α]D20 value was measured on Rudolph research analytical polarimeter, Autopol IV. Elemental analysis was performed on Elementar Analysensysteme GmbH VARIO EL III CHNOS. Melting point was determined on Boetius PMHK. MALDI TOF mass spectra were recorded on Bruker Daltonics Autoflex MALDI-TOF mass spectrometer. The melting temperatures of lysozyme and its conjugates were measured using luminescence spectrometer LS 50 (Perkin Elmer, USA). UV/Vis spectra were recorded on CINTRA 40 UV/Vis spectrometer.
3′-Methoxyavarone, 3′-(methylamino)avarone and 4′-(methylamino)avarone were synthesized by previously published methods.19,20
4′-((5-((tert-Butoxycarbonyl)amino)-5-carboxypentyl)amino)avarone (Id)
Solutions of 80 mg avarone in 24 mL ethanol and 60 mg lysine-tBoc in 84 mL water were mixed, NH4HCO3 was added to a final concentration of 50 mM and the mixture was left for 48 h at room temperature with stirring. The mixture was extracted with chloroform and the chloroform solution was dried over anhydrous Na2SO4. After evaporation of chloroform, the raw product was purified by multiple column chromatography (silica gel, 0.063-0.200 mm, Merck) with toluene, chloroform and methanol. The resulting compound was purified by preparative TLC chromatography using methanol affording 17 mg (40 %) of pure product, m.p. 129-131°C, [α]D20 = −39 (c 0.10; MeOH). Elemental analysis for C32H48N2O6 (MW=556.7). Calcd. for C (69.04%), H (8.69%), N (5.03%). Found: C (69.01%), H (8.76%), N (4.97%). 1H-NMR (200 MHz, CD3OD): δ 6.32 (s, 1H, C6′-H), 5.40 (s, 1H, C3′-H), 5.11 (bs, 1H, C3-H), 3.95 (bs, 1H, C5″-H), 3.13 (bs, 2H, C2″-H), 2.57 and 2.50 (dd, J= 6.6 Hz, 2H, C11-H), 1.51 (s, 3H, C12-H), 1.42 (s, 9H, C9″, C10″, C11″), 0.94 (d, 3H, C14-H), 0.85 (s, 3H, C15-H), 2.0-1.1 (complex signal). 13C-NMR (200 MHz, CD3OD): δc 184.76 (C5′), 184.41 (C2′), 179.50 (C6″), 157.81 (C7″), 152.40 (C4′), 149.71 (C1′), 145.12 (C6′), 144.03 (C4), 133.25 (C3′), 121.61 (C3), 80.04 (C8″), 57.05 (C5″), 43.50 (C1″), 39.53 (C11), 38.44 (C4″), 37.46 (C8), 36.04 (C5), 33.87 (C9), 30.74 (C6), 28.79 (C9″, C10″, C11″), 28.68 (C1), 27.41 (C2), 24.27 (C2″), 23.38 (C3″), 20.49 (C7), 18.30 (C15), 18.08 (C13), 17.36 (C12), 17.17 (C14). Mass spectrum (Mw+Na) ion m/z 579. UV/Vis max 225 nm (ε=2.53·105), 258 nm (ε=2.86·105), 497 nm (ε=5.53·104).
Optimization of the conditions
The reaction was studied in 20 % ethanol and in 20 % DMSO in presence of 50 mM ammonium bicarbonate in normal and in inert atmosphere (nitrogen). The extent of modification was evaluated by HPLC (HPLC Äkta purifier, Amersham Pharmacia Biotech). In the column Waters Spherisorb C18 (RP; ODS2, 4.6×250 mm; 2 m Analytical column) was injected 10 μL of the sample. The temperature of the column was 25 °C and flow rate was 200 μL/min. Eluent (A) was 0.2% formic acid/water and eluent (B) was acetonitrile. After 25 min the chromatography was stopped.
Synthesis of conjugates
A mixture of final volume of 1.2 mL containing 960 μL aqueous solution of lysozyme (Sigma-Aldrich, from chicken egg white) and 240 μL of solution of avarone and its derivatives in 96% ethanol was made, so that the final enzyme concentration was 5 mg/mL and concentration of quinone was 2 mg/mL in 20% ethanol. Into the mixture was added NH4HCO3. The final concentration of the base was 50 mM. The modification reaction lasted 48 h at room temperature with constant shaking. All samples were centrifuged for 5 min at 14,000 rpm (Eppendorf mini spin plus). Supernatants were purified on Sephadex G-50 column. Enzymes were eluted with distilled water. The fraction of modified enzyme (in V0) was collected.
Purity of the conjugates
The purity of the conjugates was checked using HPLC/MSD ESI TOF Agilent technologies system 1200 series. In the column Zorbax Eclipse Plus C18 (RP; 100×2,1mm; 1,8 μm) was injected 1 μL of the sample. The temperature of the column was 40 °C and flow rate was 400 μL/min. Eluent (A) was 0.2% formic acid/water and eluent (B) was acetonitrile. After 20 min the chromatography was stopped.
