Abstract
Water deprivation can be a major stressor to microbial life in surface and subsurface soil. In unsaturated soils, the matric potential (Ψm) is often the main component of the water potential, which measures the thermodynamic availability of water. A low matric potential usually translates into water forming thin liquid films in the soil pores. Little is known of how bacteria respond to such conditions, where, in addition to facing water deprivation that might impair their metabolism, they have to adapt their dispersal strategy as swimming motility may be compromised. Using the pressurized porous surface model (PPSM), which allows creation of thin liquid films by controlling Ψm, we examined the transcriptome dynamics of Pseudomonas putida KT2440. We identified the differentially expressed genes in cells exposed to a mild matric stress (−0.4 MPa) for 4, 24, or 72 h. The major response was detected at 4 h before gradually disappearing. Upregulation of alginate genes was notable in this early response. Flagellar genes were not downregulated, and the microarray data even suggested increasing expression as the stress prolonged. Moreover, we tested the effect of polyethylene glycol 8000 (PEG 8000), a nonpermeating solute often used to simulate Ψm, on the gene expression profile and detected a different profile than that observed by directly imposing Ψm. This study is the first transcriptome profiling of KT2440 under directly controlled Ψm and also the first to show the difference in gene expression profiles between a PEG 8000-simulated and a directly controlled Ψm.
INTRODUCTION
In environments like surface or subsurface soils, hydration conditions can change frequently, and thus, the bacteria and other organisms living in those environments can face water deprivation. The thermodynamic availability of water to bacteria is measured as the water potential, which is the expression of the energetic state of water. In soil, the two largest components of the water potential (Ψ) are the solute (Ψs) and the matric (Ψm) potentials (18). Ψs is the result of the presence of solutes in the water; Ψm is the result of adsorptive and capillary forces acting upon water held in soil pores. In a wet soil, where the pores are filled (or “saturated”) with liquid, Ψm is zero. When the soil dries and the pores drain, Ψm becomes more and more negative and, in nonsaline soils, will be the major contributor to Ψ. In Ψm-dominated environments, bacteria thus reside in thin liquid films, the thickness of which depends both on Ψm and on the geometry of the pores (34). Depending on the severity of the Ψm, bacteria experience from mild to extreme stress (desiccation, e.g., −100 MPa) (26). Bacterial responses to this stress include accumulating compatible solutes such as trehalose and sucrose (26), increasing fatty acid content of the cytoplasmic membrane (16), and producing extracellular polymeric substances (EPS) (27, 39). EPS acts as a water-binding agent (33).
In addition to direct physiological effects, low Ψm in soil also acts on bacterial motility as the liquid films present in the pores can become thin enough that water pathways between pores are disconnected (14, 25). Previously, we investigated Pseudomonas putida KT2440 motility under very mild Ψm (down to −3.5 kPa) using an experimental platform called the porous surface model (PSM) (10). In this system, microbes inoculated on a porous ceramic surface grow in thin liquid films as the medium that wets the ceramic plate is under suction, which corresponds to the Ψm experienced by the microbes. On the PSM, low Ψm values (Ψm < −2 kPa) were sufficient to arrest swimming motility by pinning the cells in liquid films of effective thicknesses less than about 1.5 μm (11). A cessation of motility was, however, not observed when the water potential was lowered using polyethylene glycol 8000 (PEG 8000) (10). This indicates that this nonpermeating solute, often used in microbial ecology to simulate Ψm (5, 17), does not modulate liquid film thickness.
Besides the physiological responses mentioned above, it is also important to understand transcriptomic responses under matric stress, as phenotypes are the manifestation of gene expression and transcription is the first step in gene expression. It is through their regulation of gene expression that bacteria adapt to changing conditions. Hence, identification of potentially important genes and understanding how they function and how their expressions are regulated at transcriptomic level will contribute significantly to our knowledge of adaptation to matric stress. Whole-genome transcriptome studies have mainly focused on the effect of intense water stress (desiccation stress with Ψm around −200 MPa) in diverse organisms such as Rhodococcus jostii (21), Bradyrhizobium japonicum (8), and Anabaena sp. strain PCC7120 (19). In temperate climates, soil bacteria rarely face such extreme stress but are more commonly confronted with Ψm in the low-MPa range (−1.5 MPa being the vegetal wilting point). Only a few studies have investigated gene expression in P. putida under such mild water stress. These studies have identified some water deprivation-related genes (35) and have demonstrated the spatiotemporal dynamics of alginate gene expression in P. putida using reporter genes (22). However, these studies did not provide genome-wide expression profiles nor did they directly control Ψm, as Ψm was simulated using PEG 8000 additions. Here, we aimed to identify the significantly differentially expressed genes (here referred to as differentially expressed genes) at a −0.4-MPa Ψm relative to a −0.5-kPa Ψm (near-saturation condition) in the model soil organism P. putida KT2440. We hypothesized that EPS-related genes would be upregulated and flagellar genes would be downregulated because investing in flagellum synthesis in the absence of a water pathway would be costly in the short term and because flagella can suppress their synthesis under low water availability in Salmonella bacteria (36). To test our hypotheses, we used an improved version of the PSM, called the pressurized porous surface model (PPSM) (15), which extends the range of possible matric potentials down to −1.5 MPa. In addition, we investigated the effect of PEG 8000 amendment to test the hypothesis that PEG 8000-simulated Ψm would result in a different expression profile than that resulting from directly applied Ψm.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas putida KT2440 cultures were grown overnight on LB plates and suspended and diluted to target cell densities in 0.9% NaCl prior to inoculation on the ceramic plates. On the PPSM, the strains were grown on a minimal liquid medium composed of 1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM Fe-EDTA, 15 mM (NH4)2SO4, 33 mM Na2HPO4, 22 mM KH2PO4, and 51 mM NaCl and 20 mM benzoate.
