Abstract
An outbreak of abortion affecting multiparous cows was associated with Hobi-like pestivirus infection. Viral RNA and antigens were detected in the tissues of two aborted fetuses. Molecular assays for other common abortogenic agents tested negative. At the genetic level, the Hobi-like pestivirus displayed the closest relatedness to Italian, Australian, and South American viruses, whereas it diverged from the prototype Thai isolate. These findings may have important implications for the pestivirus control/eradication programs in cattle herds.
TEXT
Bovine viral diarrhea viruses (BVDVs) are members of the genus Pestivirus (family Flaviviridae), responsible for a number of clinical signs, including subclinical infections, immunosuppression, acute diarrhea, respiratory disease, reproductive failures, and mucosal disease in persistently infected calves. Reproductive disorders caused by BVDVs vary according to the fetal age and include embryo death, abortion, mummification, congenital abnormalities, or stillbirths (2). Thus far, two different BVDV species have been recognized, BVDV-1 and BVDV-2, that cocirculate in cattle herds worldwide (20). In 2004, an atypical pestivirus, strain D32/00_Hobi, was isolated from a contaminated batch of fetal calf serum (FCS) (19). The virus was distantly related to BVDV-1/BVDV-2, and it was proposed as a prototype of a new pestivirus species (14, 15). Hobi-like sequences have been repeatedly detected in commercial FCS batches (17, 22, 23), whereas there are few reports on natural infections (4, 5, 21, 23) and clinical outbreaks (5). The virus has been recently detected in aborted bovine fetuses in Brazil, thus suggesting direct clinical implications (4).
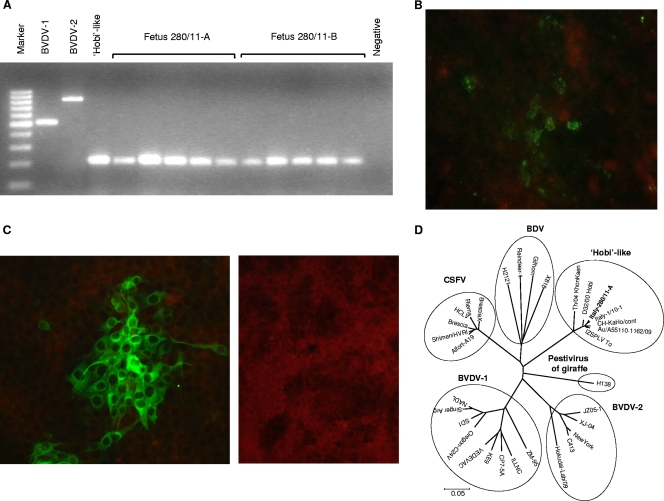
Here, we report the isolation and genetic characterization of a Hobi-like strain detected from aborted fetuses in southern Italy. The abortion outbreak occurred in June 2011 in a cattle herd where a Hobi-like pestivirus-associated respiratory disease had been recently described (5). Abortion was observed in eight multiparous cows in a group of 270 lactating Holstein cows and occurred between the fourth and sixth months of pregnancy. The animals neither showed prodromal signs nor presented postabortion complications. Two aborted fetuses (280/11-A, 280/11-B) were sent to our laboratory, and tissue samples were collected from lungs, spleens, livers, kidneys, and placentas for diagnostic investigations. Nucleic acids were purified using the DNeasy tissue kit (Qiagen) and QIAamp RNeasy minikit (Qiagen). Reverse transcription (RT)-PCR and PCR assays were performed using SuperScript one-step RT-PCR for long templates (Life Technologies) and LA PCR kit version 2.1 (TaKaRa Bio Inc.), respectively. Positive and negative controls were processed in parallel to the screened samples. The samples tested negative by PCR for Chlamydophila spp. (9), Leptospira spp. (27), Coxiella burnetii (12), Salmonella spp. (11), Toxoplasma gondii, Neospora caninum (18), Brucella spp. (1), and bovid herpesvirus 1 (26) and by RT-PCR for bluetongue virus (3, 8). Conversely, the pestivirus genome was detected with two different RT-PCR protocols (7, 24), and the virus was characterized as Hobi-like by a species-specific nested PCR (7) (Fig. 1A). Viral titers, quantified by a TaqMan-based real-time RT-PCR (13), ranged from 4.31 × 102 (kidney of fetus 280/11-B) to 5.78 × 104 (lung of fetus 280/11-B) RNA copies μl−1 of template.
