Abstract
Influenza A virus matrix protein (M1) plays an important role in virus assembly and budding. Besides a well-characterized basic amino acid-rich nuclear localization signal region at positions 101 to 105, M1 contains another basic amino acid stretch at positions 76–78 that is highly conserved among influenza A and B viruses, suggesting the importance of this stretch. To understand the role of these residues in virus replication, we mutated them to either lysine (K), alanine (A), or aspartic acid (D). We could generate viruses possessing either single or combination substitutions with K or single substitution with A at any of these positions, but not those with double substitutions with A or a single substitution with D. Viruses with the single substitution with A exhibited slower growth and had lower nucleoprotein/M1 quantitative ratio in virions compared to the wild-type virus. In cells infected with a virus possessing the single substitution with A at position 77 or 78 (R77A or R78A, respectively), the mutated M1 localized in patches at the cell periphery where nucleoprotein and hemagglutinin colocalized more often than the wild-type did. Transmission electron microscopy showed that virus possessing M1 R77A or R78A, but not the wild-type virus, was present in vesicular structures, indicating a defect in virus assembly and/or budding. The M1 mutations that did not support virus generation exhibited an aberrant M1 intracellular localization and affected protein incorporation into virus-like particles. These results indicate that the basic amino acid stretch of M1 plays a critical role in influenza virus replication.
INTRODUCTION
Matrix protein M1 of influenza A virus is the most abundant protein in the virion and has multiple functions throughout the virus replication cycle. After internalization of the virus through receptor-mediated endocytosis, acidification of the virion interior, driven by the proton channel M2 (37), leads to the dissociation of viral ribonucleoprotein (vRNP) from M1 and vRNP release into the cytoplasm (4, 15, 25, 37, 45). vRNPs are then imported into the nucleus. In the nucleus, the viral RNA (vRNA) is transcribed and replicated by the vRNA polymerase subunits PB1, PB2, and PA, along with a nucleoprotein NP. Viral mRNAs are transported into the cytoplasm and translated into proteins. The polymerase subunits and NP are imported into the nucleus, synthesize vRNAs, and form vRNPs. M1 is also imported into the nucleus during the late stages of viral replication cycle and associates with the newly formed vRNPs (4, 35, 39). The vRNP-M1 complex, in association with NS2, is exported from the nucleus to the cytoplasm (4, 5, 16, 25, 28, 33, 44) and is then transported to the plasma membrane, where assembly of the viral internal components and viral envelope proteins are completed, and virions bud from the cell surface. Thus, many of the M1 functions are mediated by its binding to vRNPs (2, 8, 11, 35, 38, 46, 48, 52, 54): M1 inhibits vRNA transcription and/or replication (2, 11, 36, 48, 54–56) and controls the nuclear export of vRNP (4, 17, 25, 26, 49–51). M1 lies beneath the viral envelope and acts as a bridge between the vRNA associated with vRNP and the integral membrane proteins by interacting with their cytoplasmic tails (9, 13, 20, 24, 41, 55).
The minimum requirement of viral proteins to drive influenza A virus assembly and budding is conflicting. Early studies using various expression systems showed that M1 alone could form virus-like particles (VLPs) (12, 21). In contrast, recent work by Chen et al. (7) and Wang et al. (47) suggests that M1 cannot form VLPs by itself and that other viral proteins are required. Wang et al. also showed that M1 does not have an inherent membrane targeting signal and is trafficked to the plasma membrane with the help of M2. M1 and M2 can form VLPs together in the absence of any other viral proteins (47). Thus, M1 is an integral part of the infectious virion and plays an important role in virus assembly and budding.
M1 is encoded by viral RNA segment 7 and consists of 252 amino acids (19). On the basis of the crystallographic structural data, M1 has been divided into three major domains: the N-terminal domain (amino acid positions 2 to 67), the middle domain (amino acid positions 88 to 164), and the C-terminal domain (amino acid positions 165 to 252) (1, 14, 43) (Fig. 1A). The amino acid residues at positions 101 to 105 were identified as a nuclear localization signal (53). Mutations in these residues cause M1 to remain in the cytoplasm and not be imported into the nucleus of virus-infected cells at a nonpermissive temperature, resulting in temperature-sensitive mutants (23). These M1 mutants are replication defective with retarded viral growth (23). Recent mutational studies also showed that mutations in this region affected the morphology of the virus (6).