SDS PAGE
SDS polyacrylamide gel was made by standard procedure21 using Hoefer SE 600 Ruby. The running gel was 10% and stacking gel was 4%. To the gel was applied 25 μL of each sample.
Characterisation of the conjugate
Mass spectrometry
The average molecular masses of intact and modified lysozyme were determined by Autoflex, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Samples deposited on gold coated target were desorbed and ionized by a 337 nm nitrogen laser which operated at 3 Hz repetition rate, using an accelerating voltage of 20 kV. The matrix was sinapic acid in 30 % acetonitrile with 0.1 % TFA. Spectra were acquired under delayed extraction conditions and in positive-ion linear mode.
Trypsin digestion
Protein spots were excised from Coomassie blue stained gels to perform in-gel trypsin digestion according to manufacturer’s directions. The obtained proteolytic peptides of each lysozyme conjugate were separately analyzed by mass spectrometry. Trypsin for proteomics was obtained from Sigma Chemical Co. (St. Louis, MO, USA).
Enzyme activity
Upon modification, activity of lysozyme and its conjugates was determinated toward cells of Micrococcus lysodeikticus (Sigma-Aldrich) by standard procedure.22 As a control, activity of lysozyme treated in the same way as during the preparation of the conjugates, but without avarone derivatives was determined.
Melting temperature
Fluorescence emission spectra were recorded using luminescence spectrometer LS 50 (Perkin Elmer, USA), equipped with a thermally controlled cell holder and a cuvette of 1 cm path length. Thermally induced unfolding of lysozyme and modified lysozymes was followed by measuring intrinsic emission fluorescence at 340 nm using excitation at 280 nm. Slit widths were set at 5 nm, for both the excitation and emission. Measurements were performed at several temperatures between 10 °C and 95 °C using heating rate of 0.5 °/min. The concentration of lysozyme and its bioconjugates was 0.04 mg/mL.
Antibacterial activity
A micro-broth dilution assay23 was used to evaluate the antimicrobial efficacy of avarone and its derivatives, lysozyme and its modifiers against Gram-positive and Gram-negative bacteria. The Gram-positive bacteria used were: Bacillus subtilis (ATCC 6633), Clostridium sporogenes (ATCC 19404), Streptosporangium longisporum (ATCC 25212), Micrococcus flavus (ATCC 10240), Sarcina lutea (ATCC 9341) and Staphylococcus aureus (ATCC 6538). The Gram-negative bacteria used were: Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 9027), Salmonella enteritidis (ATCC 13076) and Proteus vulgaris (ATCC 13315). Sterile 96-well polystyrene microtitre plates with well capacities of 300 μL were used and 100 μl of fresh Mueller Hinton broth was added to each well of the plate. One hundred microlitres of the lysozyme stock solution was added to each well of the first column. Then, 100 μL of the solution was removed from the first column and mixed thoroughly with the broth in the corresponding wells of the second column six times. Subsequently, a 100 μL aliquot was removed from each well in this column and mixed with contents of the the corresponding well of the next column. This doubling dilution was performed in rows across the plate except for the last row that was kept for use as control. The same procedure was repeated with stock solutions of lysozyme, treated lysozyme and lysozyme conjugates. Ten microlitres of bacterial cultures (108 cells per mL) were inoculated in each well of the plate. The microtitre plate was incubated at 37°C for 24 h. After that, bacterial growth was measured. The MIC was determinated as the lowest concentration that resulted in inhibition of bacterial growth.
Theoretical prediction
Molecular dynamics and building and geometry optimization of small molecules were performed using Discovery Studio® 2.5.5 software (DS, Accelrys Software Inc., 2009). Lysozyme structure was downloaded from Protein Data Bank (PDB, http://www.pdb.org/) in DS, carefully checked and then the potential covalent adducts with small molecules were made using that structure. For molecular dynamics simulations, “Standard Dynamic Cascade” protocol, from Discovery Studio® 2.5.5 protocols, was used. All calculations were done on Dell™ Precision™ 690 workstation and visualized using Planar SD2620W 26″ Widescreen Stereo/3D Monitor (Planar Systems Inc.).
RESULTS AND DISCUSSION
Theoretical prediction of the modification site
The amino acid residue(s) most likely to be modified by avarone and its derivatives were predicted by molecular dynamics simulations. The location of the residues and possible conformational changes upon modification might greatly influence the activity of the enzyme, and if the theoretical studies predicted a significant degree of inactivation, then the reaction conditions would have to be minutely adjusted to avoid unwanted modifications.