Fig 1.
Detection and characterization of Hobi-like pestivirus in aborted bovine fetuses. (A) Gel electrophoresis of products obtained from a nested PCR assay for detection and characterization of BVDVs. Marker, GeneRuler 100-bp DNA ladder (MBI Fermentas GmbH, St. Leon-Rot, Germany); BVDV-1, strain NADL; BVDV-2, strain 232/02; Hobi-like, BVDV-3 strain Italy-1/10-1; fetus 280/11-A, tissue samples (placenta, lung, spleen, liver, kidney); fetus 280/11-B, tissue samples (placenta, lung, spleen liver, kidney); negative, negative control (spleen from a BVDV-negative calf). (B) Lung of fetus 280/11-A: immunofluorescence assay. (C) Madin-Darby bovine kidney cells inoculated with the lung of fetus 280/11-A (left) or uninfected (right): immunofluorescence assay using an anti-NS3 monoclonal antibody. (D) Neighbor-joining unrooted tree based on the Npro sequences of members of the genus Pestivirus. For phylogenetic tree construction, pestivirus sequences listed in Table 1 were used. The scale bar represents the estimated numbers of nucleotide substitutions per site.
Upon immunofluorescence using an anti-NS3 monoclonal antibody (5), pestiviral antigens were detected diffusely in 10-micrometer cryostat sections of lung and spleen tissue, where the highest viral loads were revealed in real-time RT-PCR (Fig. 1B).
Virus was isolated from the lung tissues of fetus 280/11-A using Madin-Darby bovine kidney cells that were confirmed to be negative for pestivirus before sample inoculation. No cytopathic effect was observed in the inoculated cells, although virus antigen was detected in immunofluorescence (Fig. 1C) and virus RNA was detected by a Hobi-like pestivirus-specific real-time RT-PCR. Serial passages of the virus on cell culture were carried out, and the 15th passage still showed diffuse pestivirus cytoplasmic fluorescence.
Informative sequences of the pestivirus genome in the E2, 5′ untranslated region (UTR) and N-terminal autoprotease (Npro) regions (16) were amplified using previously published oligonucleotides (5, 14). The PCR products generated from strain Italy-280/11-A were subjected to direct sequencing by BaseClear B.V. (Leiden, The Netherlands), and consensus sequences were obtained using the BioEdit software package (10). The sequences (GenBank accession numbers JN703311 to JN703313) were aligned with cognate sequences using the ClustalW tool of the European Molecular Biology Laboratory (http://www.ebi.ac.uk). Upon sequence analysis, strain Italy-280/11-A appeared closely related to a pestivirus strain previously isolated in the same herd (Italy-1/10-1) in all the analyzed regions, with nucleotide (nt) identities of 99.4% (E2), 99.4% (5′ UTR), and 98.4% (Npro). Next to the prototype Italian isolate, the highest genetic relatedness was found to the Australian virus Au/A55110-1162/09 in the E2 (95.3% nt identity) and 5′ UTR (98.9% nt identity) sequences and to the Italian strain IZSPLV_To in the Npro gene (96.9% nt identity), both detected in contaminated FCS (Table 1). At the amino acid (aa) level, the closest identity was to the Brazilian strain D32/00_Hobi and the Italian isolate Italy-1/10-1 in the Npro (97.3% aa identity) and E2 protein (98.8% aa identity). Amino acid identities to BVDV-1 strains ranged from 64.9% to 69.5% in the Npro and from 59.1% to 62.2% in the E2, whereas identity to BVDV-2 was 64.2 to 67.5% in the Npro and 56.1 to 59.6% in the E2.