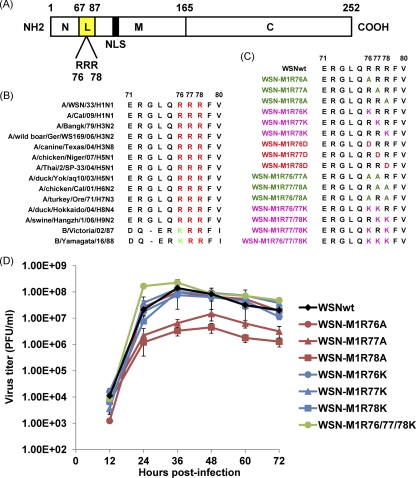
Fig 1.
Sequences of mutant M1 proteins at amino acid positions 76 to 78 and virus growth kinetics. (A) Schematic presentation of influenza A virus M1. The boundary of the three main domains is shown by the amino acid positions. N, N-terminal domain; L, the linker region (see reference 43; yellow); M, middle domain; C, C-terminal domain. The arginine residues (R) at positions 76 to 78 and the nuclear localization signal (NLS; 53) are also shown. (B) Alignment of M1 amino acid sequences of influenza A and B viruses. Shown are the amino acids at positions 71 to 80. Conserved arginine (R) and lysine (K) residues are shown in red and green, respectively. (C) M1 amino acid sequences of the mutant viruses used in the present study. Mutants are color-coded as follows: single alanine substitutions, light green; double alanine substitutions, deep green; single, double, and triple lysine substitutions, pink; single aspartic acid substitutions, red. (D) Growth kinetics of the M1 mutants. MDCK cells were infected with the indicated viruses at an MOI of 0.001. The culture supernatants collected at the indicated time points were subjected to plaque assays for virus titration. Error bars represent standard deviations of three independent experiments.
Three consecutive basic amino acid residues at positions 76 to 78 of M1, RRR, form part of the region that links the N-terminal domain to the middle domain; the residues at positions 77 and 78 could be exposed to interact with the phospholipids of the plasma membrane (43). Alignment of M1 amino acid sequences shows that these residues are highly conserved among influenza A and B viruses (Fig. 1B). The role of these residues in virus replication is not known. We hypothesized that these basic residues may form a continuously charged surface and play an important role in virus biology. Here, we examined the role of the arginine residues at positions 76 to 78 (R76, R77, and R78) of M1 in the virus replication cycle by systematically analyzing the effect of mutations at these positions in a minireplicon assay, a VLP assay, and on virus recovery.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Madin-Darby canine kidney (MDCK) cells were grown in minimal essential medium (MEM) containing 5% newborn calf serum, vitamins, essential amino acids, and antibiotics. 293 and 293T human embryonic kidney cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics. A549 human lung adenocarcinoma epithelial cells were grown in DMEM–F-12 containing 10% FBS and antibiotics. All cells were cultured at 37°C with 5% CO2.
Influenza virus A/WSN/33 (H1N1) (WSN) and its M1 mutants were generated by use of plasmid-based reverse genetics (see below) and amplified in MDCK cells grown in MEM containing 0.3% bovine serum albumin and 0.5 to 0.75 μg of TPCK [l-(tosylamido-2-phenyl)ethyl chloromethyl ketone]-treated trypsin/ml. At 48 h postinfection, the culture supernatant was harvested, clarified, divided into aliquots, and stored at −80°C. The virus titer was determined by using plaque assays in MDCK cells.
Anti-M1 mouse monoclonal antibodies (clone C111 [Takara Bio, Inc., Otsu, Japan] and clone GA2B [AbD Serotec, Oxford, United Kingdom]), an anti-NP mouse monoclonal antibody (clone AA5H [Abcam]), and an anti-M1 goat polyclonal antibody (catalog no. ab20910 [Abcam]) were used according to the manufacturer's instructions. An anti-HA mouse monoclonal antibody (clone WS 3-54) was kindly provided by Emi Takashita (National Institute of Infectious Diseases, Tokyo, Japan). An anti-WSN virus rabbit polyclonal antibody (R309) was prepared in our laboratory.
Plasmid construction.
The cDNA encoding the WSN virus M gene was amplified by use of PCR and cloned into the protein expression plasmid pCAGGS/MCS (18, 31) or the viral RNA expression plasmid pHH21 (29). Site-directed mutations were introduced into the plasmids by using the PCR-based megaprimer method (42). Primer sequences are available upon request.