The crystal structure of the hen egg white lysozyme chain A, PDB code 2W1X24 was downloaded from RCSB protein database,25 and used for molecular dynamics simulations. In previous studies it was shown that avarone, its derivatives and related quinones modified lysine residues in proteins by nucleophilic addition.17,18,26 There are 6 such residues in this structure: Lys1, Lys13, Lys33, Lys96, Lys97 and Lys116. While all of them are remote enough from the active site of the enzyme, it seems that some are less available for covalent interactions than others. Lys1, for instance, is obviously involved in ion-pair formation with Glu7. Aliphatic chain of Lys33 is somewhat buried inside the hydrophobic surroundings (Phe34, Phe38, Trp123), limiting its motility and availability. Examination of rotamer libraries27 for Lys116 showed that conformation allowing easy reaction with the quinone would have a very low probability leaving Lys96 and Lys97 as the most likely candidates for this reaction. Although both amino acids are freely accessible, Lys97 has at least two conformations with good probability that make this residue very exposed for the reactions. Also, while the amino group from Lys96 is involved in hydrogen bonding with three amide oxygens from His15–Gly16 sequence, for Lys97 only hydrogen bond with the amide oxygen of Leu75 could be easily established. Moreover, MD simulations showed that in case of Lys97 modification, there are possibilities for additional bonding between polar groups on the quinone moiety and the protein, without disturbance of the protein structure. MD simulations showed good root mean square deviation (RMSD) for the main chain of the starting structure and the resulting conformations (less than 1.000), without influence on the binding site. In conclusion, the results indicate Lys97 as the modification site, without a significant change of lysozyme conformation, although influence on solvation could not be excluded.
Optimization of the conditions for modification with avarone
Selection of the medium and conditions for modification of lysozyme had to reconcile the insolubility or denaturation of the enzyme in organic solvents and the insolubility of avarone and its derivatives in water. Therefore, solvents that are miscible with water were used, limiting the concentration of the organic solvent to 20 %. The nucleophilicity of the amino groups of the enzyme was enhanced by the addition of a relatively weak base, avoiding strong beses because of the enzyme stability and prevention of quinone polymerization. Ammonium bicarbonate was selected because it allows to record mass spectra of the crude product of modification.
In order to study the effect of the atmosphere on the modification, the reaction was studied in 20 % ethanol and in 20 % DMSO in presence of 50 mM ammonium bicarbonate in the normal and inert atmosphere (nitrogen). The time of the reaction was 48 h at room temperature. Although the reaction starts immediately, as evidenced by the change of colour of the reaction mixture, it is completed only after 48 h. Higher temperatures could not be used due to unwanted polymerisation reactions of the enzyme and of the quinones. The extent of modification was studied by HPLC. The results showed that avarone modified the enzyme in both solvents, in normal and in inert atmosphere. This is an indication that the initially formed amino hydroquinone derivative could be oxidized by the still present quinone, so that amino quinone modificate was the final product. The area of the peak with the retention time of 12.09 min, corresponding to the modified lysozyme was larger in 20 % ethanol, so that it was chosen for the reaction. As expected, the hydroquinone avarol in the inert atmosphere could not modify lysozyme, irrespective of the solvent, but in the normal atmosphere it modified the enzyme in both solvents, which indicates the oxidation of the hydroquinone by oxygen, and the subsequent addition reaction of the protein amino group with the obtained quinone. In conclusion, the optimum reaction conditions were: lysozyme concentration 5 mg/ml, quinone concentration 2 mg/ml, 20 % ethanol, 50 mM ammonium bicarbonate, room temperature, normal atmosphere, 48 h, constant stirring. The excess of the quinone was used in order to ensure the fast reoxidation of the amino hydroquinone intermediate in the nucleophilic addition.
Characterization of the modificate
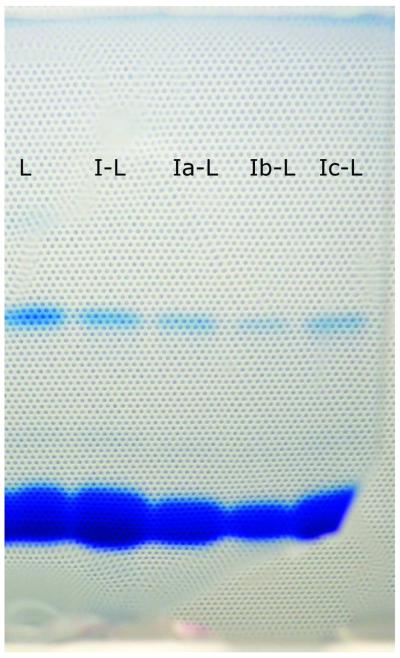
On SDS electrophoresis gel, untreated lysozyme had two bands: at 14 kDa for monomer and at 28 kDa for lysozyme dimer. Lysozyme modified with avarone and its derivatives had also two bands on gel (Fig. 1). Bands at 14 kDa were more prominent and bands at 28 kDa were weaker than bands of untreated lysozyme, indicating that the dimerization of lysozyme with avarone and its derivatives did not occur, contrary to the previous findings with β-lactoglobulin.17,18
Figure 1.
SDS electrophoregram.