Table 1.
Nucleotide and amino acid identities of Hobi-like strain Italy-280/11-A to reference pestiviruses in informative genomic regions
| Pestivirus species | Strain | Accession no. | % nt (aa) identitya |
||
|---|---|---|---|---|---|
| E2 | 5′ UTR | Npro | |||
| Hobi-like pestivirus | Italy-1/10-1 | HQ231763 | 99.4 (98.8) | 99.4 | 98.4 (96.6) |
| Th/04_KhonKaen | FJ040215 | 87.8 (88.7) | 91.8 | 89.2 (91.3) | |
| D32/00_HoBi | AY604725 (E2); AY489116 (5′ UTR); AY735486 (Npro) | 93.6 (92.1) | 97.8 | 95.8 (97.3) | |
| CH-KaHo/cont | EU385605 (E2); AY895011 (Npro) | 94.5 (91.9) | NA | 95.8 (95.3) | |
| IZSPLV_To | HM151363 (E2); HM151361 (5′ UTR); HM151362 (Npro) | 94.5 (93.3) | 98.3 | 96.9 (96.0) | |
| Au/A55110-1162/09 | FR873802 (E2); FR873797 (5′ UTR); FR873800 (Npro) | 95.3 (95.0) | 98.9 | 96.2 (96.0) | |
| BDV | H2121 (Chamois-1) | GU270877 | 57.8 (57.4) | 62.3 | 64.0 (68.8) |
| Gifhorn | GQ902940 | 58.8 (59.1) | 66.1 | 64.9 (65.5) | |
| X818 | NC_003679 | 60.4 (58.9) | 60.7 | 66.0 (68.2) | |
| Reindeer | AF144618 | 58.5 (56.6) | 65.5 | 63.6 (64.9) | |
| CSFV | Brescia X | AY578687 | 58.8 (58.0) | 67.2 | 67.1 (69.5) |
| HCLV | AF531433 | 59.0 (58.0) | 66.1 | 65.6 (69.5) | |
| Brescia | AF091661 | 59.2 (58.6) | 66.6 | 65.4 (70.1) | |
| Alfort-A19 | U90951 | 58.9 (58.0) | 66.6 | 66.0 (70.1) | |
| Shimen/HVRI | AY775178 | 58.7 (58.9) | 66.1 | 67.4 (70.1) | |
| Riems | AY259122 | 59.5 (58.0) | 66.1 | 65.4 (69.5) | |
| Pestivirus of giraffe | H138 | AF144617 | 58.7 (57.6) | 73.4 | 64.7 (66.2) |
| BVDV-1 | ILLNC | U86600 | 60.5 (59.1) | 70.4 | 63.6 (64.9) |
| ZM-95 | AF526381 | 60.9 (61.4) | 66.3 | 63.2 (67.5) | |
| Oregon-C24V | AF091605 | 60.4 (61.3) | 69.3 | 66.0 (68.2) | |
| CP7-5A | AF220247 | 61.3 (61.9) | 68.8 | 65.6 (69.5) | |
| SD1 | M96751 | 59.6 (59.9) | 69.3 | 65.1 (68.2) | |
| Singer_Arg | DQ088995 | 61.5 (60.5) | 69.8 | 64.7 (67.5) | |
| KE9 | EF101530 | 60.5 (61.0) | 70.9 | 65.6 (67.5) | |
| NADL | M31182 | 62.4 (62.2) | 69.8 | 65.6 (68.2) | |
| VEDEVAC | AJ585412 | 61.0 (60.3) | 71.5 | 66.0 (68.8) | |
| BVDV-2 | JZ05-1 | GQ888686 | 60.4 (57.0) | 75.5 | 64.0 (65.5) |
| New York′93 | AF502399 | 59.8 (57.0) | 75.0 | 65.1 (67.5) | |
| XJ-04 | FJ527854 | 60.2 (56.1) | 75.0 | 64.0 (64.2) | |
| C413 | NC_002032 | 59.5 (56.1) | 75.0 | 64.0 (65.5) | |
| Hokudai Lab/09 | AB567658 | 60.3 (59.6) | 73.4 | 65.1 (64.2) | |
NA, sequence not available.