Reverse genetics.
The wild-type WSN virus and its M1 mutants were generated by using a plasmid-based reverse genetics as described previously (29). In brief, 293T cells grown in 35-mm dishes to 70% confluence were transfected with 0.1 μg each of the eight PolI plasmids for each vRNA segment of WSN virus along with 1 μg each of the four protein expression plasmids for the viral polymerase subunits (PB1, PB2, and PA and nucleoprotein NP). At 48 h posttransfection, 0.5 to 0.75 μg of TPCK-trypsin/ml was added to the cells, which were then incubated for 1 h. The culture supernatant was then harvested, clarified, and passed onto fresh MDCK cells to monitor the virus recovery, by observing the cytopathic effect (CPE), and amplify the viruses. The identity of the recovered viruses was confirmed by sequencing of vRNA extracted from culture supernatants. The recovered viruses were named based on their mutated M1 as follows: WSN-M1R76A, WSN-M1R77A, WSN-M1R78A, WSN-M1R76K, WSN-M1R77K, WSN-M1R78K, and so on.
Reverse transcription-PCR.
vRNA was isolated from culture supernatant harvested from virus-infected MDCK cells by using an RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The extracted vRNA was reverse transcribed and amplified by using the SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) with a primer set specific for the M gene. PCR products were analyzed by means of electrophoresis on a 0.75% agarose gel. The PCR products were then sequenced to confirm the introduced mutations.
Virus growth kinetics.
MDCK cells grown in 60-mm tissue culture dishes were infected with virus at a multiplicity of infection (MOI) of 0.001. Culture supernatants were harvested at 12-h intervals, clarified, and frozen at −80°C. The virus titer in the culture supernatant at each time point was determined by use of plaque assays.
M1 expression in plasmid-transfected cells.
293T cells grown in 12-well tissue culture plates to 70% confluence were transfected with 1 μg of M1 expression plasmid. At 48 h posttransfection, cells were washed with phosphate-buffered saline (PBS) twice, lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]), and subjected to Western blotting.
VLP budding assay.
293T cells grown in 60-mm dishes to 70% confluence were transfected with 2 μg each of protein expression plasmids for M1, HA, NA, NP, M2, and NS2. At 48 h posttransfection, culture supernatant was harvested, clarified, loaded on a 20% sucrose cushion, and ultracentrifuged at 60,000 rpm for 2 h. The pelleted VLPs were rehydrated in PBS overnight at 4°C and analyzed by means of Western blotting. The transfected cells were lysed with RIPA buffer and subjected to Western blotting.
Minireplicon assay.
vRNA transcription and replication was assessed by using a minireplicon assay as described previously (34). Briefly, 293Tcells grown in 24-well tissue culture plates to 70% confluence were transfected with pPolI-WSN-NA-firefly-luciferase (22), along with the expression plasmids for the viral polymerase subunits (i.e., PA, PB1, and PB2) and an expression plasmid for NP. To assess the effect of M1 expression on vRNA transcription and replication, we included an expression plasmid for either wild-type or mutated M1 in the plasmid mixture along with the expression plasmid for NS2. pGL4.74[hRluc/TK] (Promega, Madison, WI) was also included in the plasmid mixture as an internal control for the dual-luciferase assay. At 36 h posttransfection, the firefly and Renilla luciferase activities in cells were measured on a microplate reader (Infinite M1000 [Tecan]) by using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Western blotting.
Protein samples were mixed with 2× sample buffer and loaded on precast 4 to 20% SDS-PAGE gels (Bio-Rad, Hercules, CA). After fractionation, the proteins were transferred onto nitrocellulose membranes by using an iBlot dry blotting system (Invitrogen). The membranes were blocked for 16 h at 4°C with PBS with 0.05% Tween 20 (PBS-T) containing 5% skimmed milk. Membranes were then incubated with primary antibodies for 1 h at room temperature. To detect M1, NP, and HA, anti-M1 goat polyclonal (1:2,500), anti-NP mouse monoclonal (1:1,000), and anti-HA mouse monoclonal (1:2,000) antibodies respectively, were used. To detect the major viral proteins (i.e., HA, NP, and M1), the anti-WSN virus rabbit polyclonal antibody (1:10,000) was also used. After three washes with PBS-T for 5 min each time, membranes were incubated with the anti-mouse (1:10,000), anti-goat (1:10,000), or anti-rabbit (1:20,000) secondary antibodies conjugated with horseradish peroxidase (Invitrogen) for 1 h at room temperature. After three washes with PBS-T for 15 min each time, blots were developed by using a chemiluminescence kit (Roche Applied Science, Basel, Switzerland) and visualized following autoradiography. Autoradiographs were scanned and quantitated by using ImageJ software (National Institutes of Health [NIH]).
