Abstract
Chitin, which is a biopolymer of the amino sugar glucosamine (GlcN), is highly abundant in aquatic ecosystems, and its degradation is assigned a key role in the recycling of carbon and nitrogen. In order to study the significance of chitin decomposition in two temperate freshwater lakes with contrasting trophic and redox conditions, we measured the turnover rate of the chitin analog methylumbelliferyl-N,N′-diacetylchitobioside (MUF-DC) and the presence of chitinase (chiA) genes in zooplankton, water, and sediment samples. In contrast to the eutrophic and partially anoxic lake, chiA gene fragments were detectable throughout the oligotrophic water column and chiA copy numbers per ml of water were up to 15 times higher than in the eutrophic waters. For both lakes, the highest chiA abundance was found in the euphotic zone—the main habitat of zooplankton, but also the site of production of easily degradable algal chitin. The bulk of chitinase activity was measured in zooplankton samples and the sediments, where recalcitrant chitin is deposited. Both, chiA abundance and chitinase activity correlated well with organic carbon, nitrogen, and concentrations of particulate GlcN. Our findings show that chitin, although its overall contribution to the total organic carbon is small (∼0.01 to 0.1%), constitutes an important microbial growth substrate in these temperate freshwater lakes, particularly where other easily degradable carbon sources are scarce.
INTRODUCTION
Chitin is a homopolymer of β-1,4-linked N-acetylated glucosamine (GlcNAc). It is a structural component of the cell walls of fungi and the exoskeletons of invertebrates but is also found in protozoa (45) and algae (23, 31). Due to its wide distribution, chitin is, after cellulose, the second most abundant biopolymer on earth (35). The annual production and the steady-state amount in the biosphere are on the order of 1012 to 1014 kg (30, 47). On the basis of literature data on chitin production by arthropods, the total annual chitin production in aquatic environments was estimated at 2.8 × 1010 kg chitin year−1 for freshwater ecosystems and at 1.3 × 1012 kg chitin year−1 for marine ecosystems (10). The role of chitin as a significant component of the aquatic carbon and nitrogen budget was studied extensively during the 1990s, but almost exclusively in estuarine and marine environments (7, 17, 33, 34, 48). Studies on bacterial chitin degradation in lake water are rare (4, 14, 37, 42).
Not only phyto- and zooplankton and insect carcasses, but also zooplankton molting (exuviae) and excretion of fecal pellets (peritrophic membranes) contribute to the production of huge amounts of chitinous particles in the water column (61). These chitinous particles are part of the marine or lake snow, which was shown to represent a hot spot of particulate organic matter solubilization (18, 19, 52). In the ocean, chitinolytic bacteria were found to be responsible for the hydrolysis of chitin (35, 64). After adhering to the polymeric substrate, chitinolytic bacteria express a multitude of enzymes and other proteins required for its catabolism (33). The hydrolysis of the β-(1,4)-glycosidic bonds between the GlcNAc residues is accomplished by extracellular chitinases (EC 3.2.1.14) (22). The end products of chitin degradation in the chitinolytic pathway are monomers and dimers of GlcNAc, which can be catabolized in the cytoplasm to fructose-6-P, acetate, and NH3 (3, 33).
Based on amino acid similarities, chitinases are classified into family 18 and family 19 (22). Family 19 chitinases were formerly thought to be restricted to plant origin but have since also been found in various Streptomyces species and other bacteria (4, 49, 57). However, the most information on bacterial diversity and distribution in diverse environments is available for family 18 group A chitinases (11, 24, 25, 36, 38, 55).
In the present study, we aimed to identify the main sites of chitin hydrolysis and the significance of chitin as a bacterial substrate in two temperate freshwater lakes with contrasting trophic and redox conditions. For this purpose, we analyzed the chitinase activities and the abundances of bacterial chitinase genes (chiA) in zooplankton, water from 10 different depths, and sediment samples from oligotrophic Lake Brienz (LB) and eutrophic Lake Zug (LZ). The lakes were sampled in spring and fall 2009.
MATERIALS AND METHODS
Sampling sites.
The characteristics of the study sites are listed in Table 1. LB is an oligotrophic perialpine lake located 70 km southeast of Bern, Switzerland. The lake is fully oxic throughout the year. The catchment of the lake is drained by the two main inflows, Aare and Lütschine, which together transport an annual average of 3 × 108 kg suspended material into LB (16). Both, the hydrological regime and the suspended particle load of the river Aare are influenced by hydropower operations. The continuous supply of suspended glacial particles, causing reduced light penetration, together with the scarcity of nutrients has led to an unusually low phytoplankton biomass (on average, <10 g m−2) (15, 21).
Table 1.
Characteristics of study sites
| Property | Lake Brienz | Lake Zug South Basin | Reference(s) |
|---|---|---|---|
| Position | 46°43′N, 7°58′E | 47°7′N, 8°29′E | |
| Elevation (m) | 564 | 414 | |
| Surface area (km2) | 29.8 | 16 | 21, 43 |
| Maximum depth (m) | 259 | 200 | 21, 43 |
| Volume (km3) | 5.15 | 2.0 | 21, 43 |
| Primary inflow | Aare, Lütschine | Rigiaa | |
| Primary outflow | Aare | ||
| Mean hydraulic residence time (yr) | 2.7 | 14 | 21, 43 |
| Trophic status | Oligotrophic | Eutrophic | |
| Oxygen status | Oxic | Anoxic below 140–160 m | 43 |
LZ is a eutrophic subalpine lake 30 km South of Zurich, Switzerland. The lake consists of two basins: a shallow (40- to 60-m depth) North Basin and a 200-m-deep South Basin. The South Basin is meromicitc, with mixing depths that do not exceed 100 m. Together with the eutrophic status of LZ, this leads to seasonally anoxic conditions at a depth of 140 to 160 m and permanent anoxia below that depth (41, 43).
Sampling water and sediments.
LB was sampled in mid-May and mid-September 2009 in the central part of its basin at position 46°43′18″N, 7°58′27″E. LZ was sampled at the end of March and the end of October 2009 in the South Basin at position 47°6′1″N, 8°29′4″E.
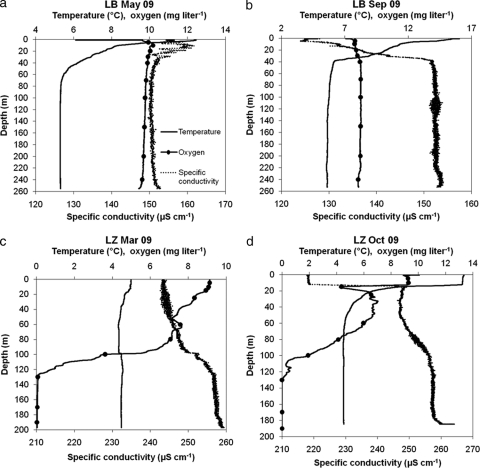
Profiles of temperature, oxygen (O2), and conductivity were taken with a conductivity-temperature-depth (CTD) profiler (Seabird SBE19; Sea-Bird Electronics, Inc., Bellevue, WA). Based on the temperature and O2 profiles determined (Fig. 1), water was sampled from 10 depths using a Niskin water sampler, poured into autoclaved 1-liter glass bottles, and transported cool and in the dark to the laboratory. We sampled LB at water depths of 5, 10, 20, 30, 40, 70, 100, 150, 200, and 240 m and LZ at 5, 10, 15, 25, 60, 80, 100, 130, 170, and 190 m.
Fig 1.
Profiles of temperature, oxygen, and conductivity of the water columns of LB sampled in May (a) and September (b) 2009 and of LZ sampled in March (c) and October (d) 2009. The dots along the oxygen profile indicate sampled water depths.
