Abstract
Vibrio cholerae responds to environmental changes by altering the protein composition of its outer membrane. In rich medium, V. cholerae expresses almost exclusively the outer membrane porin OmpU, whereas in minimal medium, OmpT is the dominant porin. The supplementation of a minimal medium with a mixture of asparagine, arginine, glutamic acid, and serine (NRES) promotes OmpU production and OmpT repression at levels similar to those seen with rich media. Here we show that the altered Omp profile is not due to an increase in the growth rate in the presence of supplemental amino acids but requires the addition of specific amino acids. The effects of the NRES mix on Omp production were mediated by ToxR, a known regulator of omp gene expression. No changes in the Omp profile were detected in a toxR mutant. Supplementation with the NRES mix resulted in significantly higher levels of ToxR, and the elevated ToxR levels were sufficient to cause a switch in Omp synthesis. The increase in the level of the ToxR protein correlated with an increase in toxR mRNA levels and was observed only when toxR was expressed from its native promoter. ToxS, which is required for ToxR activity, was necessary for NRES-mediated omp gene regulation but not for the increase in ToxR levels. The growth of V. cholerae in the presence of bile acids also resulted in Omp switching, and this required ToxR. However, unlike the NRES mix, bile acids did not increase either ToxR protein or toxR mRNA levels, suggesting a different mechanism of omp gene regulation by bile than that by amino acids.
INTRODUCTION
The Gram-negative rod Vibrio cholerae is the causative agent of the severe diarrheal disease cholera. Two serogroups of V. cholerae are associated with pandemic disease, the O1 serogroup, with its two major biotypes, classical and El Tor, and the more recently emerged serogroup O139. V. cholerae is a facultative human pathogen; all V. cholerae strains are native inhabitants of marine and estuarine environments. In order to cause disease, V. cholerae must make the transition from its natural aquatic habitat to the human host. The successful occupation of two such distinct niches requires the ability to respond rapidly to environmental changes. One way in which V. cholerae responds to environmental stimuli is by altering the protein composition of its outer membrane. Two of the major outer membrane porin proteins produced by V. cholerae are OmpT and OmpU. OmpT has greater permeability than OmpU and is thought to be the predominant porin during growth in the environment, where the concentrations of nutrients and osmolytes are expected to be low. OmpU is highly expressed during infection of the human host (24, 51). Although OmpU has a slightly larger pore size than OmpT, it has lower permeability to negatively charged compounds and confers resistance to bile acids, as well as to many antimicrobial peptides, present in the host small intestine (13, 28, 45).
In vitro, OmpT is produced under nutrient-limiting conditions, such as in minimal media, whereas OmpU is the dominant porin in rich media (26, 38). Interestingly, the Omp profile of strains grown in a minimal medium can be altered to resemble that of cells grown in rich medium by the addition of specific compounds to the growth medium. The supplementation of a minimal medium with a mixture of amino acids switches porin synthesis from primarily OmpT to almost exclusively OmpU (38). Similarly, the presence of bile acids in the growth medium causes OmpU levels to increase dramatically (46). The mechanism for Omp regulation in response to environmental signals is not currently known. Both ompT and ompU are regulated at the transcriptional level by ToxR, an important regulator of virulence genes in V. cholerae (7). ToxR activates the expression of ompU and represses the expression of ompT and has been shown to bind directly to the promoter regions of both of these genes (9, 25, 38). Although ToxR is required for the response to both bile and amino acids (38, 46), the exact role of ToxR in mediating these effects has not been determined.
ToxR is a 32.5-kDa inner membrane-spanning DNA-binding protein (39). The amino-terminal domain of ToxR is located in the cytoplasm, where it makes direct contacts with DNA via a winged helix-turn-helix motif (39, 42). The carboxy-terminal domain of ToxR is in the periplasm, where it interacts with its partner, ToxS (10, 39). ToxS is essential for ToxR activity in wild-type V. cholerae strains and was proposed to help stabilize an active conformation of ToxR and/or protect ToxR from proteolysis in the periplasm (10, 37, 44). In addition to regulating the expression of ompT and ompU, ToxR cooperates with another transcriptional regulator, TcpP, to induce the expression of the central virulence gene regulator ToxT (22). ToxT in turn activates the genes encoding the toxin-coregulated pilus (TCP) and cholera toxin (CTX) (7), both essential virulence factors of V. cholerae. In at least one classical biotype strain, O395, ToxR also directly induces the expression of the ctx genes under specific conditions, such as in the presence of bile acids (20).
Not much is known about the regulation of toxR. Although ToxR is required for virulence gene expression, the expression of toxR is not induced under conditions that favor the maximal expression of virulence genes in the laboratory (11, 31). There is evidence that the expression of toxR is influenced by the heat shock response, with increasing temperatures exerting a repressive effect on toxR expression (43). Other studies have shown that the level of ToxR is affected by the transcriptional regulator AphB in stationary phase but not in logarithmic phase (58), suggesting that toxR expression may be influenced by the growth phase. However, the mechanism of ToxR-dependent changes in omp gene expression in response to environmental cues is not well understood. We show here that toxR mRNA and ToxR protein levels increase rapidly in minimal medium upon the addition of a mixture of asparagine, arginine, glutamate, and serine (NRES mix) and that this is not due to an increase in the growth rate in the presence of supplemental amino acids. This mechanism appears to be distinct from ToxR-mediated Omp switching in the presence of bile, since no change in the amount of ToxR was observed for cells exposed to bile acids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All strains were maintained at −80°C in tryptic soy broth (TSB) plus 20% glycerol. Strains were routinely grown at 37°C in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl) (36) or in T medium (50) supplemented with 0.2% (wt/vol) sucrose, 20 μM FeSO4, and a mixture of vitamins (the recipe for the vitamin solution [100× VA] is available at http://www.genome.wisc.edu/resources/protocols/ezmedium.htm). The iron and vitamin supplements increased growth but had no effect on the Omp profile (data not shown). Amino acids (Sigma) were dissolved in water and used at a final concentration of 12.5 mM total amino acids, unless otherwise indicated. Crude intestinal mucin was harvested as follows: segments of rabbit small intestine were washed with phosphate-buffered saline (PBS), and the crude mucin was scraped from the intestines into PBS and stored at −20°C. Crude mucin was added at a final concentration of 2.5% to T medium supplemented as described above. Antibiotics were used at the following concentrations for Escherichia coli strains: 250 μg of carbenicillin per ml, 50 μg of kanamycin per ml, and 30 μg of chloramphenicol per ml. For V. cholerae strains, the concentrations used were 125 μg of carbenicillin per ml, 25 μg of kanamycin per ml, 7.5 μg of chloramphenicol per ml, 75 μg streptomycin per ml, and 40 μg polymyxin B per ml. The electroporation of V. cholerae strains was carried out as described previously (40).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference(s) |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| C6706 | V. cholerae El Tor biotype | R. A. Finkelstein |
| N16961 | V. cholerae El Tor biotype | R. A. Finkelstein |
| O395 | V. cholerae classical biotype | 33 |
| SAC119 | N16961 toxR::kan | 8 |
| ARM755 | N16961 toxS::kan | This study |
| E. coli | ||
| DH5α(λpir) | Cloning strain; host strain for pGP704 derivatives | J. Kaper |
| Plasmids | ||
| pACYC184 | Medium-copy-no. cloning vector; Cmr Tcr | 6 |
| pBAD18 | Arabinose-inducible PBAD vector; Ampr | 16 |
| pCVD442N | Suicide vector pGP704 carrying sacB and a NotI linker; Ampr Sucs | 56 |
| pHM5 | Suicide vector pGP704 carrying sacB; Ampr Sucs | 48 |
| pLAFR3 | Cosmid cloning vector; Tcr | 52 |
| pWKS30 | Low-copy-no. cloning vector; Ampr | 55 |
| pAMS29 | pCVD442N carrying toxS::kan | This study |
| pBADtox1 | pBAD18 carrying toxR, toxS, and VC0982 | This study |
| pCOStox | pLAFR3 carrying toxR, toxS, and + flanking sequence | 17; this study |
| pTox1 | pWKS30 carrying toxR, toxS, and VC0982 | This study |
PCR.