Immunofluorescence staining.
A549 cells grown on glass coverslips in 12- or 24-well tissue culture plates to 30 to 50% confluence were either transfected with 0.5 μg of M1 expression plasmid or infected with viruses at an MOI of 5. Cells were incubated for various lengths of time and then fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 15 min. They were then washed with PBS three times, permeabilized with PBS-T containing 0.2% Triton X-100 for 15 min, and blocked with PBS-T containing either 5% normal mouse serum or 10% normal goat serum for 30 min at room temperature. The cells were then incubated with an anti-M1 mouse monoclonal antibody (1:200) for 1 h at room temperature, washed with PBS-T three times, and then incubated with an anti-mouse secondary antibody conjugated with Alexa Fluor 488 (Invitrogen) at 1:1,000 for 1 h at room temperature. After being washed with PBS-T three times and counterstained with Hoechst 33258 (Invitrogen) for 10 min, the cells were rinsed in distilled water, partially dried by draining the excess liquid, and mounted on a glass slide with Immuno-Mount (Shandon, Pittsburgh, PA).
For four color immunofluorescence staining, anti-M1 and anti-NP antibodies were labeled with the fluorescent dyes Alexa Fluor 488 and Alexa Fluor 647 (Invitrogen), respectively. Virus-infected cells were incubated with the anti-M1 (1:100)- and anti-NP (1:300)-labeled antibodies together with an anti-HA mouse monoclonal antibody (1:300), followed by incubation with an anti-mouse secondary antibody conjugated with Alexa Fluor 594 (Invitrogen) at 1:1,000 and Hoechst 33258 (Invitrogen) in PBS-T for 30 min at room temperature.
Cell samples were observed under a confocal laser scanning microscope LSM510META (Carl Zeiss, Jena, Germany). Raw images were exported as TIFF files and cropped as appropriate by using Adobe Photoshop CS4 without altering the actual image attributes.
Transmission electron microscopy.
Ultrathin-section electron microscopy was performed as described previously (32). Briefly, virus-infected cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer at 12 h postinfection and postfixed with 2% osmium tetroxide in the same buffer. The cells were then dehydrated with a series of ethanol gradients followed by propylene oxide before being embedded in Epon 812 resin mixture (TAAB Laboratories Equipment, Berkshire, United Kingdom). Thin sections were stained with 2% uranyl acetate and Raynold's lead and examined under a Tecnai F20 electron microscope (FEI, Hillsboro, OR) at 80 kV.
RESULTS
Introduction of mutations into M1.
An alignment of M1 amino acid sequences showed that arginine residues at positions 76 to 78 are highly conserved among influenza A viruses, including the 2009 H1N1 pandemic and highly pathogenic H5N1 viruses (Fig. 1B). Although the arginine at position 76 of influenza A virus M1 is replaced with a lysine at the same position in the M1 of influenza B virus (Fig. 1B), this high conservation suggests that the basic amino acid residues at these positions may play an important role(s) in influenza virus replication. To examine this role, we used site-directed mutagenesis to construct protein expression plasmids for M1 in which lysine (K, conservative), alanine (A), or aspartic acid (D, nonconservative) replaced the arginines at one, two, or three positions (Fig. 1C). These plasmids expressed M1 protein of the expected size with no major effect on the expression level in transfected cells (data not shown). We also introduced the same mutations into the vRNA expression plasmids for M vRNA.
Importance of the arginines for virus recovery.
To examine whether the conservation of the amino acids at positions 76 to 78 is critical for virus viability, we attempted to generate viruses possessing mutant M1 by using plasmid-based reverse genetics (29). Virus recovery was confirmed by means of CPE (Table 1). The identity of the viruses was confirmed by sequencing of the vRNA extracted from viruses passaged in MDCK cells three times. Single, double, or triple substitutions with lysine resulted in recovery of infectious viruses with growth properties similar to those of the wild-type virus. The single substitution with alanine at one of these positions (WSN-M1R76A, WSN-M1R77A, and WSN-M1R78A) did not affect virus generation, although the generated viruses exhibited slower growth than that of the wild-type virus. In contrast, the double substitution with alanine was detrimental to virus generation. The single substitution with aspartic acid at any of these positions did not support virus production. These results indicate that basic amino acids at positions 76 to 78 of M1 are critical for virus replication.
