Abstract
Pseudomonas aeruginosa can develop resistance to polymyxin and other cationic antimicrobial peptides. Previous work has shown that mutations in the PmrAB and PhoPQ regulatory systems can confer low to moderate levels of colistin (polymyxin E) resistance in laboratory strains and clinical isolates of this organism (MICs of 8 to 64 mg/liter). To explore the role of PmrAB in high-level clinical polymyxin resistance, P. aeruginosa isolates from chronically colistin-treated cystic fibrosis patients, most with colistin MICs of >512 mg/liter, were analyzed. These cystic fibrosis isolates contained probable gain-of-function pmrB alleles that conferred polymyxin resistance to strains with a wild-type or pmrAB deletion background. Double mutant pmrB alleles that contained mutations in both the periplasmic and dimerization-phosphotransferase domains markedly augmented polymyxin resistance. Expression of mutant pmrB alleles induced transcription from the promoter of the arnB operon and stimulated addition of 4-amino-l-arabinose to lipid A, consistent with the known role of this lipid A modification in polymyxin resistance. For some highly polymyxin-resistant clinical isolates, repeated passage without antibiotic selection pressure resulted in loss of resistance, suggesting that secondary suppressors occur at a relatively high frequency and account for the instability of this phenotype. These results indicate that pmrB gain-of-function mutations can contribute to high-level polymyxin resistance in clinical strains of P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen that causes serious infections in individuals with defective host defense mechanisms. Cystic fibrosis (CF) patients, in particular, are susceptible to chronic progressive airway infections with this organism (34). In CF, the airways are variably coated or plugged with dehydrated and highly viscous mucus plaques, providing an environment conducive to the growth of P. aeruginosa and other microbial opportunists (30).
Historically, acutely ill CF patients or others infected with P. aeruginosa have been treated with combinations of broad-spectrum intravenous antibiotics, such as ceftazidime and tobramycin (47, 48, 53). Repeated use of such agents in CF patients tends to select for multidrug-resistant (MDR) P. aeruginosa strains; one such ceftazidime-resistant strain arose and spread throughout the CF patient population of Denmark during the 1980s (10, 20, 46). Diverse mechanisms of resistance and tolerance have driven the selection and evolution of this and other MDR CF strains of P. aeruginosa (3, 17, 24).
CF clinicians in Denmark and elsewhere subsequently turned to colistin (CST; polymyxin E) as a key antipseudomonal agent (18, 19, 28, 43, 56). Polymyxins (Pm) are a family of antimicrobial cyclic oligopeptides synthesized by the Gram-positive organism Bacillus polymyxa. The clinically available forms, polymyxin B (PMB) sulfate and colistimethate, a prodrug of CST, are administered to CF patients intravenously or by inhalation. Pm bind to lipopolysaccharide (LPS), the major constituent of the Gram-negative outer membrane, thus promoting membrane permeabilization and diffusion of peptide across the periplasm. Pm insertion at the inner membrane disrupts cellular respiration and results in cell lysis and death (52).
In clinical practice, CST and PMB are used most commonly to treat P. aeruginosa and Acinetobacter baumannii infections. As a consequence, Pm-resistant (Pmr) clinical isolates of these organisms that are resistant to both agents are increasingly being reported (2, 7, 14, 16, 23, 29, 33). In P. aeruginosa, Pm resistance is associated with covalent addition of 4-amino-l-arabinose (l-Ara4N) to phosphate groups within the lipid A and core oligosaccharide moieties of LPS (6, 42, 45). Genes in the arnBCADTEF-pmrE operon (also known as pmrHFIJKLME [PA3552 to PA3559]) encode enzymes responsible for the synthesis and transfer of l-Ara4N to lipid A (21, 22). This amino-sugar modification interferes with charge interactions between phosphate groups within LPS and amino groups within the cyclic Pm oligopeptide.
In P. aeruginosa and other Gram-negative organisms, the PmrAB and PhoPQ two-component regulatory systems stimulate transcription of the arnBCADTEF-pmrE operon in response to antimicrobial peptide exposure or divalent cation depletion (21, 22, 35, 36, 38). The sensor kinase PmrB activates the transcriptional response regulator PmrA through a phosphotransfer relay. Activation of PmrA can also occur as a consequence of pmrB mutation, previously observed as a cause of Pm resistance in a laboratory strain (42) and in clinical isolates (1, 5, 51). We hypothesized that pmrB mutations in highly Pmr CF strains of P. aeruginosa might similarly contribute to Pm resistance. In this study, we used CF isolates as well as laboratory strains of P. aeruginosa to define mutations in the PmrAB two-component regulatory system as an important but nonexclusive mechanism contributing to high-level clinical Pm resistance.
(Portions of the results reported here were previously presented in abstract form at the 2000 and 2005 North American Cystic Fibrosis Conferences, Baltimore, MD [7, 40].)
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genotyping.
Bacterial reference strains, clinical isolates, and their derivatives used in this study are listed in Table 1. The institutional review boards of Seattle Children's Hospital and Massachusetts General Hospital reviewed and approved the use of clinical isolates for this study. Clinical isolates were cultured from the airway secretions of CF patients as part of routine clinical care. Escherichia coli strain DH5α was used as the host strain for manipulation of recombinant plasmids. P. aeruginosa and E. coli were grown at 37°C on Luria-Bertani (LB) agar plates or in LB broth with aeration. Antibiotics were used for plasmid selection at the following concentrations: 50 mg/liter kanamycin (KAN) or 10 mg/liter gentamicin (GEN) for plasmids in E. coli DH5α and 100 mg/liter GEN for plasmids in P. aeruginosa strains PAK and PAO1 or their derivatives. All strains and isolates were stored in a 16% glycerol-LB broth solution at −80°C. Genotypes of clinical isolates were determined by pulsed-field gel electrophoresis (54) and/or multilocus sequence typing (MLST) (12), with addition of novel alleles and sequence types to the P. aeruginosa database available at http://pubmlst.org/paeruginosa.
Table 1.
Bacterial strains and clinical isolates used in this study
| Strain or isolate | Description | Source or reference |
|---|---|---|
| Laboratory reference strains and derivatives | ||
| 1067 | E. coli DH5α | 59 |
| 1927 | E. coli DB3.1 (host for Gateway vectors) | Invitrogen |
| 1026 | P. aeruginosa PAK (Pms) | S. Lory |
| 1029 | P. aeruginosa PAK pmrB6 (Pmr) | 42 |
| 1033 | P. aeruginosa PAK pmrB12 (Pmr) | 42 |
| 1369 | P. aeruginosa PAK(pJN105D::pmrAB12) | This study |
| 1812 | P. aeruginosa PAK ΔpmrAB (Pms) | This study |
| 2241 | P. aeruginosa PAK ΔarnC (Pms) | This study |
| 2251 | P. aeruginosa PAK ΔarnC(pJN105D::pmrAB12-arnC) | This study |
| 2255 | P. aeruginosa PAK ΔarnC(pJN105D::pmrAB12) | This study |
| 2735 | P. aeruginosa PAK ΔpmrAB(pJN105D::pmrAB12) | This study |
| 1555 | P. aeruginosa PAO1 (Pms) | B. Iglewski |
| 2897 | P. aeruginosa PAO1 ΔpmrAB Ω(attP::Φ(ParnB-lacZ+)) (Pms) | This study |
| Individual CF clinical isolates of P. aeruginosa and Pmr/Pms isogenic pairs | ||
| PA1109 | UK isolate (CF patient, anonymous), 1990s (Pmr) | D. VanDevanter |
| PA1125 | UK isolate (CF patient 1, Leeds), 1997 (Pmr) | 14 |
| PA1131 | UK isolate (CF patient 4, Leeds), 1999 (Pmr) | 14 |
| PA1133 | UK isolate (CF patient 5, Leeds), 2000 (Pmr) | 14 |
| PA1015 | Danish isolate (CF patient 1, Copenhagen), 1996 (Pmr) | This study |
| PA1017 | Danish isolate (CF patient 3, Aarhus), 1996 (Pmr) | This study |
| PA1603 | Danish isolate (CF patient 14, Copenhagen), 2003 (Pmr) | This study |
| PA1611 | Danish isolate (CF patient 15, Copenhagen), 2001 (Pmr) | This study |
| PA2123–PA2162 | U.S. isolates (CF patients, Seattle), 2001-2004 (Pms or Pmi) | This study |
| PA1016/PA2047 | Danish isolates (CF patient 2, Aarhus), 1996/1985 | This study |
| PA1020/PA2050 | Danish isolates (CF patient 6, Copenhagen), 1998/1995 | This study |
| PA1571/PA1575 | Danish isolates (CF patient 8, Copenhagen), 2002/1995 | This study |
| Derivatives of CF P. aeruginosa clinical isolate PA1016 | ||
| PA2034 | PA1016 ΔpmrAB (Pms) | This study |
| PA2044 | PA1016 ΔpmrAB(pJN105D::pmrAB23) | This study |
| PA2045 | PA1016 ΔpmrAB(pJN105D::pmrAB+) | This study |
| PA2330 | PA1016 ΔarnC (Pms) | This study |
| PA2367 | PA1016 ΔarnC(pJN105D::arnC) | This study |
Antimicrobial susceptibility assays.