The oligonucleotide primers for PCR were purchased from IDT Inc. (Coralville, IA) and from Invitrogen (Carlsbad, CA). PCR was performed by using Taq polymerase (Qiagen, Valencia, CA) or Platinum Pfx polymerase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Bacterial cultures grown overnight were used as the template. All clones derived from PCR products were verified by DNA sequencing.
Sequence analysis.
DNA sequencing was performed by The University of Texas Institute for Cellular and Molecular Biology DNA Core Facility using an ABI Prism 3700 DNA sequencer. Analysis of DNA sequences was carried out by using MacVector 7.2 and Clone Manager 7.04. BLAST searches and other bioinformatics analyses were done by using the National Center for Biotechnology Information (NCBI) and the National Microbial Pathogen Data Resource (NMPDR) (30) databases. Pairwise alignments were carried out by using ClustalW from within MacVector 7.2.
Construction of plasmids and chromosomal mutations.
A cosmid containing the toxRS operon (pCOStox) was identified in a library of V. cholerae CA401 genomic DNA cloned in pLAFR3 (17, 52) by PCR screening using primers specific for toxR and toxS. pCOStox was moved by bacterial conjugation into SAC119, selecting for resistance to tetracycline and polymyxin B. To create pTox1, a fragment containing toxR, toxS, and the open reading frame (ORF) VC0982 (ORF of unknown function downstream of toxS but not likely cotranscribed with toxRS) was amplified from V. cholerae strain N16961 by PCR with Platinum Pfx using primers tox.for (5′-AGGCCTCAGTCTCGCTCATAATCG) and tox.rev (5′-ATGGCCACACGCACATTCC). The resulting PCR product was cloned into pWKS30 digested with SmaI and EcoRV. To create pBADtox1, the fragment containing toxR, toxS, and the ORF VC0982 was amplified from V. cholerae strain N16961 by using Platinum Pfx with forward primers tox.Nhe.1 (5′-AAGTGCTAGCGTTGGGACAGGG) and tox.rev. The PCR product was digested with NheI and SmaI and cloned into pBAD18 digested with NheI and SmaI to yield the final construct.
To mutate the toxS gene, the NotI-SmaI fragment of pTox1 containing toxRS was cloned into pCVD442N cut with NotI and SmaI. The resulting clone was digested with XhoI, and the kan cassette from pUC4K (Pharmacia) was inserted as a SalI fragment to yield pAMS29. Allelic exchange in V. cholerae was carried out as described previously (34, 35).
SDS-PAGE and immunoblotting.
Cultures were grown overnight in LB medium and diluted 1:100 into fresh medium, as described in the figure legends. Cultures were grown to mid-log phase (optical density at 650 nm [OD650] of ∼0.3), and samples containing equivalent numbers of cells were resuspended in Laemmli solubilization buffer (23). Whole-cell extracts were resolved by SDS–10% polyacrylamide gel electrophoresis (PAGE) and visualized by Coomassie brilliant blue staining or electroblotted for 1.5 h at 45 V onto Hybond ECL nitrocellulose (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England). The positions of OmpT and OmpU were determined through comparisons of the wild-type strain with ompT and ompU mutants and by the immunodetection of OmpT and OmpU (8; data not shown). The immunodetection of ToxR was carried out by using rabbit polyclonal anti-ToxR antiserum (diluted 1:1,000) (generous gift of R. K. Taylor) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Bio-Rad Laboratories, Hercules, CA) secondary antibody (diluted 1:10,000). The anti-ToxR antiserum cross-reacts with other proteins, as is evident in many of the immunoblots shown. Although the identity of this lower band is not known, it appears consistently in all samples, regardless of growth conditions, and can therefore be used as a loading control in addition to the stained gel.
RNA isolation and real-time PCR.
Strains were grown to mid-log phase (OD650 of ∼0.3) in T medium supplemented with 0.2% sucrose, 20 μM FeSO4, and a mix of vitamins, as described above. The cultures were then divided, and amino acid solutions were added to a final concentration of 50 mM total amino acids, as indicated in the figure legends. Cultures were grown for an additional 15 min and then treated with an RNase-free solution of 95% absolute ethanol–5% phenol (pH 4.5), used at 20% (vol/vol), and kept on ice. RNA from 109 cells per sample was isolated by using the RNeasy minikit (Qiagen) according to the manufacturer's instructions for the isolation of total RNA from bacterial cells. Following column purification, each RNA sample was treated with DNase I (Invitrogen) according to the manufacturer's protocol. The RNA samples were ethanol precipitated, dried, resuspended in RNase-free water, and quantified by using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Two micrograms of RNA was reverse transcribed by using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) in a total reaction mixture volume of 20 μl, according to the manufacturer's instructions. The cDNA samples were diluted 1:10 in water, and 2.5 μl of each dilution was used as the template for real-time PCR using a TaqMan probe (6-carboxyfluorscein labeled, minor-groove binding), target-specific primers, and TaqMan Universal PCR Master Mix (Applied Biosystems). The primers and probes were designed by using Primer Express (Applied Biosystems) and synthesized by Applied Biosystems. Real-time PCR was carried out with a 7300 real-time PCR system machine from Applied Biosystems. Transcript levels in each sample were normalized internally to the constitutive rpoZ (VC2709) transcript, as described previously by Meibom et al. (32).
RESULTS
The NRES mix, but not aspartate, increases OmpU and decreases OmpT levels.