Table 1.
Virus recovery by use of reverse geneticsa
| Virus | Supernatant of plasmid-transfected cells collected at: |
Virus recovery | |||
|---|---|---|---|---|---|
| 48 hpt and CPE in MDCK cells observed at: |
72 hpt and CPE in MDCK cells observed at: |
||||
| 24 hpi | 48 hpi | 24 hpi | 48 hpi | ||
| Wild-type WSN | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R76A | + | ++ | ++ | +++ | Yes |
| WSN-M1R77A | – | ++ | – | ++ | Yes |
| WSN-M1R78A | – | – | – | + | Yes |
| WSN-M1R76D | – | – | – | – | No |
| WSN-M1R77D | – | – | – | – | No |
| WSN-M1R78D | – | – | – | – | No |
| WSN-M1R76K | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R77K | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R78K | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R76/77A | – | – | – | – | No |
| WSN-M1R76/78A | – | – | – | – | No |
| WSN-M1R77/78A | – | – | – | – | No |
| WSN-M1R76/77K | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R76/78K | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R77/78K | ++ | ++++ | +++ | ++++ | Yes |
| WSN-M1R76/77/78K | ++ | ++++ | +++ | ++++ | Yes |
293T cells were transfected with a vRNA expression plasmid for either the wild-type or mutated M1 vRNA segment along with the vRNA expression plasmids for the remaining seven vRNA segments and four protein expression plasmids for PB2, PB1, PA, and NP. Culture supernatants were harvested at 48 and 72 h post transfection and inoculated into MDCK cells. The cytopathic effect (CPE) was monitored for 48 to 72 h. +, Only a few cells showed a CPE; ++, ca. 50% of the cells showed a CPE; +++, 75% of the cells showed a CPE; ++++, 100% of the cells showed a CPE. hpt, hours post transfection;hpi, hours postinfection.
Importance of the arginines for virus growth.
To further characterize the M1 mutants that possessed the single substitution with alanine at position 76, 77, or 78, we determined the growth kinetics of the mutant viruses in MDCK cells. Consistent with the results of virus production (Table 1), both WSN-M1R77A and WSN-M1R78A grew more slowly than did the wild-type virus (Fig. 1D). There was an ∼1-log difference in virus titer at most of the time points tested. However, the growth kinetics of WSN-M1R76A, which exhibited attenuated growth during virus generation (Table 1), were comparable to those of the wild-type virus. The growth kinetics of the lysine substitution-possessing M1 mutants (WSN-M1R76K, WSN-M1R77K, and WSN-M1R78K) were comparable to or even higher (WSN-M1R76/77/78K) than those of the wild-type virus. The growth kinetics of the M1 mutants possessing the double substitutions with lysine (WSN-M1R76/77K, WSN-M1R77/78K, and WSN-M1R76/78K) were also similar to those of the wild-type virus (data not shown), supporting the contention that basic amino acids at these positions are important for virus replication.
Importance of the arginines for incorporation of viral proteins into virions.
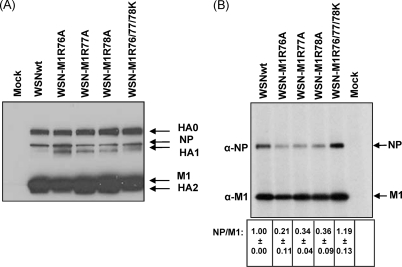
To assess the impact of the single arginine-to-alanine substitution at position 76, 77, or 78 of M1 on viral protein incorporation into virions, we examined the relative amount of viral proteins in virus-infected cells at 8 h postinfection and also in virions. The expression pattern of the major viral proteins (i.e., HA, NP, and M1) in all of the virus-infected cells was essentially similar (Fig. 2A), although the expression level of HA1 in the wild-type virus-infected cells was considerably lower than that in the M1 mutant-infected cells. Under these conditions, although the amounts of M1 in the culture supernatant were similar among the test viruses, the NP/M1 ratio in WSN-M1R76A, WSN-M1R77A, and WSN-M1R78A particles was significantly lower (P < 0.05, unpaired two-tailed t test) than that in the wild-type virus particles (Fig. 2B). In contrast, the NP/M1 quantitative ratio in WSN-M1R76/77/78K virions was comparable (P = 0.128) to that in the wild-type virus particles. These data suggest that the incorporation of NP into virions is affected by the arginine-to-alanine substitution at position 76, 77, or 78 of M1. These results also suggest that the slower growth kinetics of viruses possessing these M1 mutations may be partly attributed to the defective incorporation of viral proteins into virions.