Pm agar dilution testing was performed as described previously (11): a Nunc 96-pin replicator with 1-mm pins and an OmniTray copier were used to inoculate the surface of OmniTray plates (Nunc International, Rochester, NY) containing 2-fold serial dilutions (0.125 to 512 mg/liter) of CST sulfate salt (Sigma-Aldrich, St. Louis, MO) in Difco Mueller-Hinton agar (Becton Dickinson Diagnostic Systems, Sparks, MD). Consistent with current CLSI recommendations, strains with CST MICs of ≤2 mg/liter were considered Pm susceptible (Pms), those with a CST MIC of 4 mg/liter were considered Pm intermediate (Pmi), and those with CST MICs of ≥8 mg/liter were considered Pm resistant (Pmr). For Pmr strains, CST MICs of 8 to 32 mg/liter were considered to represent low-level resistance, those of 64 to 256 mg/liter were considered to represent moderate-level resistance, and those of ≥512 mg/liter were considered to represent high-level resistance.
For the alternative PMB plate assay, strains were inoculated into LB broth containing 1 mM MgCl2 and grown at 37°C with aeration for 16 to 20 h, to an optical density at 600 nm (OD600) of ∼3.0 to 5.0. The culture was diluted 1:50 in fresh medium and grown for 2 to 3 h at 30°C with aeration, to an OD600 of ∼0.8. Dilutions containing ∼50 to 200 CFU per 0.1 ml were spread on LB agar plates containing 1 mM MgCl2 and 0 to 800 mg/liter PMB.
For quantitative bactericidal assay (42), strains were grown and subcultured as described for the alternative PMB plate assay, diluted in Mueller-Hinton broth to a final density of 2 × 104 CFU per ml, exposed to 2-fold serial dilutions of PMB (0.5 to 512 mg/liter, as well as a drug-free control) for 30 min at 37°C, spread on LB agar after 1:10 dilution, and incubated for 16 to 20 h at 37°C for enumeration.
For all susceptibility testing methods, media used for strains with pJN105D-derived plasmids contained 0.1% l-arabinose.
Molecular methods.
Bacterial plasmids and chromosomal DNA were isolated using commercially available kits (Qiaquick and DNeasy kits [Qiagen, Valencia, CA] and Masterpure kits [Epicentre Biotechnologies, Madison, WI]). Plasmids were introduced into P. aeruginosa by electroporation (9). Sequencing of plasmids and chromosomal DNA was performed on both strands by using oligonucleotide primers (see Table S1 in the supplemental material) spaced ∼400 bp apart. PCR amplifications were performed as described previously (39), using specific oligonucleotide primers (see Tables S2 and S3).
To construct deletions, ∼1-kb chromosomal DNA segments flanking the targeted locus were PCR amplified from chromosomal DNA by using specific oligonucleotide primers (see Table S2 in the supplemental material) and were joined through splicing by overlap extension-PCR (25); deletions were marked by inclusion of a unique restriction site in the overlapping (internal) primers. For constructs made using SS series primers, the resulting ∼2-kb DNA fragment was inserted into the suicide plasmid pEX18Gm (26), using EcoRI and HindIII restriction sites. For constructs made using SM series primers, the DNA fragment was inserted into the KANr Gateway entry vector pDONR201 (Gateway cloning system; Invitrogen, Carlsbad, CA) and transferred into the Gateway destination vector pEXGmGW (58). Deletion construct plasmids introduced into P. aeruginosa were selected for chromosomal insertion on LB agar containing GEN and then counterselected for loss of the plasmid backbone on LB agar containing 5% sucrose (26). To confirm deletions, the region surrounding the target gene was PCR amplified from chromosomal DNA and digested with the restriction endonuclease BamHI (for constructs made using SS series primers) or HindIII (for constructs made using SM series primers) to detect the unique marker.
To construct expression plasmids, the target genes were PCR amplified from genomic DNA by using specific oligonucleotide primers (see Table S3 in the supplemental material), inserted into pDONR201, and transferred to pJN105D (GENr), a Gateway destination version of the l-arabinose-inducible broad-host-range expression plasmid pJN105 (44). This plasmid is derived from pBBR1, which has a copy number of 30 to 40 plasmids per cell in E. coli and Bordetella bronchiseptica (4). Expression plasmids introduced into P. aeruginosa were selected on LB agar containing GEN.
Sequencing and expression of mutant phoPQ alleles from Pmr clinical isolates were performed as described previously (39).
Transcriptional analysis.
A P. aeruginosa reporter strain for transcriptional analysis was constructed in PAO1 (B. Iglewski). A lacZ fusion to the promoter of the arnBCADTEF-pmrE operon (PA3552 to PA3559 in the PAO1 genome) (57) was constructed using the pMini-CTX::lacZ vector system (27). The region upstream of arnBCADTEF-pmrE was amplified using specific oligonucleotide primers (see Table S4 in the supplemental material) and inserted into pMini-CTX::lacZ by using EcoRI and BamHI restriction sites. The fusion was integrated into the CTX site of the chromosome, followed by removal of the plasmid backbone by use of Flp recombinase. To confirm the reporter construct, the CTX region was PCR amplified from chromosomal DNA. Reporter strains carrying expression plasmids were grown for 16 to 20 h at 37°C with aeration in LB broth supplemented with 1 mM MgCl2 and GEN. Cultures were diluted 1:100 in fresh medium supplemented with 0.1% arabinose, grown for 90 min, and assayed for β-galactosidase activity (31).
Lipid A isolation and analysis.
LPS was isolated from bacterial cells by using a rapid phenol-guanidinium thiocyanate disruption method after growth in LB broth supplemented with 1 mM MgCl2 (60). When this method failed to give clean mass spectra for specific strains (e.g., PA1133), a more labor-intensive LPS isolation method was used (13), followed by repeated extraction of the LPS with an equal volume of 2:1 chloroform-methanol (centrifuged at 10,000 × g for 10 min). Lipid A was isolated from LPS by hydrolysis (8). The lipid A structure was analyzed using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry in negative-ion mode (15). All MALDI-TOF analyses were performed on a Bruker Autoflex II mass spectrometer (Bruker Daltonics). The matrix used for lipid A analysis was 5-chloro-2-mercaptobenzothiazole (20 mg/liter in 1:1 chloroform-methanol).