Experimental system.
Both PSM and PPSM consist of a filter holder (Bontec-AS, Ballerup, Denmark) in which a ceramic plate (7.1 mm thick and 41.3 mm in diameter; 500-kPa bubbling pressure plate; Soilmoisture, Santa Barbara, CA) is located. A silicone O-ring (40 mm in inner diameter, 5 mm thick) surrounds the plate and makes the system airtight. The filter holder is connected to a reservoir of growth medium through silicone tubing. In the PSM, the matric potential is set by changing the hydraulic head between the ceramic plate and reservoir surfaces (10); in the PPSM, the matric potential is set by applying positive pressure using compressed gas (8% O2 in N2), required to create −0.4-MPa matric stress (15). Prior to inoculation, the systems are autoclaved at 121°C for 25 min.
Incubations under defined matric potentials.
Aliquots of overnight-grown P. putida KT2440 cells were inoculated (approximately 1 × 107 cells in 100 μl) on the surface of ceramic plates. The total duration of the incubation was always 5 days, after which the cells were harvested (see below). The cells were incubated for at least 48 h in the absence of matric stress (i.e., at −0.5 kPa) to allow the formation of a mature cell lawn. Then, the cells were either maintained at −0.5 kPa until the termination of experiment (control) or subjected to a −0.4-MPa Ψm for the last 4, 24, or 72 h of the 5-day incubation period. Four replicate PPSMs were incubated for each condition. In addition, incubations were performed where PEG 8000 was used to set a surrogate Ψm. To that effect, incubations were performed using the PSM setup, where the direct Ψm was set to −0.5 kPa, but the total water potential was set to −0.5 or −1.0 MPa by supplementing the growth medium with PEG 8000 (150 or 262 g/liter, respectively) (18). The incubations were again for 5 days as above, but the PEG 8000 stress was present during the full duration.
Sampling, RNA extraction, reverse transcription (RT), and labeling.
At the end of the incubation period, the (P)PSMs were quickly disassembled and the cells were flooded with stop solution (5% phenol in 100% ethanol [8]) and harvested with a cytological brush (Gynobrush; Heinz Herenz, Hamburg, Germany). Harvested cells were suspended in 2-ml Eppendorf tubes containing 200 ml of the stop solution. Immediately after, RNA extraction was performed using the Agilent MiniRNA kit (California) according to the manufacturer's protocol. RNA concentrations and integrities were checked using NanoDrop 1000 (Thermo Fisher Scientific, Delaware) and by gel electrophoresis, respectively. cDNA synthesis and labeling (CY-3 dye) were performed per the manufacturer's protocol (microarray-based prokaryotic analysis; Fairplay III; Agilent, California).
Microarray and hybridization conditions.
An Agilent whole-genome one-color oligonucleotide array of P. putida KT2440 was used as the microarray platform. The array (2 to 4 60-mer probes per gene) was designed using OligoWiz software (38). Labeled cDNA was hybridized to the array probes on the array per the manufacturer's protocol (microarray-based prokaryotic analysis; Fairplay III; Agilent, California). After hybridization, spot intensities were acquired by scanning the arrays (DNA microarray scanner; Agilent, California).
Microarray data analysis.
Gene expression data (4 biological replicates × 2 to 4 spot replicates, resulting in 8 to 16 replicates) were analyzed in the statistical software program R (www.r-project.org) using the limma package (31) available in Bioconductor (www.bioconductor.org). Data were normalized using the quantile method (40). Differentially expressed genes were determined by comparing expression values under directly applied Ψm or PEG 8000-simulated Ψm relative to values at a −0.5-kPa Ψm. Those genes with average absolute log2 fold changes higher than 1.5 and adjusted P values less than 0.01 (i.e., false discovery rate less than 1%) were identified as significantly differentially expressed genes. Finally, gene annotations were made using the annotation files available in the Comprehensive Microbial Resource website (http://cmr.jcvi.org/).
qRT-PCR.