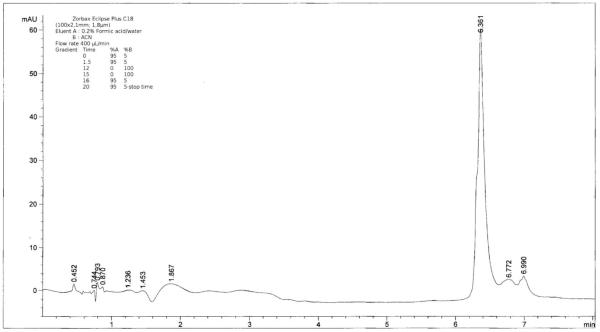
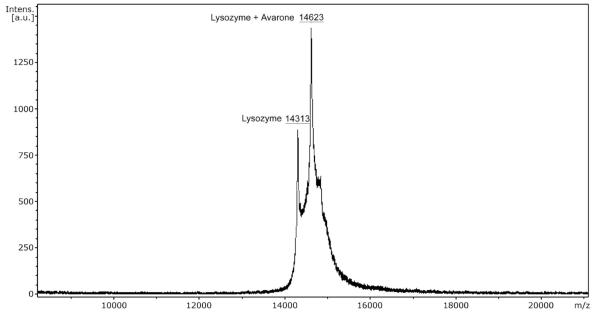
The purity of the modificates was also checked by HPLC (Fig. 2). The chromatogram showed a prominent peak of the modificate at the retention time of 6.361 min (for the avarone modificate I-L), with a shoulder due to unmodified lysozyme at 6.325 min. In order to determine how many quinone molecules were bound to lysozyme, mass spectra of native lysozyme and lysozyme modified by avarone (Fig. 3) and its derivatives (Table 1) using MALDI TOF were recorded. Spectra of the modified lysozyme show that avarone and its derivatives were covalently attached to the monomer of lysozyme. All mass spectra had two peaks: for lysozyme monomer at 14313 Da and for modified lysozyme at the mass of lysozyme plus mass of each compound minus 2 H atoms. The peak corresponding to the modified lysozyme is by far more abundant. It should be noted that under the applied conditions of the quinone compound in the large excess, only one molecule of the quinone was added to one molecule of lysozyme. This result is in stark contrast to the results obtained with simpler polyhydroxylic phenols under oxidizing conditions, which accounted for addition of up to four molecules of quinoid compounds.28
Figure 2.
HPLC chromatogram of the avarone-lysozyme conjugate.
Figure 3.
MALDI TOF mass spectrum of lysozyme and its conjugate with avarone.
Table 1.
Second peak on MALDI TOF mass spectra of lysozyme-conjugates with various derivatives of avarone
| Compound | Second peak (Da) |
|---|---|
| 3′-Methoxyavarone Ia-L | 14653 |
| 4′-(Methylamino)avarone Ib-L | 14652 |
| 3′-(Methylamino)avarone Ic-L | 14652 |
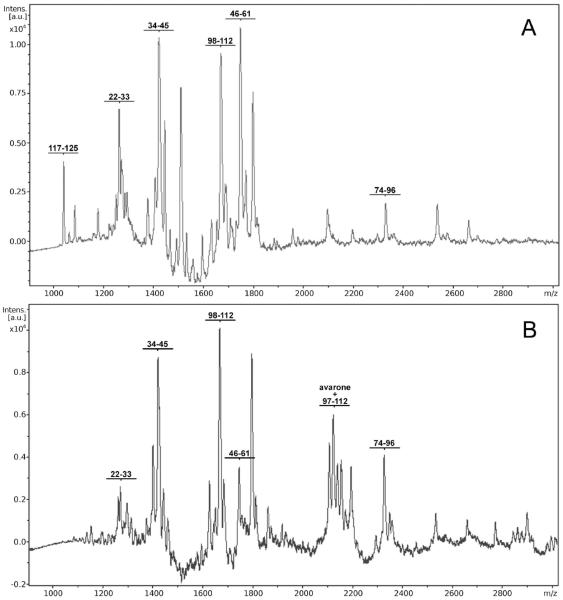
The position of the modification site was determined by tryptic digestion of both native and modified lysozyme and analysis of fragments. After tryptic digestion of native lysozyme, tryptic peptides covering 67 % of the sequence (Table 2) were obtained. In Fig. 4 MALDI TOF mass spectra of nonmodified lysozyme (Fig. 4A) and lysozyme modified by avarone (Fig. 4B), after tryptic digestion are shown. Spectra of modified lysozyme had additional peaks (Table 3) on mass corresponding to fragments of lysozyme which contain amino acid sequence 97-112 increased for the mass of avarone or its derivative without two H atoms. The presence of these peaks in all cases indicates that the site of attachment of avarone and its derivatives to lysozyme is lysine 97. Binding of avarone and its derivatives to lysine 97, because of the presence of their bulky groups and the loss of the positive charge, prevents tryptic digestion at the C-end of lysine 97, causing the appearance of new mass peaks with the mass for fragment 97-112, due to cleavage at Lys96, increased for the mass of each compound minus 2 H atoms. These results unambigously indicate that lysine 97 is the site of binding of avarone and its derivatives to lysozyme. It should be mentioned that particular reactivity of Lys97 was shown in an other study29 in which this residue was fluorescently labeled by N-hydroxysuccinimide esters. Namely, two residues were modified - Lys33 and Lys97, but the modification of the former was reversible and the modification of the latter was irreversible.