Serial in vitro passage of Pm-resistant P. aeruginosa clinical isolates.
Serial passages were carried out in the absence of antibiotic selection pressure to assess the phenotypic stability of pmrB mutants. These were initiated from mixed-stock inocula; the frozen stock was thawed briefly at 25°C, and 10 μl was transferred into 5 ml LB broth supplemented with 1 mM MgCl2. Each day zero (D0) culture was grown overnight at 37°C with aeration and used to inoculate a D1 culture by transferring 0.5 μl into 5 ml fresh LB broth supplemented with 1 mM MgCl2 but lacking PMB. An aliquot of the D0 culture was stored at −80°C after adding glycerol to a final concentration of 16%. Each culture was serially passaged every 24 h for 15 days. Aliquots of the D5, D10, and D15 cultures were stored similarly and tested for Pm susceptibility using the PMB plate assay.
Statistical analysis.
To analyze the effects of pmrB mutant alleles on arnB transcription and to assess associations between Pm resistance and the absence, presence, or predominance of specific lipid A modifications, two-sided P values were calculated using the one-sample t test and Fisher's exact test, respectively.
Nucleotide sequence accession numbers.
The new DNA sequences have been deposited in GenBank under accession no. JQ340359 to JQ340368.
RESULTS
Expression and sequencing of pmrAB alleles from highly Pm-resistant P. aeruginosa isolates infecting European cystic fibrosis patients.
We previously showed that point mutations in pmrB confer Pm resistance on a laboratory strain of P. aeruginosa (42). To explore the role of the pmrAB locus in clinical Pm resistance, we analyzed highly Pmr P. aeruginosa clinical isolates from 22 CF patients: 6 patients at clinical sites in the United Kingdom (Leeds and an anonymous site) (14) and 16 patients at clinical sites in Denmark (Copenhagen and Aarhus) (29). The Danish patients were part of a larger clinical cohort that had been treated continuously with twice-daily inhalation of CST methanesulfonate for a median of 10 years (range, 5 to 15 years) at the time that Pmr isolates were first identified in the late 1990s and for a median of 17 years (range, 10 to 21 years) at the time of a second outbreak in 2004. No decline in respiratory status was noted at the time of the initial outbreak in 1995; however, by the time of the second outbreak, patients had experienced a significant decline in lung function, with the clinical impression that they were no longer responding to inhaled CST (29). In the wake of each outbreak, CST inhalation was discontinued, and the patients were instead treated with other inhaled antibiotics; however, as of 2007, none of the patients had cleared their Pmr isolates.
The pmrAB allele from an index isolate for each patient was inserted into the broad-host-range plasmid pJN105D and expressed in PAK wild-type (WT) and PAK ΔpmrAB strain backgrounds; the WT pmrAB allele from PAK was also inserted into pJN105D, as a negative control. PMB agar plate testing of these pmrAB allelic expression strains defined 11 (50%) of the Pmr index isolates (4 from the United Kingdom and 7 from Denmark) as putative pmrAB mutants (data not shown). Colistin agar dilution testing of the pmrAB allelic expression strains revealed a lower level of Pm resistance than that of the corresponding clinical pmrAB mutants (Table 2). The allelic expression plasmids conferred Pm resistance in the ΔpmrAB strain background that was 2 to 32 times greater than that in the WT background.
Table 2.
Pm resistance conferred by episomal expression of pmrB alleles
| Strain or isolate | MLST type | CST MICa (mg/liter) | pmrB allele | CST MIC (mg/liter) conferred by pmrABb |
PmrB amino acid change(s) | |
|---|---|---|---|---|---|---|
| PAK | PAK ΔpmrAB | |||||
| PAK | 0.5 | pmrB+ | 2 | 2 | ||
| PAK pmrB6 | 32 | pmrB6 | 8 | 128 | L243Q | |
| PAK pmrB12 | 32 | pmrB12 | 32 | 256 | A248V | |
| PA1109 | ST-148 | >512 | pmrB21 | 64 | 256 | L14P/P456S |
| PA1125 | ST-388 | 256 | pmrB45 | 4 | 16 | D45 deletion |
| PA1131 | ST-389 | >512 | pmrB42 | 2 | 2 | A95T/T253M |
| PA1133 | ST-379 | >512 | pmrB22 | 8 | 128 | S257N |
| PA1015 | ST-386 | >512 | pmrB41 | 4 | 128 | R57H/A248T |
| PA1017 | ST-387 | >512 | pmrB23 | 8 | 256 | R79H/R259H |
| PA1603 | ST-398 | >512 | pmrB34 | 2 | 4 | A248T |
| PA1611 | ST-387 | >512 | pmrB26 | 16 | 256 | A54V/A248T |
| PA2047 | ST-387 | 1 | pmrB+ | 1 | 1 | |
| PA1016 | ST-387 | >512 | pmrB23 | 8 | 256 | R79H/R259H |
| PA2050 | ST-387 | 0.5 | pmrB+ | 1 | 1 | |
| PA1020 | ST-387 | >512 | pmrB24 | 8 | 256 | R135Q/M292I |
| PA1575 | ST-387 | 1 | pmrB+ | 1 | 1 | |
| PA1571 | ST-387 | >512 | pmrB25 | 16 | 256 | G188D |
Measured by agar dilution assay.
Alleles were inserted in pJN105D and expressed in the indicated strain backgrounds. CST MICs were measured by an agar dilution assay that included 0.1% l-arabinose to induce expression of the episomal pmrAB allele.
Sequencing of mutant pmrAB alleles revealed three alleles with 1-bp transitions or transversions resulting in single missense mutations in the PmrB sensor kinase, six alleles with double point mutations resulting in two missense mutations in the same PmrB molecule, and one allele with a 3-bp deletion resulting in loss of Asp 45 from PmrB (Table 2). No pmrA mutations resulting in amino acid changes were observed among resistance-conferring pmrAB alleles. Isolate PA1131 contains a non-resistance-conferring pmrAB mutant allele; this isolate has a single PmrA missense mutant allele (Arg 71 to Leu), designated pmrA2, as well as a double missense mutant allele, designated pmrB42 (Table 2).
Genotypes and phenotypes of clinical pmrAB mutants.
Analysis of chromosomal DNAs from the 11 clinical pmrAB mutants by pulsed-field gel electrophoresis indicated four distinct pulsotypes among the British isolates and three distinct pulsotypes among the Danish isolates (14), a finding confirmed by MLST (Table 2). Three of the Danish patients who were infected with pmrAB mutants had at least one isogenic Pms isolate from an earlier culture that was also available for analysis; a set of 37 Pms and 3 Pmi P. aeruginosa isolates from 23 CF patients at a U.S. clinical site served as additional controls (Table 1).
For 10 of the clinical pmrAB mutants, the CST agar dilution MIC was >512 mg/liter, in contrast to the 3 preceding Pms isogenic isolates, for which the MIC was ≤1 mg/liter (Table 2). Killing assays with the cationic antimicrobial peptides C18G and protegrin indicated cross-resistance of clinical pmrAB mutants compared to their isogenic Pms precursors (Table 3), as was also seen for the Pmr mutants PAK pmrB6 and PAK pmrB12 (42).
Table 3.