To partially validate the trends of the microarray results, quantitative reverse transcription-PCR (qRT-PCR) was performed on isolated RNA (obtained from a batch of [P]PSMs separate from the ones used for the microarray) for selected genes (algT, alg8, mucA, fliE, and flgM) with 3 replicates for each condition (4 h, 72 h, and control). The primer sets (13, 22) are listed in Table S3 in the supplemental material. cDNA synthesis and amplification were performed using the Qscript 1-Step Sybr green qRT-PCR kit (Quanta Biosciences, Maryland) in a Chromo4 thermocycler (MJ Research, Massachusetts) with a total RNA input of 50 ng according to the standard protocol of the manufacturer (Quanta Biosciences, Maryland). Data were normalized with respect to rimM (coding for the 16S rRNA processing protein RimM, which was not differentially expressed in our study as done by Li et al. [22]). Expression levels of the target genes at −0.4 MPa were calculated relative to levels at −0.5 kPa using the threshold cycle (2−ΔΔCT) method (23).
Pressure test.
We tested the effect of gas pressure on gene expression profiles of cells in liquid medium. Twenty milliliters of overnight-grown KT2440 cells was exposed to 0.4-MPa (gauge) and 0.1-MPa (atmospheric) pressures for 4 h. A 1-milliliter liquid culture was sampled for RNA extraction, and cDNA synthesis, labeling, microarray hybridization, and data analysis were performed as described above (2 biological replicates × 2 to 4 spot replicates for both 0.4-MPa and atmospheric-pressure conditions).
Microarray data accession number.
The microarray data can be accessed through GEO with accession number GSE25512 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25512).
RESULTS AND DISCUSSION
Dynamic gene expression profile under −0.4-MPa Ψm.
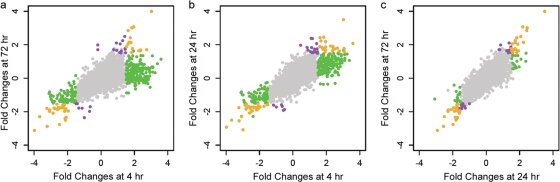
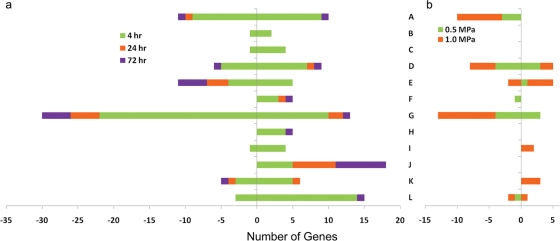
The major response toward matric stress was detected at the earliest time point (4 h), and many genes returned to their prestress level afterwards (Fig. 1a and b). The responses at 24 and 72 h were very similar in terms of numbers of genes expressed (Fig. 1c). Figure 2a shows the major categories of up- and downregulated genes for all three stress durations. (The complete list of differentially expressed genes and their fold changes is presented in Table S1 in the supplemental material.)
Fig 1.
Comparison of fold changes for all the genes in P. putida KT2440 at different times under a −0.4-MPa Ψm. Fold changes are the expression values of the genes at −0.4 MPa relative to those at −0.5 kPa (control) in log2 scale. Positive and negative values correspond to up- and downregulation, respectively. The color of the dots illustrates the conditions under which genes were detected as differentially expressed. Genes presented in gray were nondifferentially expressed. Genes in purple, green, or orange were detected only under the condition on the y axis, only under the condition on the x axis, or under both conditions, respectively. Criteria defining differential expression were absolute log2 fold change of >1.5 and adjusted P value of <0.01.
Fig 2.
Number of up- and downregulated genes according to the major role categories. (a) Ψm of −0.4 MPa. (b) Ψm equivalents of −0.5 and −1.0 MPa created by PEG 8000 amendment. Positive and negative numbers correspond to up- and downregulated genes, respectively. Categories: A, transport and binding proteins; B, transcription; C, signal transduction; D, regulatory functions; E, protein fate; F, fatty acid and phospholipid metabolism; G, energy metabolism, other; H, energy metabolism, biosynthesis and degradation of polysaccharides; I, DNA metabolism—DNA replication, repair, and recombination; J, chemotaxis and motility; K, cellular processes—adaptation to atypical condition; L, biosynthesis and degradation of surface polysaccharides and lipopolysaccharides.
We detected significant upregulation of EPS-related genes; all were related to alginate synthesis, and none were related to cellulose biosynthesis (Table 1). Surprisingly, flagellar genes were not downregulated but rather upregulated, and both their number and upregulation level increased from 4 to 72 h (Table 1). In contrast, the number and expression level of stress response-related genes (universal stress proteins, heat shock proteins, etc.) decreased with prolonged stress (Table 1). Overall, the observed gene expression trends from early to late times suggest that there may be a shift toward a nonstressed behavior.
Table 1.