Particulate organic matter (POM) from the same depths was sampled on 0.7-μm glass fiber filters (Whatman Inc., Florham Park, NJ) with in situ pumps (McLane Research Laboratories Inc., Falmouth, MA) until the filters were clogged.
Sediment cores were recovered from the two sampling sites using a gravity corer (32). The first 5 (spring) to 7 (fall) centimeters of each core were sliced at intervals of 1 centimeter. Subsamples of each layer were processed for microbiological and biogeochemical analysis (see below).
Zoo- and phytoplankton communities.
Zooplankton samples were taken with a 95-μm double-closing net (8) from 0 to 100 m (2 replicates) and preserved in 2% formaldehyde. Phytoplankton was sampled with an integrated sampler according to the method of Schröder (51) from 0 to 20 m (2 replicates). Lugol-fixed phytoplankton species were counted using the technique of Utermöhl on an inverted microscope (58). Crustacean species and their developmental stages were enumerated under a binocular dissecting microscope at ×10 to ×75. Phyto- and zooplankton biomass fresh weights were calculated from the mean cell/organism dimensions of each species (9, 21). For LB, these analyses were carried out at the Laboratory for Water and Soil Protection of the Canton of Bern.
Zooplankton chitin.
The chitin biomass in LB and LZ was calculated from the zooplankton biomass and body chitin content published by Cauchie (10), which were 4.3% and 9.8% (dry weight) for lentic branchiopoda and copepoda, respectively.
Chemical analysis.
The zooplankton, water, and sediment samples were subdivided for microbiological and chemical analyses.
The dissolved organic carbon (DOC) and dissolved nitrogen (DN) concentrations in lake water were determined after filtration through a 0.2-μm Supor membrane filter (Pall Corporation, Port Washington, NY). DOC and total organic carbon (TOC) were measured by high-temperature catalytic oxidation (720°C) with a Shimadzu TOC-V CPH (Shimadzu Scientific Instruments, Kyoto, Japan). The total nitrogen (TN) content and DN concentration were determined with a Shimadzu TOC-V CPH/TNM1.
Phosphate (PO43−) concentrations were determined following filtration through 0.45-μm cellulose acetate filters (Schleicher & Schuell GmbH, Dassel, Germany). PO43− was determined photometrically by the molybdenum blue method according to Vogler (59).
Concentrations of GlcN in zooplankton samples, in POM, and in the sediments were measured by a slightly modified method according to Zhang and Amelung (63) with a derivatization step according to Guerrant and Moss (20) and with myoinositol (Aldrich) as an internal standard. Filters were treated with 10 ml of 6 M HCl for 10 h at 100°C, which should ensure complete hydrolysis of biopolymers of GlcN, including chitin and peptidoglycan, in which it occurs in its N-acetylated form (GlcNAc). Hydrolysis causes deacetylation, and thus, concentrations of GlcN are the sums of both forms, GlcN and GlcNAc. The gas chromatography (GC) system was equipped with a flame ionization detector (HRGC 5160; Carlo Erba Instruments, Milan, Italy), a split-splitless injector, and a VF-5 MS column (60 m, 0.25-mm inner diameter, and 0.25-μm film thickness). The injector temperature was 250°C and the temperature of the detector was 300°C. Hydrogen was used as the carrier gas, with a flow rate of 2 ml min−1. The temperature profile was as follows: 120°C to 200°C at 20°C min−1, 200°C to 250°C at 2°C min−1, and 250°C to 270°C at 20°C min−1, held for 10 min at 270°C. A GlcN standard (d-glucosamine; Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) was also derivatized and used for quantification.
Chitinase activity.
Chitinase activity on water, sediment, and zooplankton sampled in fall 2009 was determined using the chitin substrate analog methylumbelliferyl-N,N′-diacetylchitobioside (MUF-DC) (Sigma-Aldrich) according to a previous method (34, 37) with slight modifications. Referring to the manufacturer's instructions, MUF-DC was dissolved in 100% dimethyl formamide (DMF) to a final concentration of 5 mM (stock solution).
In a preliminary test, the effect of the substrate concentration was determined by incubating LZ surface waters and mixed LZ sediment samples (0 to 5 cm) with six different MUF-DC concentrations (1, 5, 10, 50, 100, and 300 μM) at 20°C and 4°C, respectively. In addition, we tested DMF for inhibitory effects on Streptomyces griseus chitinase (Sigma). DMF was found to linearly inhibit activity up to 80% at 6% DMF, the concentration in the highest MUF-DC concentration (300 μM) used in the assay. Therefore, we adjusted the concentration of DMF for all assays to 6%.
Centrifuge tubes (50 ml: Greiner Bio-One, Frickenhausen, Germany) were filled with water samples mixed with formalin to a final concentration of 0.25% to prevent microbial growth (27). The influence of 0.25% formalin on the fluorescence of released MUF and on the activity of S. griseus chitinase was tested in preliminary experiments. No significant effects were found. Formalin-fixed water samples were stored cold in the dark until assayed (within 4 h after sample collection).
Sediment samples (0.5 g) and 1.48 ml of autoclaved, 0.2-μm-filtered and formalin-treated (0.25%) lake water were well mixed and assayed within 12 h of sample collection.
Water and sediment samples were amended with aliquots of the MUF-DC stock solution and incubated at 4 and 20°C, which correspond to the minimum and maximum temperatures measured in the water columns of both lakes over a year. As in situ temperatures for different samples differ from the incubation temperatures (Fig. 1) and sediments were analyzed as slurries, our data (see Fig. 3 and 4) are potential chitinase activity rates. After incubation for 1 to 3 h, the reactions were stopped in aliquots of 100 and 150 μl of water and sediment-in-water suspension by adding 10 and 15 μl of ammonium glycine buffer (pH 10.5) (12), respectively. The fluorescence of free methylumbelliferone (MUF) was measured in the water samples and sediment supernatants at 360-nm excitation and 460-nm emission using a Synergy HT microplate reader (Bio-Tek Instruments, Inc., Winooski, VT). The samples were shaken (300 rpm) between measurements.
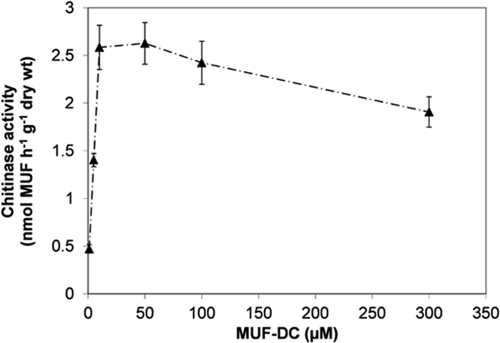
Fig 3.
Effect of substrate concentration (1, 5, 10, 50, 100, 300 μM MUF-DC) on chitinase activity in mixed LZ sediment slurries (0 to 5 cm) assayed at 4°C. The error bars indicate standard deviations of triplicate samples from one sediment core sampled in the fall.
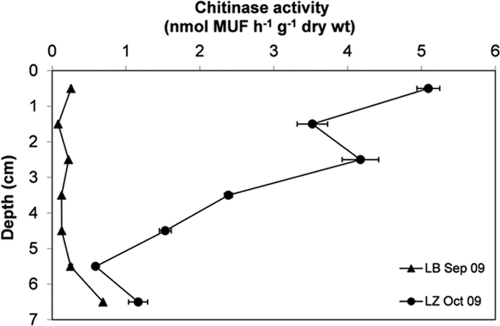
Fig 4.
Chitinase activities in the upper 7 centimeters of LB and LZ sediment relative to sediment dry weight in fall 2009 at 4°C. MUF, fluorescent methylumbelliferone released after the hydrolysis of the chitin substrate analog methylumbelliferyl-N,N′-diacetylchitobioside. The error bars represent standard deviations of triplicate measurements of one sample.