Cationic antimicrobial peptide resistance of isogenic CF isolatesa
| Strain or isolate pair (Pmr vs Pms) | PMB EC50 (mg/liter) |
FIR | C18G EC50 (mg/liter) |
FIR | Protegrin-1 EC50 (mg/liter) |
FIR | |||
|---|---|---|---|---|---|---|---|---|---|
| R strain | S strain | R strain | S strain | R strain | S strain | ||||
| PAK pmrB12 vs PAK | 8 | <0.5 | >16 | 128 | 2 | 64 | 6 | 0.125 | 48 |
| PA1016 vs PA2047 | 512 | <0.5 | >1,024 | 64 | 3 | 21 | 8 | 0.125 | 64 |
| PA1020 vs PA2050 | 512 | <0.5 | >1,024 | 32 | 4 | 8 | 12 | 0.125 | 96 |
| PA1571 vs PA1575 | 512 | <0.5 | >1,024 | 16 | 6 | 3 | 2 | 0.25 | 8 |
EC50, effective concentration that kills 50% of CFU; FIR, fold increase in resistance of Pmr (R) strain with respect to Pms (S) strain (ratio of EC50s).
The clinical pmrAB mutants displayed a small-colony phenotype, requiring a minimum of 48 h to form pinpoint nonmucoid colonies on LB agar. Several of the Pms CF isolates of P. aeruginosa that were studied as controls also displayed a small-colony phenotype, indicating that growth retardation per se does not account for the Pmr phenotype of the clinical pmrAB mutants. Of the seven clinical pmrAB mutants for which motility phenotypes were assessed, four displayed swimming and three displayed swarming, reflecting prevalence rates for these motility phenotypes that are typical of chronic CF isolates of P. aeruginosa (37).
Structural distribution and transcriptional activity of pmrAB alleles from highly Pmr clinical isolates.
The nonsilent mutations found in the pmrB alleles were clustered with respect to the predicted domain structure of PmrB (Fig. 1). One mutation localized to the cytosol immediately N-terminal to the first transmembrane (TM1) domain, six mutations localized to the periplasmic domain, one mutation localized to the HAMP domain, and five mutations localized to the four-helix bundle dimerization and phosphotransferase (DHP) domain; notably, the ATP binding domain was free of nonsilent mutations. One obvious pattern was the cooccurrence of periplasmic and DHP point mutations, seen in five double mutant alleles (Table 2). For four of these, expression of recombinant pmrB alleles containing only one of the two mutations conferred low-level Pm resistance in the ΔpmrAB strain background (Table 4). In contrast, chimeric alleles representing novel combinations of periplasmic and DHP point mutations conferred moderate-level Pm resistance.
Fig 1.

PmrB mutations in relation to predicted domain structure. The diagram shows the secretion signal (SS; amino acids [aa] 1 to 14), first transmembrane domain (TM1; aa 15 to 37), periplasmic domain (aa 38 to 160), second transmembrane domain (TM2; aa 161 to 183), HAMP linker domain (HAMP; aa 186 to 238), four-helix bundle dimerization and phosphotransferase domain (DHP; aa 239 to 304) (the active site [His 249] is marked with an asterisk), and C-terminal ATP binding domain (ATPB; aa 300 to 459), based on the SMART protein database (32). Symbols: pentagons, pmrB21 allele; hexagon, pmrB22 allele; circles, pmrB23 allele; squares, pmrB24 allele; diamond, pmrB25 allele; upright triangle and Star of David, pmrB26 allele; Star of David, pmrB34 allele; inverted triangle and Star of David, pmrB41 allele; crosses, pmrB42 allele; arrow, pmrB45 allele.
Table 4.
Recombination analysis of multimutant pmrB alleles
| Recombinant allele | Parent allele(s) | PmrB amino acid change(s) | CST MIC (mg/liter) conferred by recombinant allelea |
|---|---|---|---|
| pmrB27 | pmrB21 | L14P | 128 |
| pmrB28 | pmrB21 | P456S | 1 |
| pmrB29 | pmrB23 | R79H | 2 |
| pmrB30 | pmrB23 | R259H | 2 |
| pmrB31 | pmrB24 | R135Q | 2 |
| pmrB32 | pmrB24 | M292I | 8 |
| pmrB33 | pmrB26 | A54V | 4 |
| pmrB34 | pmrB26 | A248T | 4 |
| pmrB35 | pmrB23/pmrB26 | R79H/A248T | 256 |
| pmrB36 | pmrB26/pmrB23 | A54V/R259H | 256 |
| pmrB37 | pmrB24/pmrB23 | R135Q/R259H | 128 |
| pmrB38 | pmrB23/pmrB24 | R79H/M292I | 256 |
| pmrB39 | pmrB24/pmrB26 | R135Q/A248T | 128 |
| pmrB40 | pmrB26/pmrB24 | A54V/M292I | 256 |
Alleles were inserted into pJN105D and expressed in the P. aeruginosa PAK ΔpmrAB strain background. CST MICs were measured in an agar dilution assay that included 0.1% l-arabinose.
Two pmrB double mutant alleles represented exceptions to this pattern. The pmrB27 single mutant allele (L14P missense mutation), derived from a pmrB21 double mutant allele (isolate PA1109) that also has a P456S missense mutation, conferred moderate-level Pm resistance by itself (Table 4). In contrast, a pmrB42 double mutant allele (isolate PA1131) that has periplasmic (A95T) and DHP (T253M) missense mutations did not confer Pm resistance, regardless of whether it was expressed with its native pmrA2 mutant allele (R71L missense mutation) or a pmrA WT allele (data not shown). Coexpression of a pmrB12 or pmrB23 mutant allele with the pmrA2 mutant allele neither augmented nor diminished Pm resistance compared to coexpression with a pmrA WT allele, confirming that pmrA2 is not a resistance-conferring allele and does not act as a secondary suppressor. However, the pmrB12 mutant allele failed to confer Pm resistance in the absence of a pmrA allele (data not shown), indicating that PmrB-mediated Pm resistance is PmrA dependent.
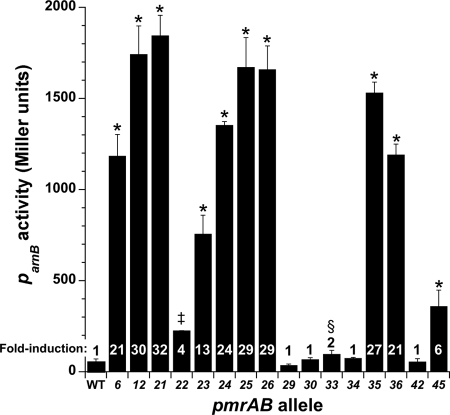
Transcriptional analysis of pmrAB allele activity, using an arnBp-lacZ transcriptional fusion as a reporter, showed that pmrAB alleles conferring low- or moderate-level Pm resistance (CST MIC of 8 to 256 mg/liter) in a ΔpmrAB background (Tables 2 and 4) increased transcription from the arnB promoter 4- to 32-fold (Fig. 2). In contrast, pmrB alleles that were associated with a Pms or Pmi phenotype induced ≤2-fold increases in arnB transcription.
Fig 2.
Transcriptional activities of WT and mutant pmrAB alleles. Alleles were expressed from pJN105D in reporter strain PAO1 ΔpmrAB Ω(attP::Φ(ParnB-lacZ+)). Fold induction is shown relative to the WT pmrAB allele. Data shown represent means for biological triplicates. Error bars show standard deviations (SD). Two-sided t tests with the WT pmrAB allele as a comparator gave P values as indicated: *, P < 0.0005; ‡, P < 0.005; and §, P < 0.01. Where not indicated, P values were >0.05.
Lipid A modifications of highly Pmr clinical pmrB mutants of P. aeruginosa.