Table 2.
Peptide fragments obtained after tryptic digestion of native lysozyme
| Peptide fragment | mass (Da) |
|---|---|
| 74-96 | 2338.7138 |
| 46-61 | 1754.8503 |
| 98-112 | 1676.8896 |
| 34-45 | 1429.4882 |
| 22-33 | 1269.4602 |
| 117-125 | 1046.1705 |
Figure 4.
MALDI TOF mass spectra after tryptic digestion of non-modified lysozyme (4A) and lysozyme-avarone conjugate (4B).
Table 3.
New peaks in the mass spectra of lysozyme conjugates with various derivatives of avarone
| Compound | New peak mass (Da) |
|---|---|
| Avarone I-L | 2115.5296 |
| 3′-Methoxyavarone Ia-L | 2145.5496 |
| 4′-(Methylamino)avarone Ib-L | 2144.5696 |
| 3′-(Methylamino)avarone Ic-L | 2144.5696 |
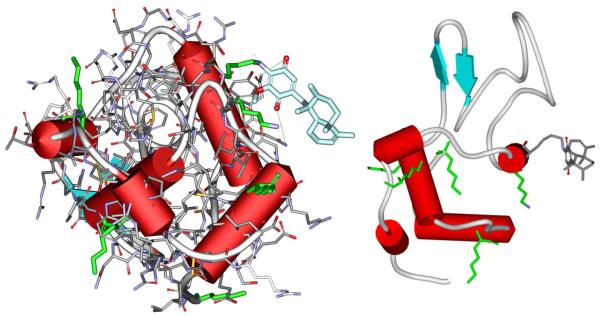
Lysine 97 which bound the quinone moiety is the residue which was predicted as the probable modification site by molecular modelling. It should be emphasized that it is located on the opposite side relative to the active site of lysozyme (Fig. 5). This means that the cleft is accesible for binding of the substrate, and that the activity of the enzyme should be preserved.
Figure 5.
Images of lysozyme conjugate with avarone bound to lysine 97.
Enzymatic activity
Based on the theoretical prediction, it could be expected that the activity of lysozyme would not be greatly reduced. Upon modification, activity of lysozyme was determined toward cells of Micrococcus lysodeikticus by standard procedure.22 For control, lysozyme was treated in the same way as during the modification reaction, but without the addition of the quinone. The enzymatic activities of lysozyme conjugates, as well as native and treated enzyme, are shown in Table 4. Lysozyme lost 23 % of its activity after exposure to the reaction conditions. The reduction of the activity after the reaction with the quinones was less pronounced, so that 59 to 67 % of the initial activity of the untreated enzyme was preserved. As expected, unsubstituted avarone (I) led to the lowest loss of activity, probably due to the least changes in the enzyme conformation, since addtional hydrogen bond donor/acceptor substituents are absent. The lowest activity was found for lysozyme modified with Ib, the only modificate in which the molecular modelling indicated an additional hydrogen bond between the methylamino group of the derivative and the backbone carbonyl of leucine 56.
Table 4.
Enzymatic activity of native lysozyme and lysozyme conjugates after modification
| lysozyme | treated lysozyme |
I-L | Ia-L | Ib-L | Ic-L | |
|---|---|---|---|---|---|---|
| activity (unit/mg) |
43000 | 32977 | 28647 | 26806 | 25267 | 27787 |
| remaining activity toward native enzyme (%) |
100 | 77 | 67 | 62 | 59 | 65 |
| remaining activity toward treated lysozyme (%) |
- | 100 | 87 | 81 | 77 | 84 |
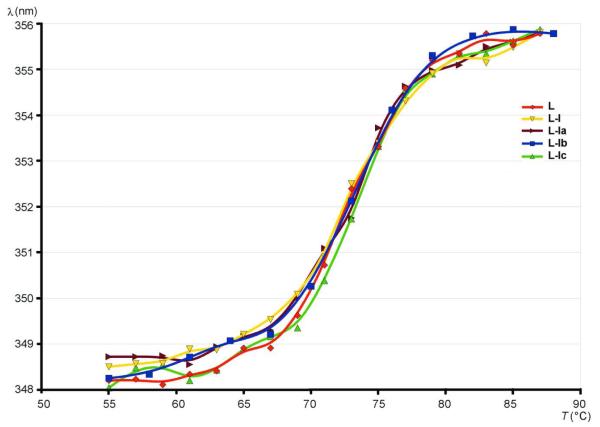
The melting temperatures of lysozyme and the modificates were measured by using fluorescence spectroscopy. As expected, the fluorescence intensity decreased with temperature30 for all the derivatives. By analysis of the dependence of the wavelength maximum of the fluorescence on temperature (Fig. 6), melting temperatures were found to be 72.74, 72.69, 73.01, 72.97 and 73.23 °C for lysozyme, and conjugates with I, Ia, Ib and Ic, respectively. Although the differences are not great, the general trend is that the melting point is somewhat increased in the conjugates, indicating a slight stabilization of structure in comparison to lysozyme. The slight conformational changes resulting from additional weak stabilizing interactions might be responsible for the reduction of activity.