Fig 2.
Viral proteins in virus-infected cells and virions. (A) Expression of viral proteins in virus-infected cells. MDCK cells were infected with the indicated viruses at an MOI of 5 and were grown in the presence of TPCK-trypsin. Cytoplasmic extracts were prepared at 8 h postinfection and analyzed by use of Western blotting with a polyclonal antibody against WSN virus. Arrows to the right indicate the positions of the viral proteins. (B) Viral proteins in virions. Viruses grown in MDCK cells in the presence of TPCK-trypsin were concentrated by ultracentrifugation through a sucrose cushion and resuspended in RIPA buffer. The protein samples were then analyzed by means of Western blotting with either anti-NP or anti-M1 monoclonal antibody. The intensity of bands for NP and M1 was quantitated by using ImageJ software (NIH). The NP/M1 quantitative ratio was calculated (the value for the wild-type virus was taken as 1), and the averages from three independent experiments (with standard deviations) are shown at the bottom of the gel image. Arrows to the right indicate the position of the viral proteins.
Importance of the arginines for intracellular trafficking of viral proteins.
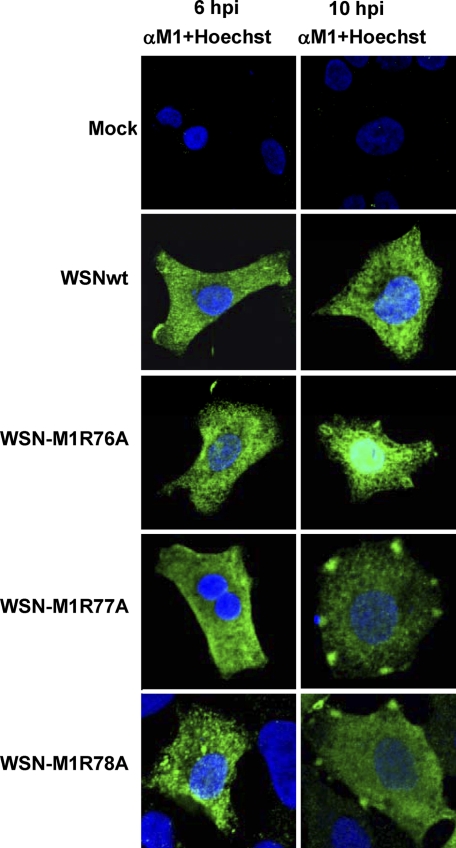
We next examined the intracellular distribution of M1 in cells infected with virus possessing the single substitution with alanine at position 76, 77, or 78 by using indirect immunofluorescence assays. A549 cells were used for immunofluorescence staining, because these cells allowed us to obtain clearer and more reproducible results than those with MDCK cells. Note that previous studies have demonstrated that both the viral protein distribution pattern (10) and the growth kinetics (17) of WSN virus in A549 cells are comparable to those in MDCK cells. At 6 h postinfection, there was no remarkable difference in the M1 localization among WSN-M1R76A-, WSN-M1R77A-, WSN-M1R78A-, and wild-type virus-infected cells (Fig. 3, left panels). In contrast, at 10 h postinfection, both M1R77A and M1R78A showed a distinctly different localization pattern from that of the wild-type M1 in virus-infected cells: in addition to cytoplasmic localization, these mutated M1s were found in patches at the cell periphery. This unique M1 accumulation was observed over time and in ca. 20% of the cells examined; the rest of the M1 mutant-infected cells showed M1 distribution comparable to that in the wild-type virus-infected cells. In the wild-type virus-infected cells, only a few (0.5 to 1%) showed the M1 patches at the cell periphery. The M1 distribution pattern in cells infected with WSN-M1R76A was similar to that of the wild-type virus, although more nuclear M1 was observed in the WSN-M1R76A-infected cells. These data suggest that M1 with the alanine substitution at position 77 or 78 is arrested at the cell periphery for longer than the wild type and, thus, differs from the wild-type in its intracellular localization especially at the cell periphery, which likely affects viral growth kinetics.