Mutations in pmrB that cause Pm resistance do so at least in part by stimulating constitutive addition of l-Ara4N to lipid A (42). To assess the presence or absence of this and other lipid A modifications (Fig. 3A), we used MALDI-TOF mass spectrometry to analyze lipid A from Pmr and Pms P. aeruginosa strains qualitatively.
Fig 3.
Lipid A structures of clinical P. aeruginosa pmrB mutants. (A) Diagram of P. aeruginosa lipid A structure. Covalent modifications include (i) acyl-oxy-acyl addition of laurate (C12:0) (blue) to 3-hydroxylaurate (3OH-C12:0) at the 2 and 2′ positions, by HtrB1 and HtrB2; (ii) 2-hydroxylation converting acyl-oxy-acyl C12:0 to 2OH-C12:0, by LpxO1 and LpxO2; (iii) removal of 3-hydroxydecanoate (3OH-C10:0) (pink) from the 3 position, by PagL; (iv) acyl-oxy-acyl addition of palmitate (C16:0) (green) to 3OH-C10:0 at the 3′ position, by PagP; and (v) addition of 4-amino-l-arabinose (l-Ara4N) (red) to the 1 and 4′ phosphates, by ArnT. (B to E) MALDI-TOF mass spectra. (B) WT strain PAK. The circled peak at m/z 1,617 corresponds to the hexa-acyl lipid A structure at left. (C) Strain PA1611. Circled peaks at m/z 1,748 and 1,855 reflect addition of l-Ara4N and C16:0, with the latter creating hepta-acyl lipid A, and the circled peak at m/z 1,986 corresponds to the hepta-acyl lipid A structure at left with l-Ara4N. (D) Strain PA1571. The circled peak at m/z 1,431 reflects deacylation of 3OH-C10:0 from the 3 position to create penta-acyl lipid A, and the circled peak at m/z 1,562 corresponds to the penta-acyl lipid A structure at left with l-Ara4N. (E) Strain PA1109. The circled peak at m/z 1,693 corresponds to the penta-acyl lipid A structure at left with two l-Ara4N moieties.
The mass spectrum for lipid A from the PAK WT strain had major peaks at mass/charge ratios (m/z) of 1,617 and 1,447 (Fig. 3B), corresponding to hexa- and penta-acylated lipid A species differing in the presence or absence of 3-hydroxydecanoate (Δm/z = −170). Minor peaks seen at m/z 1,633 and 1,463 differ from the major peaks by the 2-hydroxylation of the second laurate (Δm/z = +16). Additional minor peaks differ from the peaks listed above by dephosphorylation at the 1 or 4′ position of lipid A (Δm/z = −80).
The mass spectrum for lipid A from Pmr clinical isolate PA1611 had a major peak at m/z 1,617 but lacked a peak at m/z 1,447 (Fig. 3C), indicating a loss of the lipid A deacylase PagL. It also had a minor peak at m/z 1,601, corresponding to hexa-acylated lipid A lacking 2-hydroxylation of laurate (Δm/z = −16); a second major peak at m/z 1,855, corresponding to hepta-acylated lipid A containing palmitate (Δm/z = +238); minor peaks at m/z 1,732, 1,748, and 1,986, corresponding to addition of l-Ara4N (Δm/z = +131) to the structures at m/z 1,601, 1,617, and 1,855, respectively; and a third major peak at m/z 1,879, corresponding to addition of a second l-Ara4N moiety to the structure at m/z 1,748 (Fig. 3C).
The mass spectrum for lipid A from Pmr clinical isolate PA1571 had a major peak at m/z 1,431 (Fig. 3D), corresponding to a penta-acylated lipid A lacking 2-hydroxylation of laurate, but only a minor peak at m/z 1,601 and no peak at m/z 1,617, indicating strong lipid A deacylase activity. It also had minor peaks at m/z 1,562 and 1,693, corresponding to addition of one or two l-Ara4N moieties to the structure at m/z 1,431, and a minor peak at m/z 1,669, corresponding to addition of palmitate to the structure at m/z 1,431 (Fig. 3D). The mass spectrum for lipid A from Pmr clinical isolate PA1109 was similar to that for PA1571, except that no peak was seen at m/z 1,669 (Fig. 3E), indicating the absence of palmitoylation.
The mass spectrum for lipid A from Pmr clinical isolate PA1015 had a major peak at m/z 1,419, corresponding to a penta-acylated lipid A that is lipid IVA with a single acyl-oxy-acyl laurate. To this main peak, masses corresponding to additions of one or two l-Ara4N, palmitate, or l-Ara4N plus palmitate moieties were observed (data not shown). A minor peak corresponding to deacylation to form a tetra-acylated lipid A without and with l-Ara4N was also observed. The mass spectrum for lipid A from Pmr clinical isolate PA1133 was similar to those for both PA1571 and PA1015, in that it had peaks corresponding to penta-acylated lipid A without and with l-Ara4N addition, as did that for PA1571, and also had peaks corresponding to tetra-acylated lipid A without and with l-Ara4N addition, as did that for PA1015 (data not shown).
Mass spectra for lipid A from P. aeruginosa strains expressing resistance-conferring pmrAB alleles (i.e., single, double, or chimeric mutants) from the pJN105D plasmid in a PAK background had consistent minor peaks at m/z 1,748 and 1,764, corresponding to addition of l-Ara4N to the structures at m/z 1,617 and 1,633 (data not shown).
In light of these patterns of lipid A structural change, associations between Pm resistance and specific lipid A modifications were assessed for the 10 Pmr CF isolates with evaluable structural data compared to 40 Pms and 3 Pmi CF isolates, which served as controls (Table 5). An absolute correlation with the presence of l-Ara4N was observed (P = 5 × 10−11), as well as a very strong correlation with a lack of laurate 2-hydroxylation (P = 9 × 10−5). Pm resistance did not correlate with palmitoylation (40% versus 58%; P = 0.48) or with deacylation (P = 0.57), which occurred at a high prevalence in both Pmr and Pms CF isolates.
Table 5.
Association of specific lipid A modifications with Pm resistance of CF isolates
| Isolate group | n | No. (%) of isolatesc |
|||
|---|---|---|---|---|---|
| Aminoarabinose added to any lipid A | 2-Hydroxylation of laurate in predominant lipid A | Palmitoylation of any lipid A | Deacylation of any lipid A (removal of 3OH-C10:0) | ||
| Pmr isolatesa | 10 | 10 (100) | 1 (10) | 4 (40) | 10 (100) |
| Pms and Pmi isolatesb | 43 | 0 (0) | 34 (79) | 25 (58) | 38 (88) |
All Pmr clinical isolates listed in Table 1, except for PA1131.
Forty Pms clinical isolates and 3 Pmi clinical isolates (PA2123, PA2128, and PA2140), as listed in Table 1.
Two-sided P values for aminoarabinose, 2-hydroxylation of laurate, palmitoylation, and deacylation were 5 × 10−11, 9 × 10−5, 0.48, and 0.57, respectively.
Prolonged passage of highly Pm-resistant pmrB mutants without antibiotic selection pressure.
Although spontaneous Pmr mutants such as PAK pmrB12 had stable resistance after two passages through LB broth lacking PMB (42), it was subsequently noted that further passaging of such strains resulted in complete reversion to susceptibility (Fig. 4A). Some clinical pmrAB mutants showed complete phenotypic stability (isolate PA1109) (Fig. 4B), whereas others showed slow (isolate PA1016) (Fig. 4C) or rapid (isolate PA1020) (Fig. 4D) phenotypic reversion (i.e., loss of Pm resistance) with progressive steps of passage without antibiotic selection pressure. In contrast to the slow reversion of PA1016, another clinical isolate with the same pmrAB allele (PA1017) showed complete phenotypic stability at day 15 (data not shown), suggesting that the pmrAB locus does not represent a specific determinant of this stability.