Figure 6.
Change of fluorescence wavelength maximum with temperature.
Antibacterial activity
Since lysozyme is known to show antibacterial activity against Gram-positive bacteria, the effect of quinone-lysozyme conjugates toward bacteria was investigated. Minimum inhibitory concentration (MIC) of avarone, its derivatives and modified lysozyme toward selected Gram-positive and Gram-negative bacteria was determined and compared to unmodified lysozyme and the standard antibiotic amikacin. The results are shown in Table 5. As shown in Table 5, avarone and 3′-methoxyavarone had an activity to Gram positive bacteria similar to amikacin. The methylamino derivatives were weakly active or inactive. Lysozyme showed activity to all bacterial species tested, in particular to S. aureus. The activity of the conjugates was always similar to the activity of lysozyme or higher. The results indicate an intramolecular synergism of two components of the conjugates. Namely, the MIC values expressed as the quinone concentration were at least several times lower than MIC values of free quinones. It is particularly interesting that the conjugates with both methylamino derivatives (including the inactive 4′-derivative) showed antibacterial effects, with MIC values two to eight times lower than those of lysozyme.
Table 5.
Antibacterial activity of avarone, its derivatives, lysozyme and its conjugates with various derivatives of avarone towards Gram-positive bacteria (in parentheses are the MIC of the conjugates expressed as quinone concentration)
| MIC (μM) | ||||||
|---|---|---|---|---|---|---|
| Compound | S. lutea | S. aureus | M. flavus |
S. longisporum |
C. sporogenes |
B.subtilis |
| I | 20.03 | 40.06 | 20.03 | 20.03 | 20.03 | 80.13 |
| Ia | 36.55 | 36.55 | 36.55 | 36.55 | 18.27 | 36.55 |
| Ib | - | - | - | - | - | - |
| Ic | 469.21 | 3812.32 | 3812.32 | 3812.32 | 3812.32 | 967.74 |
| I-L | 168.23 (3.59) |
5.47 (0.12) |
168.23 (3.59) |
168.23 (3.59) |
336.46 (7.18) |
168.23 (3.59) |
| Ia-L | 95.54 (2.23) |
3.07 (0.07) |
191.09 (4.46) |
191.09 (4.46) |
191.09 (4.46) |
191.09 (4.46) |
| Ib-L | 102.38 (2.38) |
1.71 (0.04) |
402.68 (9.37) |
204.75 (4.77) |
204.75 (4.77) |
204.75 (4.77) |
| Ic-L | 95.55 (2.22) |
1.54 (0.04) |
395.85 (9.21) |
197.92 (4.61) |
197.92 (4.61) |
197.92 (4.61) |
| Treated-L | 223.57 | 27.94 | 111.79 | 447.15 | 223.57 | 111.79 |
| Lysozyme | 202.61 | 12.58 | 405.23 | 405.23 | 817.44 | 202.61 |
| Amikacin | 3.42 | 18.78 | 3.42 | 111.00 | 25.61 | 71.72 |
The results obtained with Gram negative bacteria were even more interesting (Table 6). Lysozyme was inactive to E. coli and P. vulgaris, but showed some activity to P. aeruginosa and S. enteritidis. Avarone and 3′-methoxyavarone were active (the activity of avarone was comparable to that of amikacin) and methylamino derivatives again inactive. However, all the conjugates were active, equally or more than lysozyme (except Ia-L for P. aeruginosa), and much more than the quinones. The MIC values of Ia-L for P. vulgaris and Ic-L for E. coli (for which both components of the bioconjugate were inactive) were in low micromolar range.
Table 6.
Antibacterial activity of avarone, its derivatives, lysozyme and its conjugates with various derivatives of avarone towards Gram-negative bacteria (in parentheses are the MIC of the conjugates expressed as quinone concentration)
| MIC (μM) | ||||
|---|---|---|---|---|
| Compound | E. coli | P. vulgaris | P. aeruginosa | S. enteritidis |
| I | 440.96 | 27.56 | 13.78 | 55.12 |
| Ia | 877.20 | 109.65 | 438.60 | 877.20 |
| Ib | - | - | - | - |
| Ic | - | - | - | - |
| I-L | 11.11 (0.24) |
22.22 (0.47) |
88.88 (1.90) |
11.11 (0.24) |
| Ia-L | 266.16 (6.21) |
2.14 (0.05) |
266.16 (6.21) |
68.25 (1.59) |
| Ib-L | 81.90 (1.91) |
156.98 (3.65) |
122.85 (2.86) |
307.13 (7.15) |
| Ic-L | 5.12 (0.12) |
10.24 (0.24) |
20.48 (0.48) |
40.96 (0.95) |
| Treated-L | 27.94 | 27.94 | 223.57 | 223.57 |
| Lysozyme | >740.58 | >740.58 | 188.64 | 370.29 |
| Amikacin | 8.54 | 11.95 | 85.38 | 13.66 |
The synergistic effects could be explained by different mechanisms of action, lysozyme targeting the cell wall and avarone derivatives DNA and/or proteins. Lysozyme might play the role of a vehicle that delivers the quinones to the cell. If the enzyme is attacked by bacterial hydrolytic enzymes smaller fragments containing the quinone moiety are formed, which continue to produce deleterious effects to the bacterial cell population. In order to check the effects of these hypothetical fragments, the derivative of avarone with lysine with protected α-amino group (Id), was synthesized and its antibacterial effects evaluated (Table 7). This derivative mimics a small modified peptide. The derivative was active to almost all bacteria, more active than methylamino derivatives of avarone. The activity was particularly strong to those bacteria that were the most susceptible to avarone-lysozyme conjugates, which means that the hypothesis that peptide fragments could retain a significant part of the activity is reasonable.