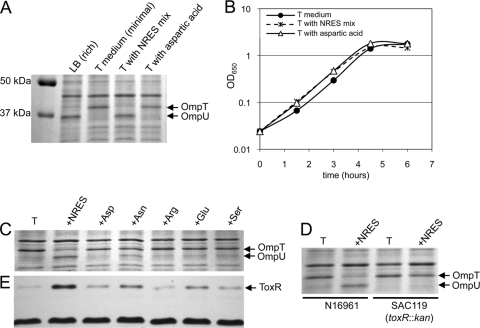
The expression of V. cholerae outer membrane porins is influenced by the medium in which the cells are grown. When El Tor strain N16961 was grown in rich medium (LB medium), OmpU was the dominant porin, whereas in minimal medium (T medium), OmpT was produced exclusively (Fig. 1A). The supplementation of the minimal medium with a mixture of asparagine, arginine, glutamate, and serine (NRES mix) caused an increase in OmpU and a decrease in OmpT levels, similar to the pattern seen with rich medium (Fig. 1A). These results are consistent with those of a previous study using the classical strain O395 (38). Because the NRES mix stimulated the growth of V. cholerae in minimal medium (Fig. 1B and data not shown), it was important to determine whether the change in the Omp pattern was a result of metabolic shifts associated with an increased growth rate or was due to a specific regulatory effect of the NRES mix. The supplementation of the medium with aspartate, an amino acid not present in the NRES mix, also stimulated the growth of V. cholerae (Fig. 1B) but did not result in changes to the Omp profile (Fig. 1A). This finding suggests that the switch from OmpT to OmpU in the presence of the NRES mix is not due to increased growth and metabolism but may be associated with a specific signal present in the NRES mix.
Fig 1.
Effects of amino acid supplementation on the Omp profile, growth rate, and ToxR levels in V. cholerae. Unless otherwise noted, experiments were carried out using wild-type El Tor strain N16961. Strains were grown overnight in LB broth and then subcultured at a 1:100 dilution in LB or T medium supplemented with the indicated amino acids at a final concentration of 12.5 mM total amino acids. Growth was monitored by measuring the optical density at 650 nm (B), and cells were harvested in mid-log phase (about 3 h postinoculation, at an OD650 of ∼0.3). Total cellular proteins were resolved by SDS–10% PAGE and stained by Coomassie blue (A, C, and D), and immunoblotting was carried out by using polyclonal anti-ToxR antisera (E). The dark band at the bottom of panel E (a cross-reactive protein of unknown identity) is included to show the equal loading of all samples.
Effect of individual amino acids on the Omp profile of V. cholerae.
When the four amino acids were tested individually, asparagine had the greatest effect on the OmpT/OmpU pattern, although no single amino acid was as effective as the NRES mix (Fig. 1C). Very little switching was observed in the presence of arginine, glutamate, or serine alone (Fig. 1C). The effect of asparagine on the Omp profile suggested that V. cholerae may respond to amines or other sources of nitrogen; however, no effects on the Omp pattern were seen when either an organic amine (spermidine or ornithine) or excess ammonium chloride was added to the medium (data not shown).
Omp switching in the presence of the NRES mix requires ToxR.
The expression of both ompT and ompU is under the control of the virulence regulator ToxR. ToxR represses ompT expression, while it increases the expression of ompU (9, 25, 38). It was shown previously in a classical strain background that toxR is required for the effects of amino acids on the Omp profile. To test whether ToxR is similarly required for the switch in Omp production in the presence of the NRES mix in an El Tor strain background, N16961 and its isogenic toxR deletion mutant, SAC119, were grown in T medium with or without the NRES supplement. No OmpU was detected in the toxR mutant strain, even in the presence of the NRES mix, consistent with an absolute requirement for ToxR in the expression of ompU (Fig. 1D). Furthermore, no reduction in the level of OmpT was seen in the toxR mutant in the presence of the NRES mix, showing that the NRES-mediated decrease in the OmpT level is also dependent upon ToxR (Fig. 1D).
ToxR protein levels increase in the presence of the NRES mix.
To understand the role of ToxR in regulating Omp production in response to environmental changes, ToxR protein levels were determined for strains grown in minimal medium supplemented with various amino acids. The level of ToxR was significantly higher in cells grown to mid-log phase in the presence of the NRES mix than in cells grown in T medium alone or in T medium supplemented with aspartic acid (Fig. 1E). When the four amino acids were tested individually, asparagine caused the greatest increase in the ToxR level (Fig. 1E). This is consistent with the presence of a small amount of OmpU in asparagine-supplemented cells but little or no detectable OmpU in cultures supplemented with other single amino acids (Fig. 1C). A small increase in the ToxR level was seen in the presence of glutamic acid, but this may not have been enough to increase OmpU to detectable amounts.
Increasing the basal level of ToxR in the cell is sufficient to cause Omp switching in minimal medium.
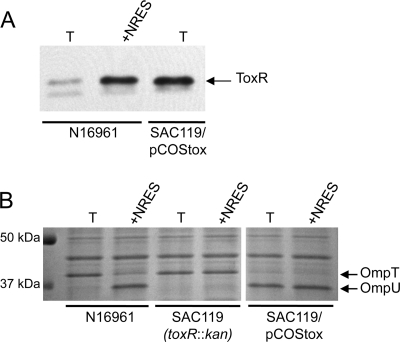
While the increase in the level of ToxR is likely the cause of Omp switching in minimal medium supplemented with the NRES mix, it is also possible that the NRES mix affects the activity of the ToxR protein at the ompU and ompT promoters. To test whether an increase in the amount of ToxR is sufficient to cause a switch from OmpT to OmpU, the toxRS operon was cloned under the control of its native promoter on a low-copy cosmid vector, pLAFR3. The amount of ToxR made from the cosmid was significantly larger than that in the wild-type strain in minimal medium and comparable to the amount of ToxR made by the wild-type strain in the presence of the NRES mix (Fig. 2A). The strain expressing toxRS from the cosmid produced exclusively OmpU, even in the absence of the NRES mix (Fig. 2B). Thus, increased levels of ToxR production from the cosmid correlated with the appearance of OmpU and the disappearance of OmpT, even without amino acid supplementation. These data strongly suggest that the mechanism underlying the switch from OmpT to OmpU in minimal medium supplemented with the NRES mix is an increase in the levels of ToxR and its partner, ToxS.
Fig 2.
An increase in the amount of ToxR is sufficient to switch Omp production from OmpT to OmpU. The wild type (N16961), the toxR mutant (SAC119), and the toxR mutant strain carrying a toxRS+ cosmid clone (SAC119/pCOStox) were grown to mid-log phase in T medium with or without 12.5 mM NRES mix. Whole-cell preparations were resolved by SDS–10% PAGE and immunoblotted by using polyclonal anti-ToxR antisera (A) or stained by Coomassie blue (B). The thick white line indicates that intervening lanes were removed from the gel image.
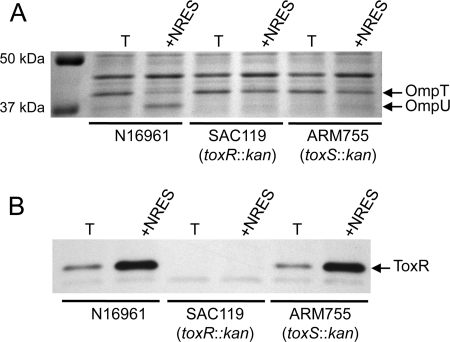
ToxS is required for ToxR activity but not for the NRES-mediated increase in ToxR protein levels.