Fig 4.
Stability of Pm resistance phenotypes after prolonged passage without antibiotic selection pressure. PMB agar plate assays were performed with day 0 cultures (triangles), day 5 cultures (circles), day 10 cultures (squares), and day 15 cultures (open diamonds) for the initially Pmr P. aeruginosa laboratory strain and clinical isolates passaged for 15 days in LB broth without Pm. (A) PAK pmrB12 mutant; (B) P. aeruginosa PA1109 (pmrB21 mutant); (C) P. aeruginosa PA1016 (pmrB23 mutant); (D) P. aeruginosa PA1020 (pmrB24 mutant). Error bars show SD. Each passage was repeated on a separate occasion with similar results.
Mass spectra for lipid A from passaged clinical isolates that lost Pm resistance varied in the absence or presence of peaks corresponding to the addition of l-Ara4N; day 15 passages of isolates PA1016 and PA1020 lacked such peaks, whereas day 15 passages of isolates PA1571 and PA1133 retained them (data not shown). This suggests that at least some of the secondary suppressor mutations that lead to phenotypic reversion occur in loci that do not affect the process through which l-Ara4N is added to lipid A.
Deletion of pmrAB or arnC in PA1016, an electrocompetent CF isolate with high-level Pm resistance.
Many Pmr clinical isolates of P. aeruginosa were not amenable to genetic manipulation because they could not be conjugated or electroporated efficiently (data not shown). In contrast, we found that isolate PA1016 could reliably be rendered electrocompetent (and thus amenable to genetic manipulation) by using standard methods (9), enabling us to assess whether its Pm resistance was dependent on its mutant pmrAB allele and on genes within the l-Ara4N biosynthetic operon.
Isolate PA1016 displayed high-level Pm resistance in a PMB killing assay (Fig. 5A and B). In contrast, a version of this isolate in which the pmrAB locus had been deleted was Pm susceptible (Fig. 5A). Expression of a plasmid (pJN105D) containing pmrAB23 (i.e., the pmrAB allele from PA1016) in the PA1016 ΔpmrAB mutant strain resulted in partial complementation; expression of a WT pmrAB allele did not. As expected, the mass spectrum for lipid A from PA1016 ΔpmrAB/pJN105D::pmrB23 had peaks corresponding to the addition of l-Ara4N, whereas lipid A from PA1016 ΔpmrAB lacked these peaks (data not shown).
Fig 5.
Pm resistance gene deletion and complementation in Pmr isolate PA1016 and Pms strain PAK. (A) PMB killing assay of isolate PA1016 (filled circles), PA1016 ΔpmrAB (open squares), PA1016 ΔpmrAB(pJN105D::pmrAB+) (open triangles), and PA1016 ΔpmrAB(pJN105D::pmrAB23) (filled diamonds). (B) PMB killing assay of isolate PA1016, PA1016 ΔarnC (open squares), and PA1016 ΔarnC(pJN105D::arnC) (filled diamonds). (C) PMB plate assay ofPAK(pJN105D::pmrAB12) (filled circles), PAK ΔarnC (open squares), PAK ΔarnC(pJN105D::pmrAB12) (open triangles), and PAK ΔarnC(pJN105D::pmrAB12-arnC) (filled diamonds). The plots reflect combined results for assays performed twice in triplicate. Error bars show SD.
Deletion of the arnC gene, encoding a putative glycosyltransferase thought to be essential for transfer of UDP-l-Ara4N to undecaprenyl phosphate (bactoprenol) as part of addition to lipid A (55), rendered isolate PA1016 susceptible to PMB (Fig. 5B). Expression of a plasmid (pJN105D) containing arnC within the PA1016 ΔarnC mutant resulted in very weak complementation. Similar to the result with isolate PA1016, expression of pJN105D::pmrAB12-arnC in a ΔarnC mutant background conferred only weak Pm resistance in the PMB plate assay relative to the resistance conferred by pJN105D::pmrAB12 in a WT background (Fig. 5C). Expression of pJN105D::pmrAB12 in the ΔarnC mutant background conferred a slight increase in Pm resistance over that of the Pms ΔarnC mutant (Fig. 5C).
DISCUSSION
This work establishes point mutations in pmrB as a regulatory mechanism of Pm resistance in CF isolates of P. aeruginosa. The pmrB mutants were cultured from CF patients at clinical centers in Denmark and the United Kingdom, where inhaled CST is often used as a long-term treatment for chronic airway infection (18, 19). Epidemic spread of Pmr P. aeruginosa occurred in Copenhagen, Denmark, and Leeds, United Kingdom (14, 29), representing a considerable concern from an infection control standpoint. However, the pmrB mutants in Copenhagen apparently arose sporadically and did not spread from patient to patient; the Copenhagen strain that did spread extensively has a resistance-conferring phoQ mutation (39). Four of the pmrB mutants from Denmark also have mutant phoPQ alleles: PA1603 has phoP24Q24, PA1571 has phoQ25, and PA1016 and PA1017 have phoQ30. However, these native phoPQ alleles appear not to confer Pm resistance (39).
All three Pmr clinical strains in Leeds are pmrB mutants; however, only one of the Leeds pmrB alleles (pmrB22) confers moderate-level Pm resistance. The isolate that carries this allele (PA1133) also has a mutant phoQ allele that appears not to confer Pm resistance (S. M. Moskowitz, M. K. Brannon, and A. K. Miller, unpublished results). The Leeds strain that spread from patient to patient (represented here by isolate PA1131) has a mutant pmrB allele (pmrB42) that does not confer Pm resistance or induce arnB transcription in a ΔpmrAB background. It also has a mutant phoPQ allele that appears not to confer Pm resistance (Moskowitz et al., unpublished results); thus, the resistance-conferring genetic determinant(s) in this strain has not been defined.
In contrast to Salmonella enterica, in which a prototypical resistance-conferring mutation occurred in the cognate response regulator PmrA (50), the few PmrA mutations that we have found in P. aeruginosa, exemplified by the pmrA2 allele in isolate PA1131, neither suppress nor augment Pm resistance in a pmrB “gain-of-resistance” mutant background. We have also sequenced the promoter region of the arnB operon in several highly Pmr clinical isolates without defining any mutations that could plausibly contribute to Pm resistance (S. M. Moskowitz and M. K. Brannon, unpublished results).
Mutations in pmrB similar or identical to some of those described here have been reported for non-CF clinical isolates of P. aeruginosa with low- to moderate-level Pm resistance. For example, the recombinant allele that we have designated pmrB32 contains a mutation that changes Met 292 to Ile and confers low-level Pm resistance when expressed in a ΔpmrAB strain background; the same residue was mutated to Thr in a non-CF clinical isolate from New York with a PMB MIC of 8 mg/liter (1). The mutant allele designated pmrB34 contains a mutation that changes Ala 248 to Thr and confers low-level Pm resistance; the same residue was mutated to Val in a laboratory mutant with moderate Pm resistance (42), and the adjacent residue (Ala 247) was mutated to Thr in a different non-CF isolate from New York with a PMB MIC of 8 mg/liter (5). The mutant allele designated pmrB45, which was found in a moderately Pmr CF isolate from the United Kingdom, also occurred in a Brazilian non-CF clinical isolate with a CST MIC of 32 mg/liter (51). These data suggest that mutations in specific residues of PmrB, particularly in the DHP domain and its catalytic H box, can represent first-step events that confer Pm resistance in P. aeruginosa.