Phylogenetic analysis was accomplished using both parsimony and neighbor-joining methods with Mega4.1 software (25), supplying statistical support with bootstrapping over 1,000 replicates. In the Npro tree, the analyzed pestiviruses clustered into six monophyletic clades consisting of BVDV-1, BVDV-2, Hobi-like pestivirus, border disease virus, classical swine fever virus (CSFV), and pestivirus of giraffe (Fig. 1D). Strain Italy-280/10-A formed a unique cluster, along with pestiviruses Italy-1/10-1 and Th/04_KhonKaen, clearly distinct from other pestivirus species. Within this monophyletic group, both the Italian strains, Italy-1/10-1 and Italy-280/11-A, were intermingled with viruses of southern American and Australian origins. This pattern of segregation was confirmed by both neighbor-joining and maximum-parsimony methods in all the analyzed genomic regions (data not shown).
Although Hobi-like pestivirus was first isolated more than 7 years ago (19), cases of natural infection have been reported only sporadically (4, 5, 21, 23), and thus far there is a single report on a clinical outbreak associated with this pestivirus (5). Hobi-like sequences were previously detected in two aborted bovine fetuses in Brazil, but clinical data were not provided, and the virus was not characterized in depth (4). In this study, we described an outbreak of abortion in cows that was associated with Hobi-like pestivirus infection. Viral RNA was detected with specific molecular assays, and the virus was isolated in cell cultures. Viral antigens were demonstrated in fetal tissues by immunofluorescence, and the virus was characterized by sequence and phylogenetic analysis of genomic informative regions. Since other abortogenic microorganisms were not detected in the fetuses, the outbreak was associated to the atypical pestivirus circulating in the herd.
The pathogenic potential of Hobi-like pestivirus has not been investigated in depth. Experimental infection of nonpregnant animals has shown that the virus is able to induce respiratory distress, fever, and moderate leukopenia in cattle and sheep, whereas pigs did not develop any clinical signs (6). Experimental infections of pregnant cows will help assess whether and to which extent this new pestivirus may impact bovine reproduction and if specific diagnostic and preventive measures are required.
ACKNOWLEDGMENT
This work was supported by grants from the University of Bari, Italy, Ricerca di Ateneo 2010, project “Epidemiologia dei pestivirus atipici (BVDV-3) in Italia.”
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1.Al Dahouk S, et al. 2007. Evaluation of genus-specific and species-specific real-time PCR assays for the identification of Brucella spp. Clin. Chem. Lab. Med. 45:1464–1470 [DOI] [PubMed] [Google Scholar]
- 2.Baker JC. 1995. The clinical manifestations of bovine viral diarrhea virus infection. Vet. Clin. North Am. Food Anim. Pract. 11:425–445 [DOI] [PubMed] [Google Scholar]
- 3.Billinis C, et al. 2001. Bluetongue virus diagnosis of clinical cases by a duplex reverse transcription-PCR: a comparison with conventional methods. J. Virol. Methods 98:77–89 [DOI] [PubMed] [Google Scholar]
- 4.Cortez A, et al. 2006. Genetic characterization of Brazilian bovine viral diarrhea virus isolates by partial nucleotide sequencing of the 5′-UTR region. Pesqui. Vet. Bras. 26:211–216 [Google Scholar]
- 5.Decaro N, et al. 2011. Atypical pestivirus and severe respiratory disease in calves, Europe. Emerg. Infect. Dis. 17:1549–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decaro N, et al. 2011. Experimental infection of cattle, sheep and pigs with ‘Hobi'-like pestivirus. Vet. Microbiol. doi:10.1016/j.vetmic.2011.08.030. [DOI] [PMC free article] [PubMed]