The activity of ToxR depends on its partner protein, ToxS, which interacts with ToxR and may help to stabilize an active conformation of ToxR (10, 37, 44). To test whether ToxS is needed for the regulation of Omp production in response to amino acids, a chromosomal toxS mutant was constructed and tested for its ability to synthesize OmpU in minimal medium in the presence of the NRES mix. Similar to the toxR mutant, the toxS mutant did not make OmpU under any condition tested, and no reduction in the OmpT level was seen in response to the amino acids (Fig. 3A). This finding is consistent with an absolute requirement for ToxS in the ToxR-dependent regulation of omp gene expression. To test whether the NRES-mediated increase in the ToxR level is dependent upon ToxS, ToxR protein levels in the toxS mutant grown with or without the amino acid mix were determined. The amount of the ToxR protein produced in the toxS mutant in response to the NRES mix was similar to the amount seen for the wild-type strain (Fig. 3B), showing that ToxS is not required for elevated ToxR levels in response to amino acids. Thus, the increase in the ToxR level is not likely to be due to an enhanced stability of the ToxR protein through interactions with ToxS.
Fig 3.
The NRES-mediated increase in the ToxR level does not require ToxS. Strains N16961 (wild type), SAC119 (toxR::kan), and ARM755 (toxS::kan) were grown overnight in LB broth and then subcultured at a 1:100 dilution in T medium with or without 12.5 mM NRES mix. Cells were harvested at mid-log phase, and whole-cell preparations were resolved by SDS–10% PAGE and stained by Coomassie blue (A) or immunoblotted by using polyclonal anti-ToxR antisera (B).
No single known or predicted protease targets ToxR in minimal medium.
ToxR may be susceptible to proteolysis by periplasmic proteases, as was demonstrated previously for another transmembrane transcriptional activator of V. cholerae virulence gene expression, TcpP (1, 29). Thus, the mechanism for increasing ToxR levels in the presence of the NRES mix could be through protection of ToxR from proteolytic cleavage and degradation. A panel of mutants in known and predicted envelope proteases was tested to see whether any of these mutants produced larger amounts of OmpU than the wild-type parental strain, signaling an increase in the level of ToxR (Table 2). Of the 14 mutants tested, none exhibited any differences in their Omp profiles in the presence or absence of the NRES mix, suggesting either that none of these proteases targets ToxR or that two or more proteases are involved in the degradation of ToxR.
Table 2.
Effects of mutations on the response to NRES
| Category | ORF | Gene | Encoded protein and/or function | Response of mutant to NRES mix | Mutant source or reference |
|---|---|---|---|---|---|
| Virulence regulatory genes | VC0983a | toxS | Regulatory protein ToxS | No Omp switching | This study |
| VC0984a | toxR | Regulatory protein ToxR | No Omp switching | 8 | |
| VC1049b | aphB | LysR family transcriptional regulator AphB | Wild type | 5 | |
| VC1651b | vieB | Response regulator VieB | Wild type | 5 | |
| VC1653b | vieS | Sensor histidine kinase VieS | Wild type | 5 | |
| VC2647b | aphA | Transcriptional regulator AphA | Wild type | 5 | |
| Chemotaxis and flagellar genes | VC1008b | motY | Sodium-driven flagellar motor protein MotY | Wild type | 5 |
| VC2065b | cheY-3 | Chemotaxis protein CheY | Wild type | 5 | |
| VC2135b | flrC | Flagellar regulatory protein C | Wild type | 5 | |
| VC2188b | flaA | Flagellin core protein A | Wild type | 5 | |
| VC2201b | cheR-2 | Chemotaxis protein methyltransferase CheR | Wild type | 5 | |
| Known and predicted periplasmic | VC0565c | degS | Periplasmic serine protease | Wild type | 29 |
| protease genes | VC0566c | degP | Periplasmic serine endopeptidase | Wild type | V. DiRita |
| VC0975b | Conserved hypothetical protein | Wild type | 5 | ||
| VC0976b | Conserved hypothetical protein | Wild type | 5 | ||
| VC1074b | Hypothetical protein | Wild type | 5 | ||
| VC1117b | htpX | Heat shock protein | Wild type | 5 | |
| VC1200b | Trypsin, putative | Wild type | 5 | ||
| VC1496b | prc | Carboxy-terminal protease | Wild type | 5 | |
| VC1709b | Zinc protease, insulinase family | Wild type | 5 | ||
| VC1994b | sspA | Protease IV | Wild type | 5 | |
| VC2253c | yaeL | Zinc metalloprotease | Wild type | 29 | |
| VCA0044b | Hypothetical protein | Wild type | 5 | ||
| VCA0063b | ptrB | Iron-regulated protease II | Wild type | 5 | |
| VCA0550b | Hypothetical protein | Wild type | 5 | ||
| Other genes tested | VC0982b | Downstream of toxS | Selenoprotein W-related protein | Wild type | 5 |
| VC0347b | hfq | RNA-binding protein | Wild type | 5 |
The mutation is in the V. cholerae N16961 background.
The mutation is in the V. cholerae C6706 background. C6706 shows a response to the NRES mix similar to that of N16961 (data not shown).
The mutation is in the V. cholerae O395 background. O395 shows a response to the NRES mix similar to that of N16961; however, the induction of OmpU synthesis is less than that in the El Tor strains, since O395 produces some OmpU in T medium alone (data not shown).
The NRES mix increases toxR mRNA levels.
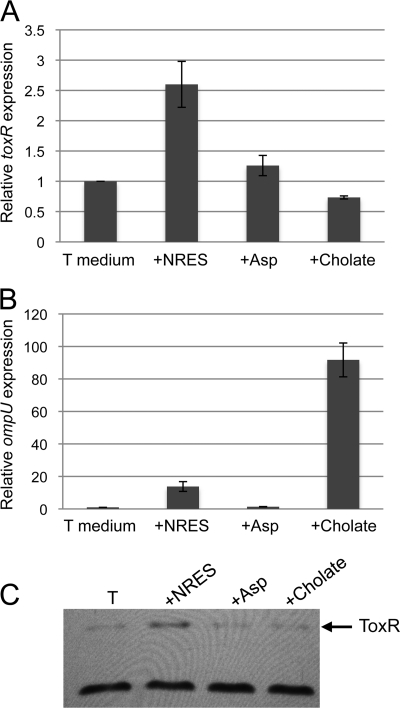
To determine whether the effect of the NRES mix on the amount of the ToxR protein occurs at the level of the toxR transcript, real-time PCR was performed to quantify toxR mRNA in cells exposed to amino acids. Wild-type V. cholerae was grown in minimal medium to mid-log phase. The culture was then divided, and either the NRES mix or aspartic acid was added. Cells were harvested after 15 min in the presence of the amino acid supplement, and RNA was extracted. A 2- to 3-fold increase in the toxR transcript level was detected after exposure to the NRES mix, but not aspartic acid, suggesting that the NRES mix has a positive effect on the transcription of toxR (Fig. 4A). The toxR transcript levels did not continue to increase with time but remained at approximately 2- to 3-fold above the level of the aspartic acid-supplemented culture after 30 min (data not shown). A concomitant increase in the level of the ToxR protein was also observed after 15 min of growth in the presence of the NRES mix but not aspartic acid (Fig. 4C). As expected, based on this increase in the ToxR protein level, a significant increase (more than 10-fold) in the amount of ompU mRNA (Fig. 4B) was detected in cells grown in the presence of the NRES mix. Aspartic acid did not cause any significant increase in the ompU mRNA level (Fig. 4B), and ompU mRNA was not detectable in a toxR mutant (data not shown). Taken together, these data support the model that the NRES-mediated regulation of ompU expression occurs through the modulation of toxR mRNA levels.