Fig 3.
Intracellular distribution of viral proteins in virus-infected cells. A549 cells were infected with the indicated viruses at an MOI of 5. Cells were fixed at 6 h (left panels) or 10 h (right panels) postinfection and subjected to an immunofluorescence assay with an anti-M1 monoclonal antibody (green). The nucleus was counterstained with Hoechst (blue). Slides were observed under a confocal laser scanning microscope with an objective with ×40 magnification, and raw images were processed as described in Materials and Methods.
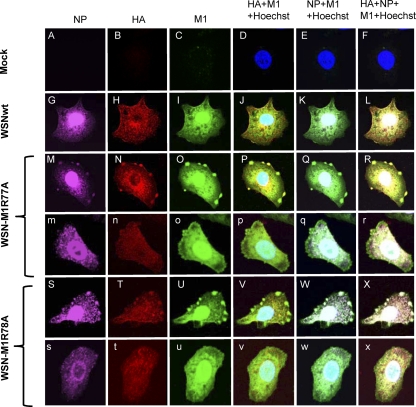
We also examined whether the other major viral proteins HA and NP colocalize with the mutant M1 in the patches at the cell periphery by using fluorescent dye-labeled antibodies (Fig. 4). In the wild-type virus-infected cells, NP (Fig. 4G) colocalized with the M1 that was distributed in the nucleus and cytoplasm (Fig. 4I). HA also colocalized with the M1 in the cytoplasm, although a portion of this glycoprotein also accumulated in the perinuclear region. Under these conditions, both HA and NP colocalized with the M1 in the patches in the cells that were infected with WSN-M1R77A (Fig. 4M to R) or WSN-M1R78A (Fig. 4S to X).
Fig 4.
Colocalization of viral proteins in virus-infected cells. A549 cells were infected with the indicated viruses at an MOI of 5. At 12 h postinfection, cells were fixed and incubated with an anti-NP (A, G, M, m, S, and s), anti-HA (B, H, N, n, T, and t), or anti-M1 (C, I, O, o, U, and u) monoclonal antibody. The nucleus was counterstained with Hoechst 33258. D, J, P, p, V, and v, merged images for HA and M1; E, K, Q, q, W, and w, merged images for NP and M1; F, L, R, r, X, and x, merged images for NP, HA, and M1.
Importance of the arginines for virus budding.
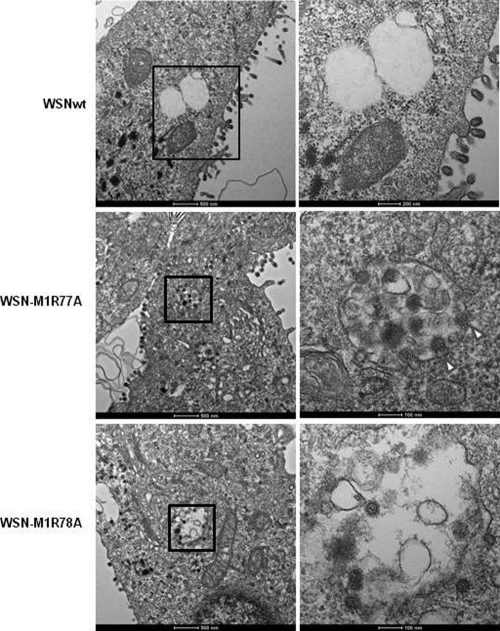
To further examine the architecture of the patches at the cell periphery, we analyzed the virus-infected cells by using transmission electron microscopy at 12 h postinfection (Fig. 5). In cells infected with the wild-type virus, spherical virions were observed on the plasma membrane. In contrast, in cells infected with either WSN-M1R77A or WSN-M1R78A, many virions were detected in intracellular vesicles and some virions appeared to be budding into these vesicles (Fig. 5, WSN-M1R77A). These events were not observed in cells infected with the wild-type virus. The morphology of the budding virions of the wild-type virus and those containing the M1 mutants were indistinguishable; all were spherical to oval-shaped. Taken together, these results indicate that the single arginine-to-alanine substitution at position 77 or 78 of M1 resulted in aberrant virus budding.
Fig 5.