- 7.Decaro N, et al. 2011. A nested PCR approach for unambiguous typing of pestiviruses infecting cattle. Mol. Cell. Probes doi:10.1016/j.mcp.2011.11.003 [DOI] [PMC free article] [PubMed]
- 8.Elia G, et al. 2008. Use of real-time RT-PCR as a rapid molecular approach for differentiation of field and vaccine strains of bluetongue virus serotypes 2 and 9. Mol. Cell. Probes 22:38–46 [DOI] [PubMed] [Google Scholar]
- 9.Everett KD, Hornung LJ, Andersen AA. 1999. Rapid detection of the Chlamydiaceae and other families in the order Chlamydiales: three PCR tests. J. Clin. Microbiol. 37:575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 11.Khan AA, Nawaz MS, Khan SA, Cerniglia CE. 2000. Detection of multidrug-resistant Salmonella typhimurium DT104 by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 182:355–360 [DOI] [PubMed] [Google Scholar]
- 12.Klee SR, et al. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Xia H, Belák S, Baule C. 2008. A TaqMan real-time RT-PCR assay for selective detection of atypical bovine pestiviruses in clinical samples and biological products. J. Virol. Methods 154:82–85 [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Kampa J, Belák S, Baule C. 2009. Virus recovery and full-length sequence analysis of atypical bovine pestivirus Th/04_KhonKaen. Vet. Microbiol. 138:62–68 [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Xia H, Baule C, Belák S. 2009. Maximum likelihood and Bayesian analyses of a combined nucleotide sequence dataset for genetic characterization of a novel pestivirus, SVA/cont-08. Arch. Virol. 154:1111–1116 [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Xia H, Wahlberg N, Belák S, Baule C. 2009. Phylogeny, classification and evolutionary insights into pestiviruses. Virology 385:351–357 [DOI] [PubMed] [Google Scholar]
- 17.Peletto S, et al. 2010. Detection and phylogenetic analysis of an atypical pestivirus, strain IZSPLV_To. Res. Vet. Sci. doi:10.1016/j.rvsc.2010.10.015. [DOI] [PubMed]
- 18.Reitt K, et al. 2007. Aetiology of bovine abortion in Switzerland from 1986 to 1995—a retrospective study with emphasis on detection of Neospora caninum and Toxoplasma gondii by PCR. J. Vet. Med. A Physiol. Pathol. Clin. Med. 54:15–22 [DOI] [PubMed] [Google Scholar]
- 19.Schirrmeier H, Strebelow G, Depner K, Hoffmann B, Beer M. 2004. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J. Gen. Virol. 85:3647–3652 [DOI] [PubMed] [Google Scholar]
- 20.Simmonds P, et al. 2011. Family Flaviviridae, p 1003–1020. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 21.Ståhl K, et al. 2007. Natural infection of cattle with an atypical ‘HoBi'-like pestivirus—implications for BVD control and for the safety of biological products. Vet. Res. 38:517–523 [DOI] [PubMed] [Google Scholar]
- 22.Ståhl K, et al. 2010. Atypical ‘HoBi'-like pestiviruses—recent findings and implications thereof. Vet. Microbiol. 142:90–93 [DOI] [PubMed] [Google Scholar]
- 23.Stalder HP, et al. 2005. Genetic heterogeneity of pestiviruses of ruminants in Switzerland. Prev. Vet. Med. 72:37–41 [DOI] [PubMed] [Google Scholar]
- 24.Sullivan DG, Akkina RK. 1995. A nested polymerase chain reaction assay to differentiate pestiviruses. Virus Res. 38:231–239 [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 26.Vilcek S. 1993. Detection of the bovine herpesvirus-1 (BHV-1) genome by PCR. J. Virol. Methods 41:245–247 [DOI] [PubMed] [Google Scholar]
- 27.Woo TH, et al. 1997. Rapid distinction between Leptospira interrogans and Leptospira biflexa by PCR amplification of 23S ribosomal DNA. FEMS Microbiol. Lett. 150:9–18 [DOI] [PubMed] [Google Scholar]