Fig 4.
Expression of toxR and ompU in response to the NRES mix or cholate. N16961 was grown to mid-log phase in T medium without amino acids or bile acid. The culture was then divided, and the NRES mix (50 mM final amino acid concentration), aspartic acid (50 mM final amino acid concentration), or 0.1% cholate was added. RNA was isolated from cells grown for 15 min in the presence of the indicated supplement. (A and B) The levels of toxR (A) and ompU (B) mRNAs were determined by real-time PCR. All transcript levels were normalized internally to rpoZ, and the expression levels are shown relative to the expression level at 15 min in T medium without amino acids or cholate. The data represent the means and standard deviations from at least three independent experiments. (C) Whole-cell extracts of cells grown for 15 min in the presence of amino acids or cholate were resolved by SDS–10% PAGE, and immunoblotting was done using polyclonal anti-ToxR antisera. The cross-reactive protein of unknown identity at the bottom of panel C is included to show the equal loading of all samples.
The native toxR promoter is required for the NRES effect.
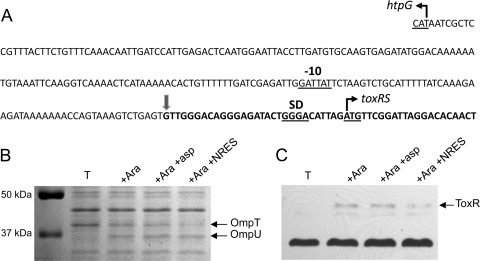
The real-time results show that the NRES mix increases either the rate of toxR transcription or the stability of the toxR mRNA, but they do not rule out the possibility that posttranscriptional effects, such as an increased efficiency of translation or a greater stability of the ToxR protein, could also contribute to the elevated ToxR levels. To confirm that the effect of the NRES mix is at the level of the mRNA, we determined whether the toxR promoter and/or the 5′ untranslated region (UTR) of the toxR mRNA is required for the response to the amino acids. A fragment containing the toxRS open reading frames was cloned under the control of the arabinose-inducible promoter in plasmid pBAD18. Included in this clone are 30 nucleotides immediately upstream of the predicted toxR translational start (39), including the putative Shine-Dalgarno sequence; thus, translation should initiate from the native toxR start codon (Fig. 5A). In the absence of arabinose, no ToxR was produced from this construct (Fig. 5C), showing that there are no promoter sequences present in the cloned insert. In cells grown in the presence of 0.02% arabinose, the ToxR protein was detectable by Western blotting (Fig. 5C). The supplementation of arabinose-induced cultures with the NRES mix did not further increase the amount of ToxR present, even when the basal level of toxRS expression was kept low to mimic the expression level in minimal medium with no amino acid supplementation (Fig. 5C). This indicates that the NRES mix does not stabilize the ToxR protein but requires the native toxR promoter to stimulate ToxR production. This finding is further evidence that the NRES mix increases ToxR levels by increasing toxR transcription or by stabilizing the toxR mRNA. If the NRES mix stabilizes the toxR transcript, this must require the 5′ UTR of the native transcript, since the NRES mix had no effect on toxR transcripts lacking the native 5′ UTR (Fig. 5C and data not shown). It should be noted that the amount of OmpU produced in cells expressing toxR from the pBAD promoter was smaller than that produced in wild-type cells, even when toxR was overexpressed by the addition of higher concentrations of arabinose (Fig. 5B and data not shown), suggesting that the plasmid-encoded ToxR may be less active than ToxR expressed from its native promoter.
Fig 5.
The NRES-dependent increase in ToxR levels requires the native toxR promoter. (A) The intergenic region between htpG and toxR is shown. A putative −10 sequence, as well as the Shine-Dalgarno (SD) sequence and the start codon for toxR predicted previously by Miller et al. (39), is underlined. Note that the −10 sequence, the Shine-Dalgarno sequence, and the translational start site have not been experimentally verified. The predicted start site and direction of the htpG and toxR open reading frames are indicated by angled arrows. The fat vertical arrow and the bold letters mark the 5′ end of the toxRS fragment included in pBADtox1, which carries the entire toxRS operon cloned under the control of the arabinose-inducible PBAD promoter. toxR mutant strain SAC119 carrying pBADtox1 was grown in T medium with 0.05% arabinose to mid-log phase with or without 12.5 mM NRES mix or aspartic acid. (B and C) Whole-cell preparations were resolved by SDS–10% PAGE (B), and immunoblotting was carried out by using polyclonal anti-ToxR antisera (C). The cross-reactive protein of unknown identity at the bottom of panel C shows the equal loading of all samples.
The NRES effect is not mediated by the AphA/B or VieS/A/B regulators.
The effect of the NRES mix on toxR mRNA levels suggested that a regulator of toxR transcription might be involved. A previous study found that the V. cholerae virulence gene regulators AphA and AphB also influence the expression of toxR (58). To test whether these regulators are involved in the response to amino acids, aphA and aphB mutants were grown to mid-logarithmic phase in minimal medium with or without amino acid supplementation, and the Omp profiles were observed. Both mutants responded similarly to the NRES mix as the wild-type parental strain, suggesting that the effects of NRES on toxR expression do not require the AphA/B transcriptional regulators (Table 2). Similarly, mutants in the VieSAB virulence gene regulatory system (54) showed no defect in the ability to respond to the NRES mix (Table 2).
The response to the NRES mix does not require an Hfq-dependent sRNA.
An increasing number of V. cholerae genes have been shown to be regulated by small RNAs (sRNAs). sRNAs can affect the stability of target mRNAs through base-pairing interactions within the 5′ UTR of the target transcript (27). Most characterized sRNAs are dependent on the RNA-binding protein Hfq (27). We tested the hypothesis that the NRES-dependent increase in the toxR mRNA level is due to the activity of an Hfq-dependent sRNA by analyzing the Omp profile of an hfq mutant. The hfq mutant showed an Omp profile similar to that of the wild-type parental strain in the presence of the NRES mix, indicating that the response to the NRES mix does not involve an Hfq-dependent sRNA (Table 2).
Chemotaxis is not essential for the NRES effect on Omp production.
V. cholerae exhibits a strong chemotactic response toward amino acids, and chemotaxis has been shown to be essential for the in vivo association of V. cholerae with the intestinal mucosa of rabbits (15). The role of chemotaxis in the NRES mix-induced increase in the ToxR level was tested. There are at least three operons containing putative chemotaxis genes in V. cholerae; however, only one set of che genes appears to be required for chemotaxis. Among the essential chemotaxis genes are cheR-2 (41) and cheY-3 (2). In this study, the cheR-2 and cheY-3 mutants responded similarly to the NRES mix as the wild-type strain, indicating that chemotaxis per se is not required for the amino acid-dependent increase in the ToxR level (Table 2). Flagellar synthesis was also not required for the NRES effect on omp gene expression (Table 2).
Temperature does not affect the NRES-mediated increase in ToxR levels.