Log2 fold change for selected genes
| Role category | Locus name | Gene symbol | Annotation | Log2 fold change |
|||
|---|---|---|---|---|---|---|---|
| 4 h | 24 h | 72 h | |||||
| Cell envelope (biosynthesis and degradation of surface polysaccharides and lipopolysaccharides) | PP_0133 | algB | Alginate biosynthesis transcriptional regulatory protein AlgB | 2.10 | |||
| PP_1277 | algA | Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase | 1.51 | ||||
| PP_1278 | algF | Alginate O-acetyltransferase | 1.90 | ||||
| PP_1279 | algJ | Alginate O-acetylation protein AlgJ | 1.66 | ||||
| PP_1280 | algI | Alginate O-acetylation protein AlgI | 1.72 | ||||
| PP_1287 | Alginate biosynthesis protein Alg8 | 1.58 | |||||
| PP_1288 | algD | GDP-mannose 6-dehydrogenase | 1.79 | ||||
| PP_1427 | algT | RNA polymerase sigma factor AlgT | 1.89 | ||||
| PP_1428 | mucA | Sigma factor AlgT (AlgU) negative regulatory protein MucA | 2.08 | ||||
| PP_1429 | mucB | Sigma factor AlgT (AlgU) regulatory protein MucB | 2.06 | ||||
| Cellular processes (chemotaxis and motility) | PP_4358 | fliM | Flagellar motor switch protein FliM | 1.63 | |||
| PP_4359 | fliL | Flagellar protein FliL | 1.62 | 2.42 | 2.85 | ||
| PP_4370 | fliE | Flagellar hook-basal body complex protein FliE | 1.84 | 2.45 | 3.01 | ||
| PP_4385 | flgG | Flagellar basal-body rod protein FlgG | 1.50 | 1.77 | |||
| PP_4386 | flgF | Flagellar basal-body rod protein FlgF | 1.75 | 1.83 | 2.42 | ||
| PP_4390 | flgC | Flagellar basal-body rod protein FlgC | 1.56 | 2.05 | |||
| PP_4391 | flgB | Flagellar basal-body rod protein FlgB | 1.74 | 2.25 | |||
| PP_4395 | flgM | Negative regulator of flagellin synthesis FlgM | 1.79 | ||||
| Transcription | PP_1623 | rpoS | RNA polymerase sigma factor RpoS | −1.98 | |||
| PP_5108 | rpoH | RNA polymerase sigma-32 factor | 2.17 | ||||
| Energy metabolism (other) | PP_3365 | Acetolactate synthase, catabolic, putative | 1.80 | 1.73 | 2.13 | ||
| PP_3578 | Phosphoglucomutase, alpha-d-glucose phosphate specific | 1.62 | |||||
| PP_3613 | l-Sorbosone dehydrogenase | 1.88 | |||||
| PP_3621 | Isoquinoline 1-oxidoreductase, alpha subunit, putative | −1.66 | |||||
| PP_3970 | Formaldehyde dehydrogenase, glutathione independent, putative | 2.19 | |||||
| PP_4011 | icd | Isocitrate dehydrogenase, NADP dependent, prokaryotic type | −1.53 | ||||
| PP_4034 | N-Carbamoyl-beta-alanine amidohydrolase, putative | −2.85 | |||||
| PP_0103 | Cytochrome c oxidase, subunit II | −2.13 | |||||
| PP_0104 | Cytochrome c oxidase, subunit I | −1.56 | |||||
| PP_0105 | Cytochrome c oxidase assembly protein | −1.71 | |||||
| PP_0106 | Cytochrome c oxidase, subunit III | −1.77 | |||||
| PP_0490 | Formate dehydrogenase, iron-sulfur subunit | −1.63 | |||||
| PP_0552 | adh | 2,3-Butanediol dehydrogenase | −2.09 | −1.84 | −2.33 | ||
| PP_0553 | acoC | Acetoin dehydrogenase, dihydrolipoamide acetyltransferase component | −2.13 | −1.81 | −2.50 | ||
| PP_0554 | acoB | Acetoin dehydrogenase, beta subunit | −2.15 | −1.76 | −2.41 | ||
| PP_0555 | acoA | Acetoin dehydrogenase, alpha subunit | −2.17 | ||||
| PP_0556 | Acetoin catabolism protein | −2.49 | −2.77 | ||||
| PP_0557 | acoR | Acetoin catabolism regulatory protein | −1.54 | ||||
| PP_0596 | Beta-alanine–pyruvate transaminase | 2.72 | |||||
| PP_0989 | gcvH-1 | Glycine cleavage system H protein | 1.65 | ||||
| PP_0999 | arcC | Carbamate kinase | −2.64 | ||||
| PP_1000 | argI | Ornithine carbamoyltransferase, catabolic | −3.08 | ||||
| PP_1001 | arcA | Arginine deiminase | −3.11 | ||||
| PP_1157 | Acetolactate synthase, catabolic, putative | 1.95 | |||||
| PP_2351 | Acetyl-CoAa synthetase, putative | −1.65 | |||||
| PP_2422 | Carboxymuconolactone decarboxylase family protein | −2.42 | |||||
| PP_2674 | qedH | Quinoprotein ethanol dehydrogenase | −1.51 | ||||
| PP_2675 | Cytochrome c-type protein | −2.36 | |||||
| PP_4401 | bkda-1 | 2-Oxoisovalerate dehydrogenase, alpha subunit | −1.60 | ||||
| PP_5033 | hutU | Urocanate hydratase | 2.08 | ||||
| PP_5338 | aspA | Aspartate ammonia-lyase | 2.37 | 1.63 | 1.69 | ||
| PP_5346 | oadA | Oxaloacetate decarboxylase, alpha subunit | −1.68 | ||||
| Energy metabolism (biosynthesis and degradation of polysaccharides) | PP_2918 | Trehalose synthase, putative | 2.05 | ||||