Zooplankton was killed by 3% hydrochloric acid and washed with autoclaved and 0.2-μm-filtered lake water. Zooplankton (0.05 g) was suspended in 2 ml 0.25% formalin-treated, autoclaved, and 0.2-μm-filtered lake water. After adding an aliquot of the MUF-DC stock solution, samples were incubated and processed as described for the sediments. The sediments were measured repeatedly up to 11 h, zooplankton up to 9 h, and water up to 100 h.
Positive controls were assayed in autoclaved, 0.2-μm-filtered, and formalin-treated (0.25%) lake water and in autoclaved sediments containing aliquots of MUF-DC stock solution and S. griseus chitinase. In addition, negative controls without S. griseus chitinase were run to test for abiotic degradation of MUF-DC.
DNA extraction.
Autoclaved glass bottles (1 liter) were filled with water samples and transported on ice and in the dark to the laboratory. About 5 liters of water from each sampled depth was filtered through a 5-μm-isopore membrane filter (Millipore, Billerica, MA) and a 0.2-μm polycarbonate filter (Whatman), each 142 mm in diameter, connected in series. The filters were frozen in liquid nitrogen immediately after filtration and stored at −80°C until DNA extraction. For extraction, a filter segment (1/4) was cut into small pieces using sterile scissors and mixed with 0.2 g of glass beads (0.1 g of 106-μm and 0.1 g of 150- to 212-μm glass beads; Sigma-Aldrich) in a 2-ml screw-cap tube (Brand GmbH & Co KG, Wertheim, Germany). Ice-cold extraction buffer (1.4 ml) (26) was added. The cells were disrupted in a FastPrep-24 bead-beating system (MP Biomedicals, Solon, OH) by beating twice for 40 s at 4 m s−1, placing the tubes on ice in between. Bead beating was followed by a freeze-thaw cycle in liquid nitrogen. The supernatant was treated with 50 μg ml−1 RNase A (Sigma-Aldrich) for 30 min at 37°C and extracted with an equal volume of phenol-chloroform-isoamylalcohol (25:24:1; pH 8; Sigma-Aldrich). After precipitation with 1 volume of isopropanol, the pelleted DNA was dissolved in Tris-EDTA (pH 8) buffer and stored at −80°C.
For extraction of sediment and zooplankton, 0.5-g and 0.05-g samples, respectively, were mixed with 1.4 ml ice-cold nucleic acid extraction buffer (26). The samples were frozen in liquid nitrogen and stored at −80°C until DNA extraction. After thawing, 0.25 g sterile 0.1-mm Zirconia beads (Biospec Products Inc., Bartlesville, OK) was added, and the samples were processed on a vortex adaptor (MoBio Laboratories, Inc., Carlsbad, CA) for 1 min at maximum speed. DNA extraction was performed as for the water filters.
The quality of DNA extracts was checked by agarose gel (1%) electrophoresis. Extracted DNA was quantified by fluorescence spectroscopy using the Quant-iT PicoGreen double-stranded DNA (dsDNA) Assay Kit (Molecular Probes, Eugene, OR) and a Synergy HT microplate reader (Bio-Tek Instruments).
The reproducibility of the applied DNA extraction protocols was tested on quadruplicate samples of sediment slurries and on four filter segments (1/4) of a single 0.2-μm polycarbonate filter, one for each lake.
Amplification and quantification of chitinase gene fragments.
We used the primer pair chif2 (GACGGCATCGACATCGATTGG) and chir (CSGTCCAGCCGCGSCCRTA), which was reported to target chitinase family 18 group A (chiA) gene fragments from a broad range of chitinolytic bacteria (62). The performance and specificity of chiA PCR primers were tested on spring water samples (LB at 10 m, LB at 240 m, LZ at 10 m, and LZ at 190 m).
Each PCR mixture (20 μl) contained 10 μl 2× Power SYBR green PCR Master Mix (Applied Biosystems, Foster City CA), 0.4 μl of each primer (10 μM; Microsynth, Balgach, Switzerland), 2 μl bovine serum albumin (BSA) (10 mg ml−1; Sigma-Aldrich), and 5 μl template diluted in nuclease-free water (Qiagen GmbH, Hilden, Germany). Amplification was performed in a 7500 Fast Real-Time PCR System (Applied Biosystems) with PCR conditions consisting of an initial denaturation step at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 65°C for 60 s, and extension at 72°C for 30 s, and finally by melting-curve analysis.
PCR resulted in products yielding a single sharp band of the expected size of ∼430 bp in gel electrophoresis (data not shown). To evaluate the specificity of the chiA PCR primers, we constructed clone libraries from the spring water samples. PCR products of chitinase gene fragments from samples were cloned into the pGEM-T vector by using the pGEM-T Easy Vector System according to the instructions of the manufacturer (Promega). Screening of the clone libraries constructed from these samples resulted in 15 different restriction fragment length polymorphism (RFLP) types among 71 screened plasmids. We sequenced 1 to 5 clones of each RFLP type, 31 clones in total (sequencing by Microsynth AG, Balgach, Switzerland). All sequences could be assigned to chitinases, indicating high specificity of the PCR protocol (unpublished data).
In preliminary experiments, the PCR amplification was assayed in 4-fold serial dilutions of total DNA extracts in nuclease-free water (Qiagen) to determine the effects of inhibiting contaminants. For instance, according to the resulting copy numbers, we assayed lake sediments in 64-fold dilutions, as they gave comparable but slightly higher (∼13%) chiA copy numbers than 16-fold-diluted templates, while results dropped off significantly for the 256-fold dilution (∼40%). Four-fold dilution of zooplankton samples and 16-fold and 64-fold dilutions of water and sediment samples, respectively, were applied in the final analysis. Therefore, different amounts of genomic DNA template were used in quantitative PCR (qPCR), i.e., less than 5 ng DNA for LB samples and ∼10 to 20 ng DNA for LZ samples. Additionally, the results of quadruplicate analyses of DNA extraction yields showed high standard deviations, up to 31%, for water filters. For sediment slurries, the standard deviation was 6%.
RESULTS
Biogeochemistry of lake water columns. (i) Oxygen.
The temperature, O2, and conductivity profiles for both lakes sampled in spring and fall are shown in Fig. 1. In both seasons, the water column of LB was fully oxic. For LZ, anoxic conditions were measured below 130 m (O2 < 0.1 mg liter−1), a shallower depth than reported previously (41, 43).
(ii) Organic carbon.
In LZ waters, the TOC concentrations were roughly three to four times higher than in LB waters (see Fig. S1a in the supplemental material) and ranged from 1.86 to 2.54 mg C liter−1 (error of measurement, 0.20 mg C liter−1). The DOC values ranged between 1.80 ± 0.20 mg C liter−1 (March 2009; 170 m) and 2.40 ± 0.20 mg C liter−1 (October 2009; 5 m). For LB, the DOC concentrations were below or close to the detection limit of 0.50 mg C liter−1 in both seasons.
(iii) Nitrogen.
The TN and DN concentrations were below the detection limit of 0.50 mg N liter−1, except for LZ waters in spring (data not shown).
(iv) Phosphate.
The PO43− concentrations were below 5 μg P liter−1 for all sampled water depths of LB, with the exception of the 5-m and 20-m surface water depths sampled in September 2009 (see Fig. S1c in the supplemental material). For LZ the PO43− concentrations increased with water depth. The concentrations ranged from 3.8 ± 0.5 μg P liter−1 (October 2009; 15 m) to 217 ± 0.5 μg P liter−1 (October 2009; 190 m).
(v) Glucosamine.