The toxR gene is divergently transcribed from a heat shock gene, htpG, and the effects of temperature on toxR expression have been studied. Using plasmid-borne transcriptional fusions, it was shown previously that increasing temperatures reduce expression from the toxR promoter in both classical and El Tor strains (43, 58). It was proposed that this is due to the displacement of RNA polymerase (σ-70) from the toxR promoter by a competing RNA polymerase (σ-32) transcribing from the htpG promoter during heat shock (43). Given that toxR is necessary for infection of the mammalian host, where it is expressed at 37°C (18, 53, 57), this result seems counterintuitive and suggests the presence of signals in vivo that counteract this repressive effect. The experiments carried out in this study were done at 37°C, raising the possibility that the NRES mix may act to suppress the negative effects of high temperatures on toxR transcription, allowing ToxR to be produced maximally at 37°C. To test this possibility, experiments were repeated at 30°C. The NRES mix stimulated ToxR production to an extent similar to that observed at 37°C (data not shown), suggesting that the NRES mix does not increase ToxR levels solely by counteracting the effects of an elevated temperature on toxR transcription.
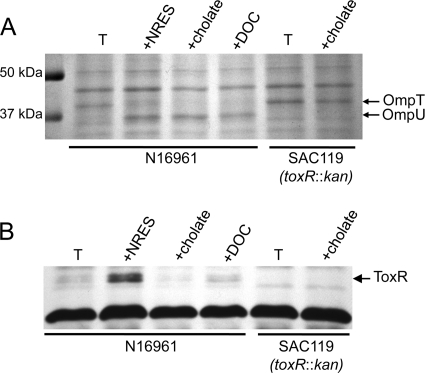
Bile acids cause Omp switching in minimal medium but do not increase ToxR levels.
It was shown in a previous study that bile induces the expression of ompU and that this effect requires ToxR (46). Thus, the effect of bile on ompU appears to be similar to the effect of the NRES mix. The role of bile in ompT regulation was not reported. We tested the effects of the bile acids cholate and deoxycholate (DOC) on the Omp profile and ToxR levels of V. cholerae strain N16961. When cultures were grown in minimal medium in the presence of either 0.1% (wt/vol) cholate or 0.01% (wt/vol) DOC, a switch from OmpT to OmpU was observed (Fig. 6A), showing that the effects of the bile acids on Omp production are similar to the effects of the NRES mix. A large increase in the level of ompU mRNA in response to cholate was also observed: a 15-min exposure to cholate induced a nearly 100-fold increase in the ompU message (Fig. 4B). No OmpU was produced in the toxR mutant strain, even in the presence of cholate (Fig. 6A), consistent with the previously reported requirement for ToxR for the bile-induced expression of ompU (46). To determine the effects of bile acids on ToxR levels, cultures were grown in minimal medium with cholate or DOC, and the ToxR protein was analyzed by Western blotting (Fig. 6B). In contrast to our observations with the NRES mix, the presence of bile acids did not increase ToxR levels. In fact, ToxR levels diminished rather than increased in the presence of cholate (Fig. 6B), and the level of toxR mRNA was similarly reduced (Fig. 4A). Thus, bile influences ToxR-dependent Omp production through a different mechanism than the amino acids.
Fig 6.
Bile acids cause a ToxR-dependent switch in Omp production but do not affect ToxR levels. Strains N16961 and SAC119 (toxR::kan) were grown overnight in LB broth and then subcultured at a 1:100 dilution in T medium with or without either cholate (0.1%) or deoxycholate (0.01%). Whole-cell preparations of mid-log-phase cells were resolved by SDS–10% PAGE and stained by using Coomassie blue (A) or immunoblotted by using polyclonal anti-ToxR antisera (B). The cross-reactive protein of unknown identity at the bottom of panel B is included to show the equal loading of all samples.
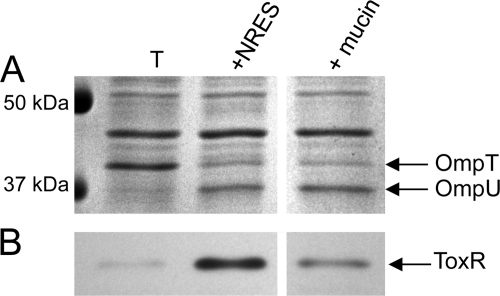
Intestinal mucin causes Omp switching but only a small increase in ToxR levels.
The increase in the expression level of OmpU in response to the NRES mix or bile is particularly interesting because it mimics the Omp pattern in the host environment, where OmpU is highly expressed (24, 51). In the small intestine, V. cholerae is found in close association with mucin (15, 47), which may contain nutrients as well as the appropriate signals for the regulation of genes affecting host colonization. To test the effect of mucin on the Omp profile of V. cholerae, crude intestinal mucin was harvested from rabbit small intestines and used to supplement minimal medium. Intestinal mucin dramatically increased the growth of V. cholerae in minimal medium, suggesting that the rabbit small intestine is a high-nutrient environment (data not shown). Like the NRES mix and bile acids, crude mucin caused a clear shift in the Omp profile, with OmpU replacing OmpT (Fig. 7A). Interestingly, only a modest increase in the ToxR level was detected in cells grown in the presence of mucin (Fig. 7B). The small increase in the ToxR level may be due to the presence of amino acids in crude mucin; however, the large effect of mucin on Omp production in the absence of significant ToxR induction suggests an effect more similar to the effects of bile, and this may reflect the presence of bile components in the crude intestinal mucin extracts. Furthermore, since the cells grew very robustly in the presence of intestinal mucin (data not shown), the data support the conclusion that an increase in the growth rate alone is not sufficient to stimulate ToxR production.
Fig 7.
Effect of intestinal mucin on the Omp profile and ToxR levels of V. cholerae. Strain N16961 was grown overnight in LB broth and then subcultured at a 1:100 dilution in LB or T medium. The NRES mix was added at a final concentration of 12.5 mM total amino acids. Crude rabbit small intestinal mucin was added at 2.5% (vol/vol). Growth in the presence of the different supplements was monitored (data not shown), and cells were harvested at mid-log phase. Whole-cell preparations were resolved by SDS–10% PAGE and stained by Coomassie blue (A) or immunoblotted by using polyclonal anti-ToxR antisera (B). The thick white line indicates that intervening lanes were removed from the gel image.
DISCUSSION
V. cholerae virulence gene expression within the human host is precisely timed to ensure colonization and toxin production in the appropriate microenvironments within the small intestine. To achieve this tightly regulated colonization and virulence factor production, V. cholerae must respond to a variety of host-specific signals, including changes in temperature, pH, osmolarity, and nutrient availability. The exact nature of these in vivo signals is not well understood. Efforts to induce virulence gene expression in vitro by mimicking the in vivo conditions of the human small intestine (temperature of 37°C, high osmolarity, and alkaline pH) have not always been successful, since the conditions that promote maximal toxin production in the laboratory (“inducing conditions” for classical strains and “AKI” conditions for El Tor strains) are largely unrelated to any conditions encountered in the host (reviewed in reference 7).