| PP_4050 | glgA | Glycogen synthase | 1.83 | ||||
| PP_4051 | Alpha-amylase family protein | 1.57 | |||||
| PP_4060 | Alpha-amylase family protein | 1.80 | |||||
| Cellular processes (adaptation to atypical condition) | PP_0089 | osmC | Hydroperoxide resistance protein OsmC | 2.67 | 1.80 | ||
| DNA metabolism (DNA replication, repair, and recombination) | PP_3255 | Ku protein | 2.33 | ||||
| PP_3260 | DNA ligase, ATP dependent, putative | 2.04 | |||||
| PP_3268 | MutT/nudix family protein | 2.72 | |||||
| PP_3967 | MutT/nudix family protein | 1.68 | |||||
| PP_4010 | cspD | Cold shock protein CspD | −2.08 | ||||
| Fatty acid and phospholipid metabolism | PP_0368 | Acyl-CoA dehydrogenase, putative | 1.84 | ||||
| PP_1996 | accD | Acetyl-CoA carboxylase, carboxyl transferase, beta subunit | 1.59 | ||||
| PP_3264 | Phospholipase family protein | 2.73 | |||||
| PP_4379 | 3-Oxoacyl-(acyl carrier protein) synthase III | 1.52 | |||||
| PP_5266 | Acetyl-CoA hydrolase/transferase family protein | 2.05 | |||||
| Cellular processes (adaptation to atypical condition) | PP_2326 | Universal stress protein family protein | −1.64 | ||||
| PP_3156 | Universal stress protein family | −2.55 | −1.74 | −1.75 | |||
| Protein fate | PP_3312 | Heat shock protein, putative | −1.73 | ||||
| PP_3313 | Heat shock protein, putative | −2.43 | −1.95 | −1.77 | |||
| PP_3314 | Heat shock protein, HSP20 family | −2.55 | −1.80 | −1.83 | |||
| DNA metabolism (other) | PP_0975 | hupN | DNA-binding protein HU, form N | 3.08 | 1.58 | ||
CoA, coenzyme A.
Expression of alginate genes.
We detected a transient expression of many alginate-related genes; all were upregulated at 4 h but none at later periods (Table 1). qRT-PCR also showed similar expression trends for algT, mucA, and alg8 genes, except for an increased upregulation of alg8 at 72 h (see Fig. S1 in the supplemental material). Similarly, Li et al. (22) observed transient alginate gene expression under water deprivation (down to −1.5 MPa) in P. putida mt-2 by using an alginate bioreporter. We found that the first three genes in the algT operon (algT, mucA, and mucB) were upregulated. In P. aeruginosa, the algT operon determines the alginate-producing phenotype (41), and many studies involving other Pseudomonas strains have mentioned the importance of AlgT under various environmental stress conditions from desiccation to heat shock (2, 20, 28). In P. aeruginosa, AlgT is required for algB transcription and AlgB is required for the algD transcription (41). We also detected that algB and half of the genes in the algD operon were upregulated (Table 1). It is likely that this coordinated expression of many alginate genes, being part of the major alginate operons, may bring about alginate production. It is known that alginate creates a hydrated environment under water deprivation in P. putida (6). This may explain the transient gene expression that we observed, as the creation of a hydrated environment might alleviate perceived water stress and subsequently halt overexpression of the alginate operons.
Expression of flagellar genes.
In contrast to our expectation and to the literature for P. putida under water deprivation (35), we did not observe downregulation of flagellar genes in the cells exposed to −0.4 MPa. The microarray data even point toward upregulation of a number of flagellar genes. The qRT-PCR (see Fig. S1 in the supplemental material) did not indicate significant downregulation for the selected genes either, but it also did not show significant upregulation as in the microarray. The expressed genes (Table 1) are all located first or second in 5 of the 17 flagellar operons as known in P. aeruginosa: fliEFGHIJ (class 2), fliLMNOPQRflhB (class 2), flgBCDE (class 3), flgFGHIJKL (class 3), and flgMN (class 2 and 4) (9). It is questionable whether flagella were actually overproduced, as not all flagellar genes, and notably none of the class IV genes, showed upregulated transcription. However, it has been suggested that activation of flagellar genes may not necessarily indicate flagellar synthesis but may indicate a stress tolerance response where the flagellar export apparatus is used to export other proteins unrelated to flagellar assembly (30).
Expression of sigma factors and other stress-related genes.