For both lakes, concentrations of particulate GlcN were highest in the euphotic zone and decreased with depth (see Fig. S1d in the supplemental material). The highest value of 70.6 nmol liter−1 was measured at the 5-m water depth of LZ sampled in March 2009. For LB, the GlcN concentrations were roughly 1 order of magnitude lower than for LZ.
Biogeochemistry of sediment profiles.
For LB sediments, the TOC, TN, and GlcN contents were always lowest for the 1- to 2-cm layer, in which the sediment becomes anoxic (46), and increased again with depth (Table 2). Biogeochemical parameters in LZ sediment (Table 3) were roughly 1 order of magnitude higher than in LB sediments, with the exception of the 6- to 7-cm layer sampled in fall, which showed similar TOC, TN, and GlcN contents in both lakes.
Table 2.
Biogeochemistry of sediments from LB sampled in May and September 2009a
| Depth (cm) | Core LB, May 2009 |
Core LB, September 2009 |
||||
|---|---|---|---|---|---|---|
| TOC (% dry wt) | TN (% dry wt) | GlcN (μmol g dry wt−1) | TOC (% dry wt) | TN (% dry wt) | GlcN (μmol g dry wt−1) | |
| 0–1 | 0.59 | 0.05 | 1.30 | 0.67 | 0.04 | 0.48 |
| 1–2 | 0.39 | 0.04 | 0.62 | 0.48 | 0.04 | 0.38 |
| 2–3 | 0.48 | 0.04 | 0.97 | 0.70 | 0.06 | 0.97 |
| 3–4 | 0.61 | 0.06 | 0.84 | 0.65 | 0.06 | 0.73 |
| 4–5 | 0.67 | 0.06 | 0.92 | 0.70 | 0.06 | 1.04 |
| 5–6 | 0.82 | 0.08 | 0.75 | |||
| 6–7 | 1.23 | 0.13 | 2.58 | |||
Error of measurement (standard deviation) of one sample: TOC, ±0.1%; TN, ±0.02%; GlcN, ±10%.
Table 3.
Biogeochemistry of sediments from LZ sampled in March and October 2009a
| Depth (cm) | Core LZ, March 2009 |
Core LZ, October 2009 |
||||
|---|---|---|---|---|---|---|
| TOC (% dry wt) | TN (% dry wt) | GlcN (μmol g dry wt−1) | TOC (% dry wt) | TN (% dry wt) | GlcN (μmol g dry wt−1) | |
| 0–1 | 4.72 | 0.72 | 14.2 | 3.27 | 0.44 | 14.9 |
| 1–2 | 2.85 | 0.39 | 6.56 | 3.08 | 0.38 | 13.7 |
| 2–3 | 4.99 | 0.73 | 12.3 | 4.12 | 0.49 | 19.1 |
| 3–4 | 4.94 | 0.74 | 13.3 | 2.64 | 0.33 | 7.29 |
| 4–5 | 3.60 | 0.48 | 10.5 | 2.80 | 0.39 | 5.55 |
| 5–6 | 2.11 | 0.30 | 8.23 | |||
| 6–7 | 1.31 | 0.17 | 4.36 | |||
Error of measurement (standard deviation) of one sample: TOC, ±0.1%; TN, ±0.02%; GlcN, ±10%.
Zoo- and phytoplankton community compositions.
The phytoplankton communities in both lakes were composed of chrysophytes (golden algae), diatoms, cryptophytes, dinoflagellates, chlorophytes (green algae), haptophytes (only in LB in May 2009), euglenoids (only in LZ in October 2009), and cyanobacteria. The phytoplankton biomass was dominated by diatoms, except for LZ in March 2009, when the predominant organisms were cyanobacteria (60%). Diatoms accounted for 40% (May 2009) and 66% (September 2009) of the biomass in LB and for 16% (March 2009) and 71% in LZ (October 2009).
In both lakes, the predominant zooplankton species were cladocerans (Daphnia sp., Diaphanosoma brachyurum, Leptodora kindtii, and Bosmina sp. [only in LZ in March 2009]) and calanoid copepods (Diaptomidae). The vast majority of the zooplankton biomass consisted of copepods. For the spring sampling campaign, they accounted for over 99% of the zooplankton biomass in both lakes, and for the fall sampling campaign, for 61% and 80% in LB and LZ, respectively.
Zoo- and phytoplankton biomass.
In March/May 2009, the zoo- and phytoplankton biomasses contributed equally to the total plankton biomass in both lakes (Fig. 2). In LZ, the plankton biomass (25.9 g m−2) was more than double that of LB (11.7 g m−2). In October 2009, the vast majority of the LZ plankton biomass (33.9 g m−2) was phytoplankton (>90%). The late fall phytoplankton bloom is in agreement with chlorophyll a measurements from the same year (Environmental Agency of Canton Zug, unpublished data). The low proportion of zooplankton biomass also goes along with the predominance of large grazing-resistant forms of diatoms, such as Asterionella formosa and Fragilaria crotonensis, and the scarcity of food sources for zooplankton, like small algae and cyanobacteria (data not shown). In contrast, the zooplankton biomass from LB collected in September 2009 accounted for 72% of the total plankton biomass of 28.1 g m−2. Judging from LB plankton-monitoring data for the year 2009 (Environmental Agency of Canton Bern, unpublished data), the zoo- and phytoplankton biomasses generally were approximately equal, with the exception of January and late summer (July to September), when the zooplankton biomass was significantly greater (up to 74% of the total plankton biomass).
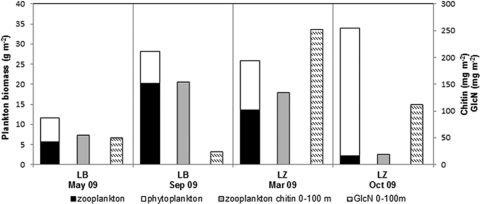
Fig 2.
Zoo- and phytoplankton fresh biomass, zooplankton chitin estimate, and particulate GlcN summarized over 0- to 100-m water depths for LB sampled in May and September 2009 and for LZ sampled in March and October 2009.
Zooplankton chitin.
In March/May 2009, chitin estimated from zooplankton abundances was 134 mg m−2 for LZ and therefore more than 2 times higher than in LB (55.3 mg m−2) (Fig. 2). In contrast, in fall, zooplankton chitin in LB (154 mg m−2) was about 8 times higher than in LZ (18.7 mg m−2).
Zooplankton chitin biomass compared to TOC and GlcN concentrations.
For the spring sampling campaign, relating the chitin estimates from the zooplankton to the carbon pool, zooplankton chitin contributed 0.04% of the TOC integrated over the upper 100 m of the water column of LB (63.5 g C m−2) and 0.03% of the TOC pool of LZ (205 g C m−2). For the fall sampling campaign, the contribution of zooplankton chitin to the TOC was almost 30 times higher in LB (0.1270%) than in LZ (0.0045%).
In LZ, the particulate GlcN concentration integrated over the 0- to 100-m water column was about 2-fold (March 2009) and 6-fold (October 2009) higher than the zooplankton chitin biomass (Fig. 2). In LB, it was as high as the zooplankton chitin biomass in May 2009, but it was only one-sixth of the zooplankton chitin biomass in September 2009. This discrepancy could be caused by the higher inorganic glacial particle load from the inflows in the second half of the year (15, 16), which caused higher turbidity in the surface waters (1). As a consequence, the primary production maximum was shifted to a lower water depth of 2.5 m compared to May 2009, when it was observed at a water depth of 5 m (D. Carstens, K.E. Köllner, H. Bürgmann, B. Wehrli, and C.J. Schubert, submitted for publication). Since the shallowest in situ pumping was performed at 5 m, the fall sampling probably missed a significant proportion or even the maximum of the zooplankton biomass in the surface waters of LB.
Zooplankton chemistry.