The expression of the virulence gene regulon in V. cholerae is controlled by the master regulatory switch ToxR; however, ToxR levels do not change in response to either inducing or AKI conditions (11, 21, 31). This has led to the hypothesis that toxR expression is constitutive and that the response to environmental stimuli does not occur through the modulation of ToxR levels. Rather, it was proposed that environmental signals primarily affect the expression of the gene encoding the downstream regulator ToxT. This proposal is supported by the observation that the toxT expression level is increased under both inducing and AKI conditions (12, 19, 21, 31, 49), and the constitutive expression of toxT can alleviate the repressive effects of certain nonpermissive conditions on virulence gene expression (11, 12, 21). In this study, we show that ToxR synthesis is not constitutive under all conditions but is sensitive to specific components of the growth medium. The amount of ToxR was significantly larger in cells grown in a rich medium than in a minimal medium, and a high level of ToxR was produced in minimal medium supplemented with the NRES mix. This increase in the ToxR level correlated with changes in the Omp profile, consistent with previously reported observations that the addition of these four amino acids to minimal medium effects a switch in Omp production from OmpT to OmpU (38). Furthermore, increasing the basal level of ToxR by expressing toxR from a cosmid was sufficient to cause Omp switching in minimal medium in the absence of any added amino acids (Fig. 2A and B), showing that omp gene regulation is sensitive to ToxR levels. This finding indicates that the effect of the NRES mix on the Omp profile is due to an increase in the amount, rather than the activity, of ToxR.
The Omp profile of cells grown in minimal medium without amino acids was indistinguishable from the Omp profile of a toxR mutant (Fig. 1D). Thus, although ToxR is clearly produced in the absence of the NRES mix (Fig. 1E), this basal level of ToxR is not enough to regulate Omp gene expression, suggesting that there may be a threshold level required for ToxR activity at the omp promoters. Consistent with this hypothesis is the observation that supplementation with asparagine alone, which caused only a modest increase in ToxR levels (Fig. 1D), resulted in an intermediate Omp phenotype, with both OmpT and OmpU being visible in the outer membrane (Fig. 1C).
The NRES mix positively affected the amount of toxR mRNA, suggesting that the mechanism underlying the increase in the ToxR level is either the transcriptional activation of toxR or the increased stability of the toxR mRNA. This is further supported by our observation that elements within the native toxR promoter are needed for the response to the NRES mix. Interestingly, the divergently transcribed heat shock gene htpG was downregulated in the presence of the NRES mix but not in the presence of aspartic acid (A. R. Mey, unpublished results). This could be due to competition for the binding of RNA polymerase at the overlapping promoters, pointing to effects of the NRES mix at the level of toxR transcription rather than mRNA stability. Studies are under way to identify any potential regulators of toxR expression that may be responsive to amino acid stimulation.
The correlation between the increase in the toxR mRNA level and the increase in the ToxR protein level suggests that even a relatively modest increase in the toxR transcript level in response to the amino acid mix may be sufficient to account for the high levels of the ToxR protein. It was similarly noted in a previous study of toxT expression that a less-than-2-fold decrease in the toxT transcription level was associated with significantly reduced CTX and TCP production (45). This finding indicates that the expression of the major V. cholerae virulence gene regulators is highly sensitive and is tightly controlled at the transcriptional level in order to maintain the proper expression of virulence factors.
ToxR shares many structural and functional features with the V. cholerae virulence regulator TcpP (reviewed in reference 7), which is known to be susceptible to proteolysis by the envelope protease YaeL in the absence of its partner, TcpH (1, 29). Protection from proteolysis is critical for maintaining steady-state levels of TcpP (1). In contrast, ToxR does not appear to rely on its partner, ToxS, for stability. A mutation in toxS did not produce lower levels of ToxR under the conditions tested (Fig. 3B), consistent with data from previous studies showing that ToxR, unlike TcpP, is stable in the absence of its partner in exponentially growing cells (1, 44). Interestingly, a recent study by Xu et al. (58) found that by late stationary phase, ToxR was no longer detectable in a toxS mutant, despite the presence of toxR mRNA. This finding suggests that ToxS does play a role in protecting ToxR from degradation under certain conditions; however, those conditions are very different from the ones used in this study. We found that the toxS mutant strain, like its wild-type parental strain, responded to the NRES mix by increasing the level of ToxR. This finding argues against a model in which ToxS increases the stability of the ToxR protein in the presence of the NRES mix. Although ToxR levels increased in the absence of ToxS, there was no ToxR activity at the omp promoters, showing that ToxR has an absolute requirement for ToxS that cannot be overcome by simply increasing the amount of ToxR. Although we do not have the tools to assess the amount of ToxS made in the presence of the NRES mix, we noted that the toxS transcript level showed a similar 2- to 3-fold increase in response to the NRES mix as toxR mRNA (Mey, unpublished), as expected based on the previously reported operon structure of the toxRS genes (37). This suggests that the NRES-mediated increase in the ToxR protein level is paralleled by an increase in the ToxS level, further underscoring the importance of ToxS in forming transcriptionally active ToxR. No increase in ToxR protein levels was observed in response to the NRES mix when toxR and toxS were expressed from a heterologous promoter, indicating that the ToxR protein itself is not stabilized by the amino acids under the conditions tested.
The NRES mix was originally discovered as a combination of amino acids that stimulated the highest level of CTX production in a defined medium (4). How V. cholerae senses and responds to these particular amino acids is not clear. Of the four amino acids, asparagine had the greatest effect on ToxR-mediated Omp production. This is consistent with previously reported data showing that asparagine by itself induced the highest level of CTX synthesis compared with the other three amino acids in the NRES mix (3). Nevertheless, a more robust response was seen when asparagine was added together with at least one other amino acid. Asparagine and serine are consumed efficiently during logarithmic growth, whereas arginine and glutamate are not (3). It is conceivable that the ToxR-mediated response to amino acid stimulation may involve the transport and/or metabolism of at least one of the amino acids during exponential growth.
Exposure to bile salts had an effect on the outer membrane profile of V. cholerae similar to that of exposure to the NRES mix. In the presence of bile salts, OmpU levels increased dramatically, while OmpT levels dropped to an almost undetectable level (Fig. 6A). In a previous study by Provenzano et al. (46), no effects of bile acids on OmpT levels were observed. This is likely because the cells were grown in LB medium, in which OmpT levels are already very low (Fig. 1A). Thus, any further decrease in the ompT expression level would be difficult to detect under those conditions. In contrast to the NRES mix, the bile acids had no effect on the amount of ToxR produced, even though ToxR was required for the response to bile (Fig. 6B). In fact, cholate had a slight repressive effect on both ToxR protein and toxR mRNA levels (Fig. 4A and 6B). This finding suggests that the mechanism underlying the switch from OmpT to OmpU in response to bile is fundamentally different from the mechanism governing the response to the amino acids. Instead of increasing the amount of ToxR produced, bile may increase the activity of the ToxR protein. Alternatively, exposure to bile may increase the amount or activity of a coregulator of ompU expression, since cells grown in the presence of bile acids synthesized very high levels of ompU mRNA. It is also noteworthy that although both bile acids and amino acids eventually caused a complete switch in the OmpT/OmpU profile (Fig. 6A), the bile acids caused a much more rapid increase in ompU expression. Within 15 min of exposure, the effect of cholate on the level of ompU mRNA was an order of magnitude greater than the effect of the NRES mix (Fig. 4B). This finding suggests that the kinetics of ompU induction may be different for these two signals, consistent with two different mechanisms of ompU activation.