Several mutant pmrB alleles found in highly Pmr CF isolates had two nonsilent mutations, one in the periplasmic domain and another in the DHP domain. These double mutant alleles conferred moderate Pm resistance (CST MIC of 128 to 256 mg/liter) in a ΔpmrAB background. Recombinant pmrB alleles with only one of the two mutations conferred much less Pm resistance, in most cases only 1 to 2 dilutions above that conferred by a WT allele, indicating that periplasmic and DHP missense mutations have synergistic effects resulting in marked activation of the PmrAB system.
Interestingly, recombinant double mutant pmrB alleles (i.e., novel pairs of periplasmic and DHP mutations) conferred moderate levels of Pm resistance similar to the levels observed for the original double mutant alleles. This suggests that any of the specific point mutations observed in the periplasmic domain can act as a second-step event in a DHP domain mutant, altering the periplasmic domain conformation such that kinase activity is enhanced and/or phosphatase activity is inhibited in the altered DHP domain. For example, in the pmrB34 allele, an Ala 248-to-Thr mutation in the DHP domain appears as a first-step PmrB mutation. In the pmrB26 and pmrB41 alleles, this DHP domain mutation is combined with a periplasmic domain mutation (Ala 54 to Val and Arg 57 to His, respectively), representing a second mutational step.
The allelic expression plasmids conferred Pm resistance that was 2 to 32 times higher in a ΔpmrAB strain background than in a WT background. Similarly, Abraham and Kwon observed that episomal expression of WT pmrAB suppresses the Pm resistance of the pmrB M292T allele (1). These observations are consistent with the hypothesis that expression of a WT pmrAB allele can partially or completely suppress the phenotypic effects of a mutant allele, regardless of whether the expression of either is chromosomal or episomal. This is probably a functional consequence of PmrB dimerization: whereas mutant-mutant homodimers likely lack PmrB phosphatase activity, the mutant-WT heterodimers formed when both alleles are expressed (49) would be expected to preserve some PmrB phosphatase activity and thus limit the degree of Pm resistance conferred.
However, even in a ΔpmrAB strain background, the Pm resistance of pmrAB allelic expression strains was less than that of the corresponding Pmr clinical isolates in which the mutant pmrAB alleles were identified. One possible explanation is that the PmrAB system is autoregulated (i.e., is subject to transcriptional positive feedback) under the control of its native promoter (as in the clinical isolates); this positive feedback may be enhanced further by induction of mutant PmrB in the presence of Pm (38). Mutations in additional loci, such as genes that influence the structure of the lipid A and core oligosaccharide moieties of LPS, may further augment Pm resistance in the clinical isolates.
A wide variety of structural modifications can be seen in lipid A from CF isolates of P. aeruginosa (41), and most, if not all, of these variations were observed among the highly Pmr isolates that were studied here. The patterns of lipid A modification among the Pmr isolates did not correlate with specific pmrB alleles. However, for unpassaged pmrB mutants and their corresponding Pms controls, we observed an absolute correlation between l-Ara4N addition to lipid A and clinical Pm resistance. Upon passage, some of the pmrB mutants lost Pm resistance but maintained l-Ara4N addition. Taken together, these observations suggest that l-Ara4N addition is necessary but not sufficient for Pm resistance. As previously observed in a preliminary analysis (40), loss of lauroyl 2-hydroxylation also correlated strongly with clinical Pm resistance. It is not known whether the dephosphorylated minor species frequently observed in lipid A analyses actually contribute to Pm resistance or are merely a technical artifact of lipid A preparation.
In addition to demonstrating variations in lipid A structure, we showed that some clinical pmrB mutants of P. aeruginosa have a relatively stable Pm resistance phenotype even after 15 days of passage without antibiotic selection pressure, while others may completely lose resistance within 5 to 10 days. Thus, in some instances, pmrB mutation appears to impose fitness costs, such that mutants are outcompeted by revertants (or WT strains) when both are present in an ecological niche or in vitro culture without Pm selection pressure. Such phenotypic instability has also been observed for some clinical phoQ mutants (S. M. Moskowitz and M. Pier, unpublished results). This loss of resistance suggests the emergence of secondary suppressor mutations. In clinical phoQ mutants, phoP mutations may act as partial or complete secondary suppressors of Pm resistance (39). We also observed phenotypic instability while analyzing Pmr strains through targeted gene deletions, which are typically constructed in the absence of Pm selection pressure. Loss of Pm resistance in a targeted gene deletion mutant that episomal expression fails to complement can sometimes be attributable to emergence of secondary suppressor mutations during strain construction. As a consequence, we have adopted the practice of first constructing targeted gene deletions in a WT strain background and then introducing the Pm resistance-conferring allele or mutation via deletion or allelic replacement as the last step prior to strain testing.
Abraham and Kwon showed that episomal expression of arnBCADTEF completely complements the genetic disruption of arnB in a mildly Pmr clinical pmrB mutant (1). In contrast, although we were able to show through construction of clean deletions in CF isolate PA1016 that its Pm resistance is dependent on pmrAB and arnC, episomal expression of the native alleles (pmrAB23 and arnC, respectively) only partially restored Pm resistance. Similar partial complementation was also observed in a WT strain background. This implies that inducible episomal expression of ArnC is not as robust as its native chromosomal expression, limiting Pm resistance under the conditions used here to assess this phenotype.
In summary, alterations in the PmrAB system caused by specific mutations in the pmrB gene represent an important but nonexclusive regulatory mechanism of clinical Pm resistance. Such mutations can cause low to moderate Pm resistance but also contribute to high-level resistance. Specific residues in the periplasmic and DHP domains of PmrB are mutated repeatedly in Pmr strains and appear to act synergistically to confer markedly increased resistance on periplasmic-DHP domain double mutants. Both single and double mutant pmrB alleles drive expression of the arnB operon, resulting in l-Ara4N addition to lipid A as an important biochemical mechanism of Pm resistance. Additional changes in LPS, such as loss of lauroyl 2-hydroxylation in lipid A, are associated with high-level Pm resistance, though a causative role is not yet established. Conversely, some highly Pmr isolates gradually lose resistance when passaged repeatedly without antibiotic selection pressure. Such loss of resistance does not consistently correlate with loss of l-Ara4N addition, because some secondary suppressor mutations evidently interfere with resistance in ways that leave this lipid A modification intact. Defining such secondary suppressors is likely to provide new insights into the regulatory and biochemical mechanisms that contribute to high-level Pm resistance in P. aeruginosa.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lillian Yeung, Laurel Stevens, Ulla Johansen, Pia Poss, Helle Nordbjerg, Kristin Adams, Bob Ernst, Jessica Foster, and Jane Burns for technical assistance, Dutch VanDevanter for providing clinical isolate PA1109, and Alina Gutu and Sun Ho Kim for reviewing drafts of the manuscript.
This work was supported by Public Health Service grant K08HL067903 to S.M.M. from the National Heart, Lung, and Blood Institute, grant R01AI067653 to S.M.M. from the National Institute of Allergy and Infectious Diseases, and grant R01AI030479 to S.I.M. from the National Institute of Allergy and Infectious Diseases. This work was also supported by grant MOSKOW01A1 to S.M.M. from the CF Foundation.