The GlcN content of zooplankton sampled in fall accounted for 69.6 ± 4.5 and 94.8 ± 7.1 μmol g dry weight−1 in LB and LZ, respectively, and was, therefore, about one-third higher in the zooplankton of LZ. The results for the TOC, TN, and GlcN contents of zooplankton samples are summarized in Table 4.
Table 4.
Zooplankton chemistry, chitinase activity, and chiA copies on zooplankton samplesa
| Site | TOC (% dry wt) | TN (% dry wt) | GlcN (μmol g dry wt−1) | Chitinase activity at 4°C (nmol MUFb h−1 g dry wt−1) | Chitinase gene copy no. |
|
|---|---|---|---|---|---|---|
| (chiA copies mg dry wt−1) | (chiA copies ng DNA−1) | |||||
| LB | 52.1 ± 0.7 | 8.77 ± 0.09 | 69. 6 ± 4.5 | 26.0 ± 4.4 | 1600 ± 705 | 1.39 ± 0.61 |
| LZ | 42.4 ± 0.7 | 9.00 ± 0.24 | 94. 8 ± 7.1 | 50.4 ± 9.3 | 5726 ± 578 | 13.9 ± 1.4 |
The errors given are standard deviations of triplicate measurements of one sample.
MUF, fluorescent methylumbelliferone released after the hydrolysis of the chitin substrate analog methylumbelliferyl-N,N′-diacetylchitobioside.
Chitinase activity.
The effect of the substrate concentration on chitinase activity was tested in LZ surface waters and mixed sediment slurries. For the sediment samples, MUF-DC turnover was found to be highest (2.63 ± 0.22 nmol h−1 g dry sediment−1) at a substrate concentration of 50 μM (Fig. 3) and dropped at 100 and 300 μM. For the water samples, the chitinase activity was below the limit of detection for all substrate concentrations, even after 4 days of incubation. For negative controls, no turnover rates could be detected, which implies that only biotic degradation of MUF-DC was detected during the incubation.
We assayed the sediments and zooplankton from both lakes sampled in fall 2009 with 50 μM MUF-DC at 4°C and 20°C. MUF-DC turnover rates at 20°C were roughly one-third higher than at 4°C for the zooplankton samples from both lakes and up to 3- and 8-fold higher for LB and LZ sediments, respectively. For reasons of simplicity, only the data for 4°C are discussed and shown in Table 4 and Fig. 4.
(i) Zooplankton.
Chitinase activity on LZ zooplankton was about double that measured for LB zooplankton on a dry-weight basis (Table 4).
(ii) Sediment.
Chitinase activity (at 4°C) in LB sediments ranged from 0.08 (1 to 2 cm) to 0.69 (6 to 7 cm) nmol MUF h−1 g dry weight−1 and in LZ sediments from 0.59 (5 to 6 cm) to 5.10 (0 to 1 cm) nmol MUF h−1 g dry weight−1 (Fig. 4). Comparing the two lake systems, LZ's chitinase activity per gram dry sediment was up to 40 times higher than that measured in LB sediments but decreased with depth and converged on LB values in the 5- to 6- and 6- to 7-cm layers (Fig. 4). However, normalized to the GlcN concentrations, the chitinase activities were in the same range for both lakes, with no clear depth-related trend (data not shown).
Normalized to the TOC, TN, and GlcN contents, the chitinase activities were on the same order of magnitude as the values measured for zooplankton (data not shown).
Chitinase gene copies. (i) Water.
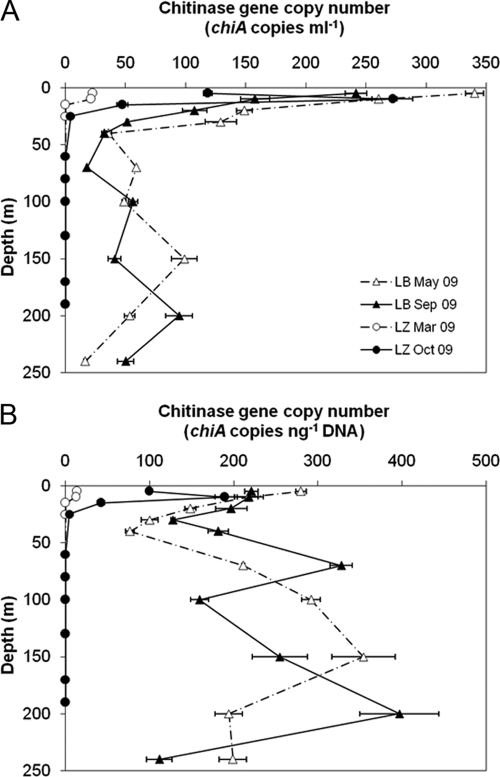
For the 0.2- to 5-μm water fractions of LZ, specific chiA fragments could be amplified only for the 5-m and 10-m water depths sampled in spring and for the 5-m to 25-m water depths sampled in fall (Fig. 5). In comparison, the chiA gene copies detected per ml of water were 2 to 15 times higher in the surface waters of LB, with the exception of the 10-m water depth of LZ in October 2009 (272 ± 17 chiA copies ml−1). The maximum chiA concentration in LB waters (340 ± 7 chiA copies ml−1) was measured at the 5-m water depth in May 2009. In LB, the chiA gene was detected at all depths. For the ≥5-μm fractions, we got no specific chiA signals for LB. chiA was detected in the surface water depths of LZ in this fraction. However, concentrations were up to 20 times lower than for the 0.2- to 5-μm fractions (data not shown).
Fig 5.
chiA gene copy numbers in the water columns of LB and LZ. (A) chiA copies per ml of water. (B) chiA copies per ng extracted DNA. The error bars represent standard deviations of triplicate measurements of one sample.
(ii) Sediment.
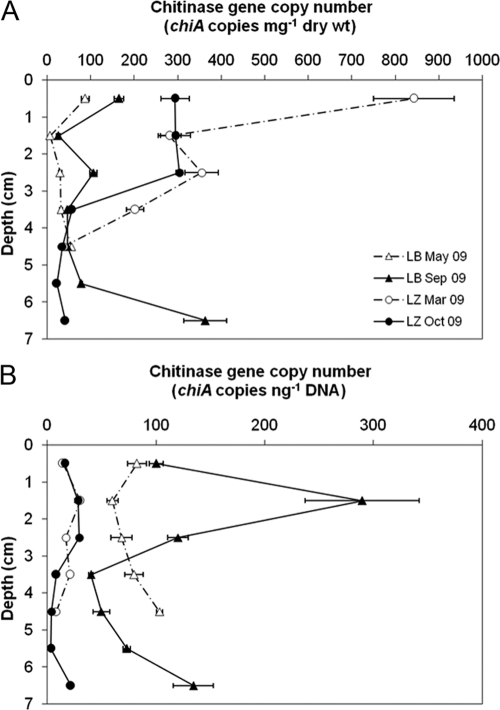
In LB sediment, the lowest chiA content was measured in the 1- to 2-cm layer, increasing again with depth below this layer, as observed for the chitinase activity (Fig. 4 and 6A). In May 2009, the highest chiA content of 87.0 ± 9.2 chiA copies mg dry weight−1 was measured in the 0- to 1-cm sediment layer and in September 2009 in the 6- to 7-cm layer (363 ± 49 chiA copies mg dry weight−1). The chiA content in the 0- to 1-cm layer of LZ sediment was about 2 (October 2009) to 10 (March 2009) times higher than that of LB on a dry weight basis. Normalized to the TOC, TN, and GlcN contents of the sediment, the chiA numbers were higher in LB, with few exceptions (data not shown). For water and sediment samples, chiA concentrations normalized to GlcN and TOC are available in Fig. S2 and S3 in the supplemental material.