A few sigma factors, known as transcription specificity providers (24), were also differentially expressed in this study (Table 1). The RNA polymerase sigma factor, rpoS, was downregulated at 4 h. rpoS is associated with environmental fitness in Pseudomonas fluorescens (32), and many water deprivation-related genes are under putative control of RpoS in P. putida (35). Downregulation of rpoS observed in our study conflicts with the studies above. However, there is speculation that in P. aeruginosa RpoS and AlgT (AlgU) may be in competition (4). There may be a similar behavior in P. putida, as we observed significant upregulation of algT next to the downregulation of rpoS. Another sigma factor, RNA polymerase sigma-32 factor rpoH, was upregulated at 4 h. It is reported that in P. aeruginosa AlgT (AlgU) causes rpoH to be expressed (29). Our data suggest a similar mechanism in P. putida. Most of the downregulated genes are associated with energy metabolism, with the highest number at 4 h (Fig. 2), suggesting that the metabolic activities are either turned off or slowed down due to the stress. Alvarez et al. (1) reported that under water stress Rhodococcus opacus slows its metabolic activities down, especially in the first hours of stress. Some of the upregulated genes function in the subcategory of energy metabolism related to biodegradation and synthesis of polysaccharides. The simultaneous upregulation of alginate synthesis genes suggests that this energy could be dedicated to alginate synthesis. The hydroperoxide resistance protein gene osmC was upregulated at 4 and 24 h (Table 1). This was also expected since accumulation of reactive oxygen species (ROS) is another common consequence of water deprivation (26). Chang et al. (7) observed that ROS accumulation was significantly higher when Ψm dropped below −0.5 MPa. The decrease in differential expression of osmC from 4 to 24 h and its return to basal levels at 72 h again suggest a shift from stressed to nonstressed conditions. In fact, Chang et al. (7) further reported that alginate helps to cope with oxidative stress in P. putida mt-2. As ROS can damage DNA and proteins (26), the observed upregulation of DNA replication, repair, and recombination genes is well anticipated. Trehalose synthase (PP2918) was upregulated at 4 h but was not detected at 24 and 72 h (Table 1); trehalose accumulation and synthesis have been long associated with desiccation tolerance and osmoprotection in bacteria and yeast (26, 37). Some of the fatty acid and phospholipid metabolism-related genes were upregulated at 4, 24, and 72 h (Table 1). This could be related to the observation that bacteria change their fatty acid composition under stress (16). Some of the universal stress proteins and heat shock proteins were downregulated at 4, 24, and 72 h, although these genes are associated with the general stress response (12). Most of the differentially expressed genes with a regulatory function were detected at 4 h (Fig. 2). hupN (DNA-binding protein HU, form N) was one of the top upregulated genes at 4 h, but its level of upregulation decreased at 24 h, and it was not detected at 72 h (Table 1). HupN has a role in DNA bending, and it has been mentioned that the environmental fitness of P. putida depends on how environmental signals are integrated and DNA bending is used as signal transmission (3).
Pressure test.
There were no significantly expressed genes when the culture was exposed to 0.4-MPa (gauge) versus 0.1-MPa (atmospheric) headspace pressure (results not shown). Hence, the effects observed on the PPSM operated at −0.4 MPa can be interpreted solely as a result of the applied Ψm, not as a result of the gas pressure.
Gene expression profile under −0.5- and −1.0-MPa Ψm equivalents of PEG 8000 amendment.
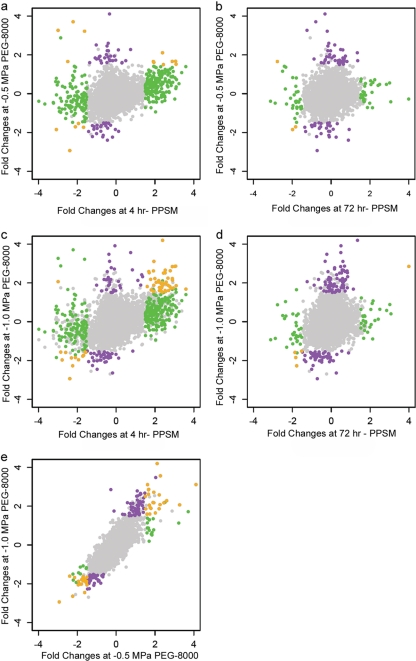
Figure 2b shows the major categories of the differentially expressed genes under the PEG 8000-simulated matric stress (the complete list is in Table S2 in the supplemental material). There was either no response or a very low response to PEG 8000-simulated Ψm compared to −0.4-MPa Ψm (Fig. 2a). Although the 5-day PEG 8000-simulated Ψm is more comparable to the 72-h Ψm in terms of stress duration, the number of shared differentially expressed genes was higher for 4-h Ψm than for 72-h Ψm (Fig. 3a and c versus b and d). For a better comparison, we also checked the proportions of the shared genes to the number of differentially expressed genes for 4- and 72-h Ψm. At a −0.5-MPa PEG 8000-simulated Ψm, the proportions of the shared genes were similar for 4- and 72-h Ψm (0.054 versus 0.056, respectively), whereas at a −1.0-MPa PEG 8000-simulated Ψm, the proportion was higher for 4-h Ψm than for 72-h Ψm (0.13 versus 0.074, respectively). Clearly, in all cases the number of shared differentially expressed genes was very low. It is possible that decreasing the PEG 8000-simulated Ψm below −1.0 MPa may increase the number of shared genes between PEG 8000-simulated and directly applied Ψm, as the number of differentially expressed genes increases with decreasing PEG 8000-simulated Ψm (Fig. 3e). The shared differentially expressed genes in PEG 8000-simulated and directly applied Ψm cases were mostly conserved hypothetical genes, without any known function. Most importantly, unlike in the −0.4-MPa Ψm case, we did not detect any flagellar genes in the PEG 8000-simulated Ψm case. Neither did we detect any alginate synthesis-related genes, although van de Mortel and Halverson (35) detected algA upregulation under a −1.5-MPa PEG 8000-simulated Ψm after 24 h. Despite the fact that the stress durations were different for PEG 8000-simulated (5 days) and directly applied Ψm (4- and 72-h) cases, our results suggest that a PEG 8000-simulated matric potential does not affect the gene expression profile of KT2440 in the same way as the directly applied matric potential does. This suggests that bacteria sense water deprivation differently depending on which water potential component (osmotic and matric) is causing the stress, an idea which deserves further investigation.