The presence of amino acids or bile acids in the growth medium not only influences the Omp profile of V. cholerae but also stimulates the synthesis of CTX (3, 4, 14, 20, 38). Within the host environment, nutrient availability and the presence of antimicrobial compounds such as bile could represent important indicators of entry into the appropriate environment for colonization and toxin production. The picture of how different environmental signals are perceived and integrated into an overall response that maintains the proper expression of virulence genes is far from complete; however, ToxR synthesis and activity in response to amino acids and bile acids suggest an important in vivo mechanism for ensuring protection against antimicrobial compounds in the lumen of the small intestine through the activation of ompU and the repression of ompT, as well as for ensuring the expression of genes required for colonization and CTX production in the right place at the right time.
ACKNOWLEDGMENTS
We are indebted to Ron Taylor for his generous gift of the ToxR antiserum and to Glenn Otto and Carolyn Fisher for preparing the rabbit intestinal mucin. We thank Victor DiRita for providing V. cholerae mutant strains and for helpful suggestions, and we thank Liz Wyckoff for insightful discussions and critical reading of the manuscript.
This work was supported by grants AI091957 and AI50669 from the National Institutes of Health.
Footnotes
Published ahead of print 5 December 2011
REFERENCES
- 1. Beck NA, Krukonis ES, DiRita VJ. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 186:8309–8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 101:5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Callahan LT, III, Richardson SH. 1973. Biochemistry of Vibrio cholerae virulence. III. Nutritional requirements for toxin production and the effects of pH on toxin elaboration in chemically defined media. Infect. Immun. 7:567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Callahan LT, III, Ryder RC, Richardson SH. 1971. Biochemistry of Vibrio cholerae virulence. II. Skin permeability factor-cholera enterotoxin production in a chemically defined medium. Infect. Immun. 4:611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 105:8736–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang ACY, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cotter PA, DiRita VJ. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519–565 [DOI] [PubMed] [Google Scholar]
- 8. Craig SA, Carpenter CD, Mey AR, Wyckoff EE, Payne SM. 2011. Positive regulation of the Vibrio cholerae porin OmpT by iron and Fur. J. Bacteriol. 193:6505–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crawford JA, Kaper JB, DiRita VJ. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235–246 [DOI] [PubMed] [Google Scholar]
- 10. DiRita VJ, Mekalanos JJ. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29–37 [DOI] [PubMed] [Google Scholar]
- 11. DiRita VJ, Neely M, Taylor RK, Bruss PM. 1996. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. U. S. A. 93:7991–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DiRita VJ, Parsot C, Jander G, Mekalanos JJ. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 88:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duret G, Delcour AH. 2010. Size and dynamics of the Vibrio cholerae porins OmpU and OmpT probed by polymer exclusion. Biophys. J. 98:1820–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finkelstein RA, LoSpalluto JJ. 1970. Production of highly purified choleragen and choleragenoid. J. Infect. Dis. 121(Suppl):63. [DOI] [PubMed] [Google Scholar]
- 15. Freter R, O'Brien PC, Macsai MS. 1979. Effect of chemotaxis on the interaction of cholera vibrios with intestinal mucosa. Am. J. Clin. Nutr. 32:128–132 [DOI] [PubMed] [Google Scholar]
- 16. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henderson DP, Payne SM. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7:461–469 [DOI] [PubMed] [Google Scholar]
- 18. Herrington DA, et al. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higgins DE, Nazareno E, DiRita VJ. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hung DT, Mekalanos JJ. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. U. S. A. 102:3028–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanjilal S, Citorik R, LaRocque RC, Ramoni MF, Calderwood SB. 2010. A systems biology approach to modeling Vibrio cholerae gene expression under virulence-inducing conditions. J. Bacteriol. 192:4300–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krukonis ES, Yu RR, DiRita VJ. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67–84 [DOI] [PubMed] [Google Scholar]
- 23. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:59–64 [DOI] [PubMed] [Google Scholar]
- 24. LaRocque RC, et al. 2008. Proteomic analysis of Vibrio cholerae in human stool. Infect. Immun. 76:4145–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li CC, Crawford JA, DiRita VJ, Kaper JB. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189–203 [DOI] [PubMed] [Google Scholar]
- 26. Li CC, Merrell DS, Camilli A, Kaper JB. 2002. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 43:1577–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Majdalani N, Vanderpool CK, Gottesman S. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 40:93–113 [DOI] [PubMed] [Google Scholar]
- 28. Mathur J, Waldor MK. 2004. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 72:3577–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matson JS, DiRita VJ. 2005. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 102:16403–16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McNeil LK, et al. 2007. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 35:D347–D353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medrano AI, DiRita VJ, Castillo G, Sanchez J. 1999. Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator toxT in response to culture conditions. Infect. Immun. 67:2178–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meibom KL, et al. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mekalanos JJ, et al. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551–557 [DOI] [PubMed] [Google Scholar]
- 34. Mey AR, Craig SA, Payne SM. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73:5706–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mey AR, Payne SM. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835–849 [DOI] [PubMed] [Google Scholar]
- 36. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Miller VL, DiRita VJ, Mekalanos JJ. 1989. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 171:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller VL, Taylor RK, Mekalanos JJ. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271–279 [DOI] [PubMed] [Google Scholar]
- 40. Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493–1507 [DOI] [PubMed] [Google Scholar]
- 41. O'Toole R, et al. 1999. The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J. Bacteriol. 181:4308–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ottemann KM, DiRita VJ, Mekalanos JJ. 1992. ToxR proteins with substitutions in residues conserved with OmpR fail to activate transcription from the cholera toxin promoter. J. Bacteriol. 174:6807–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parsot C, Mekalanos JJ. 1990. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc. Natl. Acad. Sci. U. S. A. 87:9898–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfau JD, Taylor RK. 1998. Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J. Bacteriol. 180:4724–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Provenzano D, Klose KE. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 97:10220–10224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Provenzano D, Schuhmacher DA, Barker JL, Klose KE. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 68:1491–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ritchie JM, Rui H, Bronson RT, Waldor MK. 2010. Back to the future: studying cholera pathogenesis using infant rabbits. mBio 1(1):e00047–00010 doi:10.1128/mBio.00047-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Runyen-Janecky LJ, Hong M, Payne SM. 1999. Virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schuhmacher DA, Klose KE. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simon EH, Tessman I. 1963. Thymidine-requiring mutants of phage T4. Proc. Natl. Acad. Sci. U. S. A. 50:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sperandio V, Giron JA, Silveira WD, Kaper JB. 1995. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 63:4433–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stasakawicz B, Dahlbeck D, Keen N, Napoli C. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84:2833–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tischler AD, Lee SH, Camilli A. 2002. The Vibrio cholerae vieSAB locus encodes a pathway contributing to cholera toxin production. J. Bacteriol. 184:4104–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 56. Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188:6515–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu Q, Dziejman M, Mekalanos JJ. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U. S. A. 100:1286–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu X, Stern AM, Liu Z, Kan B, Zhu J. 2010. Virulence regulator AphB enhances toxR transcription in Vibrio cholerae. BMC Microbiol. 10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]