Footnotes
Published ahead of print 21 November 2011
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Abraham N, Kwon DH. 2009. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 298: 249–254 [DOI] [PubMed] [Google Scholar]
- 2. Adams MD, et al. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53: 3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alonso A, Campanario E, Martinez JL. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145: 2857–2862 [DOI] [PubMed] [Google Scholar]
- 4. Antoine R, Locht C. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6: 1785–1799 [DOI] [PubMed] [Google Scholar]
- 5. Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53: 5150–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boll M, Radziejewska-Lebrecht J, Warth C, Krajewska-Pietrasik D, Mayer H. 1994. 4-Amino-4-deoxy-l-arabinose in LPS of enterobacterial R-mutants and its possible role for their polymyxin reactivity. FEMS Immunol. Med. Microbiol. 8: 329–341 [DOI] [PubMed] [Google Scholar]
- 7. Brannon MK, et al. 2005. Colistin-treated cystic fibrosis patients harbor polymyxin-resistant strains of Pseudomonas aeruginosa, some with mutation of PmrAB, a two-component regulator of lipid A structure. Pediatr. Pulmonol. Suppl. 28: 289 [Google Scholar]
- 8. Caroff M, Tacken A, Szabo L. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175: 273–282 [DOI] [PubMed] [Google Scholar]
- 9. Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64: 391–397 [DOI] [PubMed] [Google Scholar]
- 10. Ciofu O, Giwercman B, Pedersen SS, Høiby N. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS 102: 674–680 [PubMed] [Google Scholar]
- 11. Clinical Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. M7-A7. CLSI, Wayne, PA [Google Scholar]
- 12. Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42: 5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darveau RP, Hancock RE. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denton M, et al. 2002. Transmission of colistin-resistant Pseudomonas aeruginosa between patients attending a pediatric cystic fibrosis center. Pediatr. Pulmonol. 34: 257–261 [DOI] [PubMed] [Google Scholar]
- 15. Ernst RK, et al. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286: 1561–1565 [DOI] [PubMed] [Google Scholar]
- 16. Falagas ME, Bliziotis IA. 2007. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int. J. Antimicrob. Agents 29: 630–636 [DOI] [PubMed] [Google Scholar]
- 17. Foweraker JE, Laughton CR, Brown DF, Bilton D. 2009. Comparison of methods to test antibiotic combinations against heterogeneous populations of multiresistant Pseudomonas aeruginosa from patients with acute infective exacerbations in cystic fibrosis. Antimicrob. Agents Chemother. 53: 4809–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frederiksen B, Koch C, Høiby N. 1997. Antibiotic treatment of initial colonization with Pseudomonas aeruginosa postpones chronic infection and prevents deterioration of pulmonary function in cystic fibrosis. Pediatr. Pulmonol. 23: 330–335 [DOI] [PubMed] [Google Scholar]
- 19. Frederiksen B, Lanng S, Koch C, Høiby N. 1996. Improved survival in the Danish center-treated cystic fibrosis patients: results of aggressive treatment. Pediatr. Pulmonol. 21: 153–158 [DOI] [PubMed] [Google Scholar]
- 20. Giwercman B, Lambert PA, Rosdahl VT, Shand GH, Høiby N. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed beta-lactamase producing strains. J. Antimicrob. Chemother. 26: 247–259 [DOI] [PubMed] [Google Scholar]
- 21. Gunn JS, et al. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27: 1171–1182 [DOI] [PubMed] [Google Scholar]
- 22. Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68: 6139–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob. Agents Chemother. 52: 351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51: 4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho SN, Horton RM. June 1991. Method for gene splicing by overlap extension using the polymerase chain reaction. US patent 5023171 [PubMed] [Google Scholar]
- 26. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212: 77–86 [DOI] [PubMed] [Google Scholar]
- 27. Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43: 59–72 [DOI] [PubMed] [Google Scholar]
- 28. Jensen T, et al. 1987. Colistin inhalation therapy in cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. J. Antimicrob. Chemother. 19: 831–838 [DOI] [PubMed] [Google Scholar]
- 29. Johansen HK, Moskowitz SM, Ciofu O, Pressler T, Høiby N. 2008. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J. Cyst. Fibros. 7: 391–397 [DOI] [PubMed] [Google Scholar]
- 30. Knowles MR, Boucher RC. 2002. Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Invest. 109: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kulasekara HD, et al. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55: 368–380 [DOI] [PubMed] [Google Scholar]
- 32. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37: D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J, et al. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50: 2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2: 1051–1060 [DOI] [PubMed] [Google Scholar]
- 35. Macfarlane EL, Kwasnicka A, Hancock RE. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146: 2543–2554 [DOI] [PubMed] [Google Scholar]
- 36. Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34: 305–316 [DOI] [PubMed] [Google Scholar]
- 37. Mahenthiralingam E, Campbell ME, Speert DP. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50: 205–217 [DOI] [PubMed] [Google Scholar]
- 39. Miller AK, et al. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 55: 5761–5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moskowitz SM, et al. 2000. Polymyxin resistance and lipid A structure of Pseudomonas aeruginosa isolated from colistin-treated and colistin-naïve cystic fibrosis patients. Pediatr. Pulmonol. Suppl. 20: 272 [Google Scholar]
- 41. Moskowitz SM, Ernst RK. 2010. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell. Biochem. 53: 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186: 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moskowitz SM, et al. 2008. Shifting patterns of inhaled antibiotic use in cystic fibrosis. Pediatr. Pulmonol. 43: 874–881 [DOI] [PubMed] [Google Scholar]
- 44. Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227: 197–203 [DOI] [PubMed] [Google Scholar]
- 45. Nummila K, Kilpelainen I, Zahringer U, Vaara M, Helander IM. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16: 271–278 [DOI] [PubMed] [Google Scholar]
- 46. Pedersen SS, Koch C, Høiby N, Rosendal K. 1986. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J. Antimicrob. Chemother. 17: 505–516 [DOI] [PubMed] [Google Scholar]
- 47. Pedersen SS, et al. 1987. Combined imipenem/cilastatin and tobramycin therapy of multiresistant Pseudomonas aeruginosa in cystic fibrosis. J. Antimicrob. Chemother. 19: 101–107 [DOI] [PubMed] [Google Scholar]
- 48. Permin H, et al. 1983. Ceftazidime treatment of chronic Pseudomonas aeruginosa respiratory tract infection in cystic fibrosis. J. Antimicrob. Chemother. 12(Suppl A):313–323 [DOI] [PubMed] [Google Scholar]
- 49. Pioszak AA, Ninfa AJ. 2003. Mechanism of the PII-activated phosphatase activity of Escherichia coli NRII (NtrB): how the different domains of NRII collaborate to act as a phosphatase. Biochemistry 42: 8885–8899 [DOI] [PubMed] [Google Scholar]
- 50. Roland KL, Martin LE, Esther CR, Spitznagel JK. 1993. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 175: 4154–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schurek KN, et al. 2009. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53: 4345–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Storm DR, Rosenthal KS, Swanson PE. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46: 723–763 [DOI] [PubMed] [Google Scholar]
- 53. Szaff M, Høiby N, Flensborg EW. 1983. Frequent antibiotic therapy improves survival of cystic fibrosis patients with chronic Pseudomonas aeruginosa infection. Acta Paediatr. Scand. 72: 651–657 [DOI] [PubMed] [Google Scholar]
- 54. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33: 2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Trent MS, et al. 2001. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J. Biol. Chem. 276: 43132–43144 [DOI] [PubMed] [Google Scholar]
- 56. Valerius NH, Koch C, Høiby N. 1991. Prevention of chronic Pseudomonas aeruginosa colonisation in cystic fibrosis by early treatment. Lancet 338: 725–726 [DOI] [PubMed] [Google Scholar]
- 57. Winsor GL, et al. 2009. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37: D483–D488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4: 253–263 [DOI] [PubMed] [Google Scholar]
- 59. Woodcock DM, et al. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17: 3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125: 651–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.