Fig 3.
Comparison of fold changes for all the genes in P. putida KT2440 under a −0.4-MPa Ψm versus PEG 8000-simulated stress. Fold changes are the expression values of the genes under a stress condition (imposed either by PPSM or by PEG 8000) relative to those at −0.5 kPa (control) in log2 scale. Positive and negative values correspond to up- and downregulation, respectively. (a) −0.5-MPa PEG 8000 at 5 days versus −0.4-MPa PPSM at 4 h; (b) −0.5-MPa PEG 8000 at 5 days versus −0.4-MPa PPSM at 72 h; (c) −1.0-MPa PEG 8000 at 5 days versus −0.4-MPa PPSM at 4 h; (d) −1.0-MPa PEG 8000 at 5 days versus −0.4-MPa PPSM at 72 h; (e) −1.0-MPa PEG 8000 at 5 days versus −0.5-MPa PEG 8000 at 5 days. The color of the dots illustrates the conditions under which genes were detected as differentially expressed. Genes presented in gray were nondifferentially expressed. Genes in purple, green, or orange were detected only under the condition on the y axis, only under the condition on the x axis, or under both conditions, respectively. Criteria defining differential expression were absolute log2 fold change of >1.5 and adjusted P value of <0.01.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Villum Kann Rasmussen Foundation Center of Excellence, Center for Environmental and Agricultural Microbiology (CREAM). B.F.S. acknowledges funding from a Marie Curie Excellence Grant (MEXT-CT-2005-024004, RaMAda), and A.D. acknowledges funding from a Danish Council for Strategic Research grant (2104-08-0012, MIRESOWA).
Footnotes
Published ahead of print 2 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alvarez HM, et al. 2004. Physiological and morphological responses of the soil bacterium Rhodococcus opacus strain PD630 to water stress. FEMS Microbiol. Ecol. 50:75–86 [DOI] [PubMed] [Google Scholar]
- 2. Aspedon A, Palmer K, Whiteley M. 2006. Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J. Bacteriol. 188:2721–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartels F, Fernandez S, Holtel A, Timmis KN, de Lorenzo V. 2001. The essential HupB and HupN proteins of Pseudomonas putida provide redundant and nonspecific DNA-bending functions. J. Biol. Chem. 276:16641–16648 [DOI] [PubMed] [Google Scholar]
- 4. Behrends V, Ryall B, Wang X, Bundy JG, Williams HD. 2010. Metabolic profiling of Pseudomonas aeruginosa demonstrates that the anti-sigma factor MucA modulates osmotic stress tolerance. Mol. Biosyst. 6:562–569 [DOI] [PubMed] [Google Scholar]
- 5. Chang WS, Halverson LJ. 2003. Reduced water availability influences the dynamics, development, and ultrastructural properties of Pseudomonas putida biofilms. J. Bacteriol. 185:6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang WS, et al. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 189:8290–8299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang WS, Li X, Halverson LJ. 2009. Influence of water limitation on endogenous oxidative stress and cell death within unsaturated Pseudomonas putida biofilms. Environ. Microbiol. 11:1482–1492 [DOI] [PubMed] [Google Scholar]
- 8. Cytryn EJ, et al. 2007. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 189:6751–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dasgupta N, et al. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809–824 [DOI] [PubMed] [Google Scholar]
- 10. Dechesne A, Or D, Gulez G, Smets BF. 2008. The porous surface model: a novel experimental system for online quantitative observation of microbial processes under unsaturated conditions. Appl. Environ. Microbiol. 74:5195–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dechesne A, Wang G, Gulez G, Or D, Smets BF. 2010. Hydration-controlled bacterial motility and surface dispersal. Proc. Natl. Acad. Sci. U. S. A. 107:14369–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dos Santos VA, Heim S, Moore ERB, Straetz M, Timmis KN. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6:1264–1286 [DOI] [PubMed] [Google Scholar]
- 13. Fonseca P, Moreno R, Rojo F. 2008. Genomic analysis of the role of RNase R in the turnover of Pseudomonas putida mRNAs. J. Bacteriol. 190:6258–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griffin DM, Quail G. 1968. Movement of bacteria in moist, particulate systems. Aust. J. Biol. Sci. 21:579–582 [DOI] [PubMed] [Google Scholar]
- 15. Gulez G, Dechesne A, Smets BF. 2010. The pressurized porous surface model: an improved tool to study bacterial behavior under a wide range of environmentally relevant matric potentials. J. Microbiol. Methods 82:324–326 [DOI] [PubMed] [Google Scholar]