Table 7.
Antibacterial activity of 4′-((5-((tert-butoxycarbonyl)amino)-5-carboxypentyl)amino)avarone (Id)
| Bacteria | MIC (μM) | Amikacin |
|---|---|---|
| S. lutea | 1124.10 | 3.42 |
| S. aureus | < 4.32 | 18.78 |
| M. flavus | 1124.10 | 3.42 |
| S. longisporum | 2248.20 | 111.00 |
| C. sporogenes | 562.05 | 25.61 |
| B. subtilis | 2248.20 | 71.72 |
| E. coli | <4.32 | 8.54 |
| P. vulgaris | 2248.20 | 11.95 |
| P. aeruginosa | 2248.20 | 85.38 |
| S. enteritidis | >8992.81 | 13.66 |
CONCLUSION
A series of conjugates of lysozyme with the derivatives of avarone were prepared. In all of them only Lys97 added to the quinone moiety. The obtained conjugates retained most of the enzymatic activity and in many cases had an enhanced antibacterial activity compared to both the quinones and lysozyme. The intramolecular synergism might be explained by combined effects of the agents, whereby lysozyme might both target the cell wall, and serve as a vehicle for delivering the quinone which acts on DNA and proteins. Further studies of the biological effects of the conjugates especially regarding the cytotoxicity and toxicity seem justified.
Supplementary Material
ACKNOWLEDGEMENT
The authors are grateful to the Ministry of Education and Science of the Rebublic of Serbia for financial support (Project 172055). Authors wish to thank the DTPCC (Developmental Therapeutics Program Computer Center) at NCI-Frederick for using their resources for computer simulations. U.A. acknowledges a grant from Deutscher Akademischer Austausch Dienst (DAAD).
Footnotes
Supporting information
Mass spectra of the conjugates before and after tryptic digestion.
REFERENCES
- (1).Calleweart L, Michiels CW. Lysozyme in the animal kingdom. J. Biosci. 2010;35:127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- (2).Subroto T, Sufiati S, Beintema JJ. Papaya (Carica papaya) lysozyme is a member of the family 19 (basic, class II) chitinases. J. Mol. Evol. 1999;49:819–821. doi: 10.1007/pl00000075. [DOI] [PubMed] [Google Scholar]
- (3).Gao YG, Zhang S, Krentz S, Darius S, Power J, Lagarde G. Inhibition of spoilage lactic acid bacteria by lysozyme during wine alcoholic fermentation. Aust. J. Grape Wine Res. 2002;8:76–83. [Google Scholar]
- (4).Maidment C, Dyson A, Beard J. A study into measuring the antibacterial activity of lysozyme-containing foods. Nutr. Food Sci. 2009;39:29–35. [Google Scholar]
- (5).Cunningham FE, Proctor VA, Goetsch SJ. Egg-white lysozyme as a food preservative: an overview. World Poultry Sci. J. 1991;47:141–163. [Google Scholar]
- (6).Proctor VA, Cunningham FE. The chemistry of lysozyme and its use as a food preservative and a pharmaceutical. Crit. Rev. Food Sci. Nutr. 1988;26:359–395. doi: 10.1080/10408398809527473. [DOI] [PubMed] [Google Scholar]
- (7).Bratlid D, Moe PJ. Serum lysozyme activity in children with acute leukemia. Eur. J. Pediatr. 1978;127:263–268. doi: 10.1007/BF00493542. [DOI] [PubMed] [Google Scholar]
- (8).Maroufi B, Ranjbar B, Khajeh K, Manesh HN, Yaghoubi H. Structural studies of hen egg-white lysozyme dimer: Comparison with monomer. Biochim. Biophys. Acta. 2008;1784:1043–1049. doi: 10.1016/j.bbapap.2008.03.010. [DOI] [PubMed] [Google Scholar]
- (9).Radziejewska RC, Lesnierowski G, Kijowski J. Antibacterial activity of hen egg white lysozyme modified by thermochemical technique. Eur. Food Res. Technol. 2009;228:841–845. [Google Scholar]
- (10).Radziejewska RC, Leśnierowski G, Kijowski J. Properties and application of egg white lysozyme and its modified preparation - a review. Pol. J. Food Nutr. Sci. 2008;58:5–10. [Google Scholar]
- (11).Van der Linden DS, Short D, Dittmann A, Yu PL. Synergistic effects of ovine-derived cathelicidins and other antimicrobials against Escherichia coli O157:H7 and Staphylococcus aureus 1056 MRSA. Biotechnol.Lett. 2009;31:1265–1267. doi: 10.1007/s10529-009-0010-9. [DOI] [PubMed] [Google Scholar]