Fig 6.
chiA concentration per mg of dry sediment (A) and per ng extracted DNA (B) of LB and LZ. The error bars represent standard deviations of triplicate measurements of one sample.
Since the efficiency of any nucleic acid extraction method from environmental samples is less than 100% and we did not use an internal standard to correct for this, the absolute copy numbers of chiA were likely underestimated. Variable extraction efficiencies may affect comparisons between different sample types. Therefore, chiA copy numbers normalized to the amount of DNA used in the PCR are shown in Fig. 5B and 6B. For the water column of LB, chiA gene copy numbers per ng DNA exceeded the numbers detected for the surface waters (Fig. 5B). In the sediments, chiA gene copy numbers were higher for all sediment layers of LB than for the sediments of LZ (Fig. 6B).
(iii) Zooplankton.
We determined the chiA gene copy numbers on zooplankton samples, considering them a main source of chitin in our lake ecosystem and therefore a hot spot of bacterial chitin hydrolysis. The number of chiA copies detected in LZ zooplankton samples was more than 3-fold higher than in LB zooplankton samples on a dry weight basis (Table 4). Normalized to the amount of DNA used in the PCR, it was 10-fold higher.
Normalized to the GlcN concentrations, the chiA copy numbers associated with zooplankton were approximately on the same order of magnitude as the chiA copies in the sediment of LZ but up to 10-fold lower than the values detected in the sediment from LB (see Fig. S3a in the supplemental material). Compared to the results for the water columns, the chiA concentrations normalized to GlcN were 100- to 1,000-fold lower in zooplankton samples from LZ and LB, respectively (see Fig. S2a in the supplemental material).
Correlations between chitinase activity, chiA abundance, and biogeochemical parameters. (i) Water.
For both sampling campaigns, the chiA copy numbers detected in the water column of LB and the GlcN concentration were highly significantly correlated (P < 0.01; n = 10) (Table 5).
Table 5.
Pearson correlation coefficient and significance of correlation between chiA gene copy number, chitinase activity, and biogeochemical data for water and sediment samples of LB and LZ
| Parameter | Correlated with: | Pearson correlation coefficient (r2) |
|||
|---|---|---|---|---|---|
| LB May 2009 | LB September 2009 | LZ March 2009 | LZ October 2009 | ||
| Water | |||||
| GlcN (nmol liter−1) | Chitinase gene copy no. (chiA copies liter−1) | 0.72b | 0.64b | ||
| Sediment | |||||
| Chitinase activity at 4°C (nmol MUF h−1 g−1 dry wt) | Chitinase gene copy no. (chiA copies mg dry wt−1) | 0.96b | 0.86b | ||
| GlcN (nmol g dry wt−1) | 0.85a | 0.73a | 0.22 | 0.86b | |
| TOC (% dry wt) | 0.48 | 0.79b | 0.14 | 0.60a | |
| TN (% dry wt) | 0.21 | 0.68a | 0.20 | 0.52 | |
| GlcN (nmol g dry wt−1) | Chitinase activity at 4°C (nmol MUF h−1 g dry wt−1) | 0.82b | 0.72a | ||
| TOC (% dry wt) | 0.91b | 0.63a | |||
| TN (% dry wt) | 0.84b | 0.58a | |||
P < 0.05.
P < 0.01.
(ii) Sediment.
For both lake sediments, the chiA content and the chitinase activity correlated highly significantly (P < 0.01; n = 7) (Table 5). For the LB core sampled in the fall, significant correlations were found between the chiA copy number and the GlcN, TOC, and TN contents. In spring, these correlations were not significant, except for the correlation between the chiA copy number and GlcN. Chitinase activity and GlcN, TOC, and TN concentrations also correlated significantly for the fall cores from both lakes, but in LZ, the correlation was not as highly significant as in LB sediments.
DISCUSSION
Sources of chitin and glucosamine in freshwater lakes.
Chitin was previously reported to be produced in large amounts in aquatic ecosystems (10, 47). The mean chitin standing biomass from marine planktonic crustaceans (copepods, cladocerans, and decapod larvae) was calculated to be 26.3 mg m−2 (30), which is on the same order of magnitude as the zooplankton chitin biomass we estimated for our two different lacustrine ecosystems. Our results also agree with the results from a study on chitin dynamics in a mesotrophic bog and a hypereutrophic lake, in which the chitin biomass (from crustaceans) fluctuated between 2 and 200 mg m−2 (42). The available data thus point to similar chitin biomasses in marine and lacustrine ecosystems, with considerable local and seasonal variability.
However, zooplankton is not the only chitin source in an aquatic ecosystem. It is known that chitin is also produced by protozoa, fungi, and algae, especially by diatoms (2, 23, 45). Diatom chitin, known as chitan, can amount to a significant constituent of the cellular biomass. For the diatom Thalassiosira fluviatilis, for example, chitan was found to represent 31 to 38% of the total cell mass (40). In contrast, Smucker reported only 7% and found significant differences between various diatom species (ranging from 2 to 10% [dry weight]) (54). However, he also attributed this discrepancy to chitan losses due to the extraction method that had been used. To our knowledge, chitin content estimates for diverse algal species are not available in the literature, and a calculation of phytoplankton chitin, analogous to the one provided for zooplankton chitin, is thus currently not feasible. As a rough estimate, assuming a chitan content between 5% and 30% of dry diatom biomass and a mean dry weight of 300 pg per diatom cell (49), diatom chitin would constitute 102 to 610 mg m−2 for LB in May 2009, 177 to 1,061 mg m−2 for LB in September 2009, 261 to 1,568 mg m−2 for LZ in March 2009, and 503 to 3,015 mg m−2 for LZ in October 2009. Thus, significant amounts of chitin, exceeding the zooplankton contribution, may have been produced by this source. This may also explain our observations in LZ, where the particulate GlcN pool (0 to 100 m) was indeed found to be higher than the zooplankton chitin (Fig. 2).
GlcN is not only the main constituent of the biopolymer chitin, but together with muramic acid (MurA), it also forms the disaccharide backbone of the bacterial cell wall polymer peptidoglycan. Based on measurements of MurA concentrations, Carstens et al. determined the contribution of bacterial cells to the particulate GlcN (Carstens et al., submitted). Bacterium-derived GlcN accounted for up to 26% and 34% of total GlcN in the euphotic zones of LZ and LB, respectively. Compared to the water column of LZ the proportions of bacterial GlcN were higher in the water column of LB, with a maximum of 94% at 200 m sampled in fall 2009.
Chitinolytic activity and populations in freshwater lakes.
Several studies on chitinase activity in aquatic environments have been published, mainly for marine and estuarine water and sediments, which applied a wide range of substrate concentrations, ranging from 20 nM to 5 mM (6, 27, 28, 34, 44, 53, 60). This makes the determination of substrate saturation curves for the environment under study crucial. We found an optimum substrate concentration of 50 μM for the lake sediments, whereas no chitinase activity could be measured in the water at any substrate concentration. Using a comparable approach (with 20 μM MUF-GlcNAc), Martinez et al. could not detect any chitinase activity in seawater (39). In contrast, in alkaline hypersaline Mono Lake, turnover rates for 10 μM MUF-DC in water and sediment samples were 1,000-fold higher than the chitinase activities of the sediments analyzed in the present study (37). However, Mono Lake is an environment extremely rich in chitin from shrimp exuvia and carcasses. Its lack of detection in the water samples of our lake ecosystems indicates that chitinase activity was mostly associated with particles. In aquatic environments, aggregates are known as hot spots of exoenzymatic activities (18, 53). Chitinase activity, in particular, was previously reported to be mainly associated with particulate fractions, e.g., it was found associated with the >3-μm or the 2- to 100-μm particle size class (29, 50). Similarly, we found significant activity in the water only on the zooplankton samples, i.e., the >95-μm fraction. Particle-associated microbes have the advantage of benefiting directly from the soluble oligomers produced by chitin hydrolysis. However, planktonic bacteria can also profit from chitin hydrolysis products, which has been shown recently for members of planktonic freshwater Actinobacteria (5).