- 16. Halverson LJ, Firestone MK. 2000. Differential effects of permeating and non-permeating solutes on the fatty acid composition of Pseudomonas putida. Appl. Environ. Microbiol. 66:2414–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holden PA, Halverson LJ, Firestone MK. 1997. Water stress effects on toluene biodegradation by Pseudomonas putida. Biodegradation 8:143–151 [DOI] [PubMed] [Google Scholar]
- 18. Holden PA. 2001. Biofilms in unsaturated environments. Methods Enzymol. 337:125–143 [DOI] [PubMed] [Google Scholar]
- 19. Katoh H, Asthana RK, Ohmori M. 2004. Gene expression in the cyanobacterium Anabaena sp. PCC7120 under desiccation. Microb. Ecol. 47:164–174 [DOI] [PubMed] [Google Scholar]
- 20. Keith LMW, Bender CL. 1999. AlgT (σ22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J. Bacteriol. 181:7176–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. LeBlanc JC, Goncalves ER, Mohn WW. 2008. Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 74:2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, Nielsen L, Nolan C, Halverson LJ. 2010. Transient alginate gene expression by Pseudomonas putida biofilm residents under water-limiting conditions reflects adaptation to the local environment. Environ. Microbiol. 12:1578–1590 [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2d̂dCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 24. Martinez-Bueno MA, Tobes R, Rey M, Ramos JL. 2002. Detection of multiple extracytoplasmic function (ECF) sigma factors in the genome of Pseudomonas putida KT2440 and their counterparts in Pseudomonas aeruginosa PA01. Environ. Microbiol. 4:842–855 [DOI] [PubMed] [Google Scholar]
- 25. Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting microbial habitats and activity in unsaturated porous media—a review. Adv. Water Resour. 30:1505–1527 [Google Scholar]
- 26. Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roberson EB, Firestone MK. 1992. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl. Environ. Microbiol. 58:1284–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schnider-Keel U, Lejbolle KB, Baehler E, Haas D, Keel C. 2001. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 67:5683–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schurr MJ, Deretic V. 1997. Microbial pathogenesis in cystic fibrosis: coordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 24:411–420 [DOI] [PubMed] [Google Scholar]
- 30. Segura A, Hurtado A, Duque E, Ramos JL. 2004. Transcriptional phase variation at the flhB gene of Pseudomonas putida DOT-T1E is involved in response to environmental changes and suggests the participation of the flagellar export system in solvent tolerance. J. Bacteriol. 186:1905–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3(1):article 3. http://www.bepress.com/sagmb/vol3/iss1/art3 [DOI] [PubMed] [Google Scholar]
- 32. Stockwell VO, Loper JE. 2005. The sigma factor RpoS is required for stress tolerance and environmental fitness of Pseudomonas fluorescens Pf-5. Microbiology 151:3001–3009 [DOI] [PubMed] [Google Scholar]
- 33. Sutherland IW. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9 [DOI] [PubMed] [Google Scholar]
- 34. Tuller M, Or D, Dudley LM. 1999. Adsorption and capillary condensation in porous media: liquid retention and interfacial configurations in angular pores. Water Resour. Res. 35:1949–1964 [Google Scholar]
- 35. van de Mortel M, Halverson LJ. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735–750 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey RM. 2005. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24:2034–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Welsh DT, Herbert RA. 1999. Osmotically induced intracellular trehalose, but not glycine betaine accumulation promotes desiccation tolerance in Escherichia coli. FEMS Microbiol. Lett. 174:57–63 [DOI] [PubMed] [Google Scholar]
- 38. Wernersson R, Junkcer AS, Nielsen HB. 2007. Probe selection for DNA microarrays using OligoWiz. Nat. Protoc. 2:2677–2691 [DOI] [PubMed] [Google Scholar]
- 39. Wilkinson JF. 1958. The extracellular polysaccharides of bacteria. Bacteriol. Rev. 22:46–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Workman C, et al. 2002. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3:research0048–research0048.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.