- (12).Matters D, Cooper HJ, McDonnell L, Iniesta J, Heptinstall J, Derrick P, Walton D, Peterson I. Mass spectrometry in demonstrating the site-specific nitration of hen egg white lysozyme by an improved electrochemical method. Anal. Biochem. 2006;356:171–181. doi: 10.1016/j.ab.2006.06.033. [DOI] [PubMed] [Google Scholar]
- (13).Zhang HM, Tang BP, Wang YQ. The interaction of lysozyme with caffeine, theophylline and theobromine in solution. Mol. Biol. Rep. 2010;37:3127–3136. doi: 10.1007/s11033-009-9891-x. [DOI] [PubMed] [Google Scholar]
- (14).Sladić DM, Gašić MJ. Reactivity and biological activity of the marine sesquiterpene hydroquinone avarol and related compounds from sponges of the order Dictyoceratida. Molecules. 2006;11:1–33. doi: 10.3390/11010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Minale L, Riccio R, Sodano G. Avarol, a novel sesquiterpenoid hydroquinone with a rearranged drimane skeleton from the sponge Dysidea avara. Tetrahedron Lett. 1974;38:3401–3404. [Google Scholar]
- (16).De Rosa S, Minale L, Riccio R, Sodano G. The absolute configuration of avarol, a rearranged sesquiterpenoid hydroquinone from a marine sponge. J. Chem. Soc. Perkin Trans. 1976;1:1408–1414. [Google Scholar]
- (17).Novaković I, Vujčić ZM, Božić TT, Božić NM, Milosavić NB, Sladić DM. Chemical modification of β-lactoglobulin by quinones. J. Serb. Chem. Soc. 2003;68:243–248. [Google Scholar]
- (18).Sladić DM, Novaković I, Vujčić ZM, Božić TT, Božić NM, Milić D, Šolaja BA, Gašić MJ. Protein covalent modification of biologically active quinones. J. Serb. Chem. Soc. 2004;69:901–907. [Google Scholar]
- (19).Trifunović I, Sladić D, Dogović N, Gašić MJ. Carbon-13 chemical shift assigment in substituted 1,4-cyclohexadienones. J. Serb. Chem. Soc. 1987;52:559–563. [Google Scholar]
- (20).Cozzolino R, De Giulio A, De Rosa S, Strazzullo G, Gašić MJ, Sladić D, Zlatović M. Biological Activities of Avarol Derivatives, 1. Amino Derivatives. J. Nat. Prod. 1990;53:699–702. [Google Scholar]
- (21).Laemlli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- (22).Shugar D. Biochim. Biophys. Acta. 1952;8:302–309. doi: 10.1016/0006-3002(52)90045-0. [DOI] [PubMed] [Google Scholar]
- (23).NCCLS (National Committee for Clinical Laboratory Standards) Approval standard document M7-A5. Villanova, PA, USA: 2000. [Google Scholar]
- (24).Cianci M, Helliwell JR, Suzuki A. The interdependence of wavelength, redundancy and dose in sulfur SAD experiments. Acta Crystallogr. Sect. D. 2008;64:1196–1209. doi: 10.1107/S0907444908030503. [DOI] [PubMed] [Google Scholar]
- (25). http://www.rcsb.org/
- (26).Pajić I, Vujčić Z, Vujčić M, Novaković I, Sladić D, Gašić MJ. Chemical modification of the lectin of the marine coral Gerardia savaglia by marine quinone avarone. J. Serb. Chem. Soc. 2007;72:1271–1274. [Google Scholar]
- (27).Ponder JW, Richards FM. Tertiary Templates in Proteins. Use of Packing Criteria in the Enumeration of Allowed Sequences for Different Structural Classes. J. Mol. Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- (28).Rawel HM, Kroll J, Rohn S. Reactions of phenolic substances with lysozyme - physicochemical characterisation and proteolytic digestion of the derivatives. Food Chem. 2001;72:59–71. [Google Scholar]
- (29).Teske CA, Simon R, Niebisch A, Hubbuch J. Changes in Retention Behavior of Fluorescently Labeled Proteins During Ion-Exchange Chromatography Caused by Different Protein Surface Labeling Positions. Biotechnol. Bioeng. 2007;98:193–200. doi: 10.1002/bit.21374. [DOI] [PubMed] [Google Scholar]
- (30).Zielenkiewich W, Swierzewski R, Attanasio F, Rialdi G. Thermochemical, volumetric and spectroscopic properties of lysozyme-poly(ethylene)glycol system. J. Therm. Anal. Calorim. 2006;83:587–595. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.