By chiA-specific quantitative PCR, we could confirm the presence of chitinolytic bacteria in zooplankton and sediment and in the 0.2- to 5-μm water fractions. The chiA abundance normalized to any parameter (DNA, GlcN, or TOC) that was associated with the 0.2- to 5-μm water fraction was significantly higher than that detected in the zooplankton samples, which we had assumed to contain high concentrations of chitinolytic bacteria. Access to zooplankton chitin (α-chitin) is probably hindered by cross-linked structural components, such as glycans and proteins. These have to be degraded by different microbial communities initializing the decomposition of zooplankton carcasses (17, 56, 57). However, we did not analyze the fraction of eukaryotic DNA in the DNA extracts to normalize the results to only bacterial DNA, which could change the described ratios.
The bacterial degradation of chitin is known to be highly regulated (33). Therefore, the detection of bacterial chiA gene copy numbers indicates only the presence of bacteria capable of chitin degradation and is not a direct measure of active chitin hydrolysis, i.e., chitinase activity. However, in the present study, chiA gene abundance in sediments was highly correlated with chitinase activity, and the increased abundance of the chiA gene in the water column of LB, where chitin made a greater contribution to the carbon pool, also indicates a relationship between gene abundance and chitin degradation. The higher prominence of chitin as a bacterial substrate in oligotrophic compared to eutrophic lakes has also been shown in culture-dependent analyses of chitinolytic bacteria in Polish lakes (13, 14).
In conclusion, significant correlations between chiA gene abundance, chitinase activity, and biogeochemical data evidenced the contribution of chitin to the carbon and nitrogen budget in the lake sediments, in particular for the oligotrophic system of LB (Table 5). We therefore assign chitin a role as a significant microbial growth substrate in temperate freshwater lakes, especially where other easily degradable carbon sources are scarce.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the people who helped with sampling and chemical analysis: Michael Schurter, Alois Zwyssig, Tina Wunderlin, Torsten Diem, Gijs Nobbe, Leticia Stojkovski, Mathias Kirf, Beat Kienholz, Ruth Stierli, and Richard Illi (Analytik- und Ausbildungslabor laboratory). We thank Markus Zeh and Katrin Guthruf, Environmental Agency of Canton Bern, for providing the plankton-monitoring data from Lake Brienz and Hans Rudolf Bürgi, Eawag, Department of Aquatic Ecology, Dübendorf, Switzerland, for the updated list of phytoplankton species volumina. We thank Peter Keller, Environmental Agency of Canton Zug, for providing monitoring data for Lake Zug.
The present study is part of an interdisciplinary project combining biogeochemistry and microbiology funded by the Swiss National Science Foundation (SNF) (grant K-23Kl-118111) and Eawag, the Swiss Federal Institute of Aquatic Science and Technology.
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. AWA Amt für Wasser und Abfall, Gewässer- und Bodenschutzlabor GBL 2009. Brienzersee Tiefenprofile 2009. AWA Amt für Wasser und Abfall, Gewässer- und Bodenschutzlabor GBL, Bern, Switzerland: http://www.bve [Google Scholar]
- 2. Bartnicki-Garcia S, Lippman E. 1982. Fungal cell wall composition, p 229–252 In Laskin AJ, Lechevalier HA. (ed), CRC handbook of microbiology, 2nd ed, vol IV, Microbial composition: carbohydrates, lipids and minerals CRC Press, Boca Raton, FL [Google Scholar]
- 3. Bassler BL, Yu C, Lee YC, Roseman S. 1991. Chitin utilization by marine bacteria: degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276–24286 [PubMed] [Google Scholar]
- 4. Beier S. 2010. Bacterial degradation, use of chitin in aquatic habitats. PhD thesis Uppsala University, Uppsala, Sweden [Google Scholar]
- 5. Beier S, Bertilsson S. 2011. Uncoupling of chitinase activity and uptake of hydrolysis products in freshwater bacterioplankton. Limnol. Oceanogr. 56:1179–1188 [Google Scholar]
- 6. Bowman JP, McCammon SA, Gibson JAE, Robertson L, Nichols PD. 2003. Prokaryotic metabolic activity and community structure in Antarctic continental shelf sediments. Appl. Environ. Microbiol. 69:2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyer JN. 1994. Aerobic and anaerobic degradation and mineralization of 14C-chitin by water column and sediment inocula of the York River estuary, Virginia. Appl. Environ. Microbiol. 60:174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bürgi H, Züllig H. 1983. Eine neue Netzgarnitur mit Kipp-Schliessmechanismus für quantitative Zooplanktonfänge in Seen. Schweiz. Z. Hydrol. 45:505–507 [Google Scholar]
- 9. Bürgi HR, Weber P, Bachmann H. 1985. Seasonal variations in the trophic structure of phyto-and zooplankton communities in lakes in different trophic states. Schweiz. Z. Hydrol. 47:197–224 [Google Scholar]
- 10. Cauchie HM. 2002. Chitin production by arthropods in the hydrosphere. Hydrobiologia 470:63–96 [Google Scholar]
- 11. Cottrell MT, Wood DN, Yu L, Kirchman DL. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the α- and γ-subclasses of the proteobacteria. Appl. Environ. Microbiol. 66:1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daniels LB, Glew RH. 1984. β-d-Glucosidases in tissue, p 217–226 In Bergmeyer HU. (ed), Methods of enzymatic analysis. John Wiley & Sons, New York, NY [Google Scholar]
- 13. Donderski W. 1984. Chitinolytic bacteria in water and bottom sediments of two lakes of different trophy. Acta Microbiol. Pol. 33:163–170 [PubMed] [Google Scholar]
- 14. Donderski W, Brzezinska MS. 2003. The utilization of N-acetyloglucosamine and chitin as sources of carbon and nitrogen by planktonic and benthic bacteria in Lake Jeziorak. Pol. J. Environ. Stud. 12:685–692 [Google Scholar]
- 15. Finger D, et al. 2007. Effects of alpine hydropower operations on primary production in a downstream lake. Aquat. Sci. 69:240–256 [Google Scholar]
- 16. Finger D, Schmid M, Wüest A. 2006. Effects of upstream hydropower operation on riverine particle transport and turbidity in downstream lakes. Water Resour. Res. 42:W08429 [Google Scholar]
- 17. Gooday GW. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387–430 [Google Scholar]
- 18. Grossart HP, Simon M. 1998. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat. Microb. Ecol. 15:127–140 [Google Scholar]
- 19. Grossart HP, Simon M. 1998. Significance of limnetic organic aggregates (lake snow) for the sinking flux of particulate organic matter in a large lake. Aquat. Microb. Ecol. 15:115–125 [Google Scholar]
- 20. Guerrant GO, Wayne Moss C. 1984. Determination of monosaccharides as aldononitrile, O-methyloxime, alditol, and cyclitol acetate derivatives by gas chromatography. Anal. Chem. 56:633–638. [Google Scholar]
- 21. Guthruf K, Maurer V, Pokorni B, Zeh M. Entwicklung des Phyto- und Crustaceenzooplanktons Brienzersee. AWA Amt für Wasser und Abfall, Gewässer- und Bodenschutzlabor GBL, Bern, Switzerland. 2009 [Google Scholar]
- 22. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herth W. 1978. A special chitin-fibril-synthesizing apparatus in the centric diatom cyclotella. Naturwissenschaften 65:260 [Google Scholar]
- 24. Hjort K, et al. 2010. Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen-suppressive soil. FEMS Microbiol. Ecol. 71:197–207 [DOI] [PubMed] [Google Scholar]
- 25. Hobel CFV, Marteinsson VT, Hreggvidsson GÓ, Kristjánsson JK. 2005. Investigation of the microbial ecology of intertidal hot springs by using diversity analysis of 16S rRNA and chitinase genes. Appl. Environ. Microbiol. 71:2771–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hönerlage W, Hahn D, Zeyer J. 1995. Detection of mRNA of nprM in Bacillus megaterium ATCC 14581 grown in soil by whole-cell hybridization. Arch. Microbiol. 163:235–241 [Google Scholar]
- 27. Hood MA. 1991. Comparison of four methods for measuring chitinase activity and the application of the 4-MUF assay in aquatic environments. J. Microbiol. Methods 13:151–160 [Google Scholar]
- 28. Hoppe HG. 1983. Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl substrates. Mar. Ecol. Prog. Ser. 11:299–308 [Google Scholar]
- 29. Hoppe HG, Giesenhagen HC, Gocke K. 1998. Changing patterns of bacterial substrate decomposition in a eutrophication gradient. Aquat. Microb. Ecol. 15:1–13 [Google Scholar]
- 30. Jeuniaux C, Voss-Foucart MF. 1991. Chitin biomass and production in the marine environment. Biochem. Syst. Ecol. 19:347–356 [Google Scholar]
- 31. Kapaun E, Reisser W. 1995. A chitin-like glycan in the cell wall of a Chlorella sp. (Chlorococcales, Chlorophyceae). Planta 197:577–582 [Google Scholar]
- 32. Kelts K, Briegel U, Ghilardi K, Hsu K. 1986. The limnogeology-ETH coring system. Schweiz. Z. Hydrol. 48:104–115 [Google Scholar]
- 33. Keyhani NO, Roseman S. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108–122 [DOI] [PubMed] [Google Scholar]
- 34. Kirchman DL, White J. 1999. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 18:187–196 [Google Scholar]
- 35. Kirchner M. 1995. Microbial colonization of copepod body surfaces and chitin degradation in the sea. Helgol. Meeresunters. 49:201–212 [Google Scholar]
- 36. LeCleir GR, Buchan A, Hollibaugh JT. 2004. Chitinase gene sequences retrieved from diverse aquatic habitats reveal environment-specific distributions. Appl. Environ. Microbiol. 70:6977–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. LeCleir GR, Hollibaugh JT. 2006. Chitinolytic bacteria from alkaline hypersaline Mono Lake, California, U. S. A. Aquat. Microb. Ecol. 42:255–264 [Google Scholar]
- 38. Lindsay EA, Colloff MJ, Gibb NL, Wakelin SA. 2010. The abundance of microbial functional genes in grassy woodlands is influenced more by soil nutrient enrichment than by recent weed invasion or livestock exclusion. Appl. Environ. Microbiol. 76:5547–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martinez J, Smith DC, Steward GF, Azam F. 1996. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10:223–230 [Google Scholar]
- 40. McLachlan J, Falk MAM. 1965. Studies on the chitan (chitin:poly-N-acetylglucosamine) fibers of the diatom Thalassiosira fluviatilis Hustedt. Can. J. Bot. 43:707–713 [Google Scholar]
- 41. Meckler AN, Schubert CJ, Cowie GL, Peiffer S, Dittrich M. 2004. New organic matter degradation proxies: valid in lake systems? Limnol. Oceanogr. 49:2023–2033 [Google Scholar]
- 42. Miyamoto S, Yamamoto H, Seki H. 1991. Chitin dynamics in the freshwater environment. Biochem. Syst. Ecol. 19:371–377 [Google Scholar]
- 43. Moor CH, Schaller T, Sturm M. 1996. Recent changes in stable lead isotope ratios in sediments of Lake Zug, Switzerland. Environ. Sci. Technol. 30:2928–2933 [Google Scholar]
- 44. Mudryk ZJ, Skórczewski P. 2004. Extracellular enzyme activity at the air-water interface of an estuarine lake. Estuarine Coastal Shelf Sci. 59:59–67 [Google Scholar]
- 45. Mulisch M. 1993. Chitin in protistan organisms: distribution, synthesis and deposition. Eur. J. Protistol. 29:1–18 [DOI] [PubMed] [Google Scholar]
- 46. Müller B, et al. 2007. Present and past bio-available phosphorus budget in the ultra-oligotrophic Lake Brienz. Aquat. Sci. 69:227–239 [Google Scholar]
- 47. Poulicek M, Gaill F, Goffinet G. 1998. Chitin biodegradation in marine environments. ACS Symp. Ser. 707:163–210 [Google Scholar]
- 48. Poulicek M, Jeuniaux C. 1991. Chitin biodegradation in marine environments: an experimental approach. Biochem. Syst. Ecol. 19:385–394 [Google Scholar]
- 49. Reynolds CS. 1984. The ecology of freshwater phytoplankton. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 50. Richardot M, et al. 1999. Proteolytic and glycolytic activities in size-fractionated surface water samples from an oligotrophic reservoir in relation to plankton communities. Aquat. Sci. 61:279–292 [Google Scholar]
- 51. Schröder R. 1969. Ein summierender Wasserschöpfer. Arch. Hydrobiol. 66:241–243 [Google Scholar]
- 52. Simon M, Lenhard A, Tilzer MM. 1993. Bacterial production and the sinking flux of particulate organic carbon in a large and deep lake in comparison to oceanic environments. Mar. Microb. Food Webs 7:161–176 [Google Scholar]
- 53. Smith DC, Simon M, Alldredge AL, Azam F. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139–142 [Google Scholar]
- 54. Smucker RA. 1991. Chitin primary production. Biochem. Syst. Ecol. 19:357–369 [Google Scholar]
- 55. Svitil AL, Ní Chadhain SM, Moore JA, Kirchman DL. 1997. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 63:408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tang KW, Bickel SL, Dziallas C, Grossart HP. 2009. Microbial activities accompanying decomposition of cladoceran and copepod carcasses under different environmental conditions. Aquat. Microb. Ecol. 57:89–100 [Google Scholar]
- 57. Tang KW, Hutalle KML, Grossart HP. 2006. Microbial abundance, composition and enzymatic activity during decomposition of copepod carcasses. Aquat. Microb. Ecol. 45:219–227 [Google Scholar]
- 58. Utermöhl H. 1958. Zur Vervollkommnung der quantitativen phytoplankton-methodik. Mitt. Int. Ver. Limnol. 9:1–38 [Google Scholar]
- 59. Vogler P. 1965. Beiträge zur Phosphatanalytic in der Limnologie. Fortschr. Wasserchem. Grenzgeb. 2:109–119 [Google Scholar]
- 60. Vrba J, Šimek k, Pernthaler J, Psenner R. 1996. Evaluation of extracellular, high-affinity β-N-acetylglucosaminidase measurements from freshwater lakes: an enzyme assay to estimate protistan grazing on bacteria and picocyanobacteria. Microb. Ecol. 32:81–99 [PubMed] [Google Scholar]
- 61. Weiss P, Schweitzer B, Amann R, Simon M. 1996. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl. Environ. Microbiol. 62:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xiao X, et al. 2005. Chitinase genes in lake sediments of Ardley Island, Antarctica. Appl. Environ. Microbiol. 71:7904–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang X, Amelung W. 1996. Gas chromatographic determination of muramic acid, glucosamine, mannosamine, and galactosamine in soils. Soil Biol. Biochem. 28:1201–1206 [Google Scholar]
- 64. Zobell CE, Rittenberg SC. 1938. The occurrence and characteristics of chitinoclastic bacteria in the sea. J. Bacteriol. 35:275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.