Ultrastructural analysis of virus-infected cells. MDCK cells infected with the indicated viruses were examined under a transmission electron microscope at 12 h postinfection. Right panels show enlargements (2.5× magnification for WSNwt and 5× magnification for WSN-M1R77A and WSN-M1R78A) of the intracellular vesicles containing virions shown in the left panels. Arrowheads indicate budding virions.
Impact on virus replication of the M1 mutations that did not support virus recovery cycle.
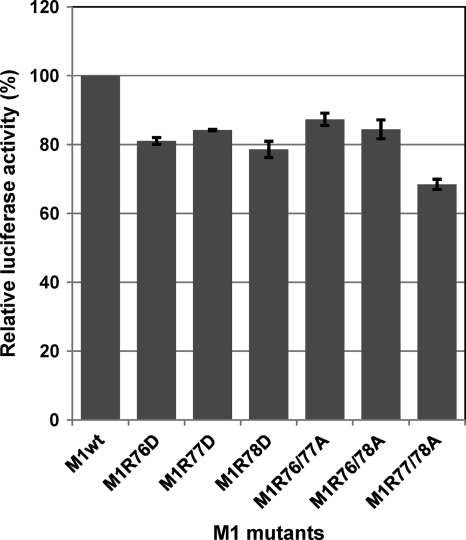
To further assess the role of the arginines at positions 76 to 78 of M1 in virus replication, we next focused on the M1 mutations that did not allow the generation of infectious viruses (Table 1): the single substitution with aspartic acid and the double substitutions with alanine. Since studies have demonstrated that M1 inhibits vRNA transcription and/or replication (2, 11, 36, 48), the impact of these substitutions on vRNA transcription and replication were assessed by using minigenome assays in the presence of M1 and NS2 (NS2 interacts with M1 and regulates M1's inhibitory effect on vRNA transcription and replication [40]). The results showed that the impact of these M1 substitutions was limited, with levels of vRNA transcription and replication in the presence of these mutant M1s being 70 to 90% of those of the wild-type M1 (Fig. 6). Mutant M1s with the single alanine substitution at these positions also showed a level of vRNA transcription and replication comparable to that of the wild-type M1 (data not shown). These results indicate that the effect of the arginines on vRNA transcription and/or replication is limited.
Fig 6.
Effect of the mutations on vRNA transcription and replication. 293T cells were transfected with expression plasmids for PB2, PB1, PA, NP, NS2 and Renilla luciferase (internal control) and plasmid for the expression of an influenza viral minigenome encoding the firefly luciferase gene along with either a wild-type or mutant M1-expressing plasmid. At 48 h posttransfection, the cells were subjected to the dual-luciferase assay. The value for the wild-type M1 was set as 100%. The data from three independent experiments are shown. Error bars represent standard deviations of three independent experiments.
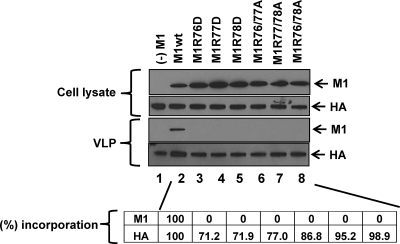
To test whether these M1 mutations support VLP formation, 293T cells were transfected with an expression plasmid for either the wild-type or mutated M1, along with the expression plasmids for HA, NA, NP, M2, and NS2, all of which are known to support VLP formation (7, 27, 30, 47). We then used Western blotting to detect viral proteins in the culture supernatants. Wild-type M1, as well as HA, was detected in the culture supernatants (Fig. 7, lane 2). In contrast, none of the M1 mutants tested (i.e., M1R76D, M1R77D, M1R78D, M1R76/77A, M1R77/78A, and M1R76/78A) was detected, although HA was detectable (Fig. 7, lanes 3 to 8). Comparable amounts of M1 and HA were expressed in all of the plasmid-transfected cell samples (Fig. 7, upper panels). Note that even in the absence of the M1-expressing plasmid (Fig. 8, left column), HA was detected in the culture supernatant from the transfected cells, because HA and NA are sufficient for VLP formation and budding (7). These results indicate that the incorporation of the tested M1 mutants into VLPs was severely impaired.
Fig 7.
Effect of the mutations on M1 incorporation into VLPs. 293T cells were transfected with an expression plasmid for either wild-type or mutated M1 along with expression plasmids for HA, NA, NP, M2, and NS2. At 48 h posttransfection, culture supernatants were subjected to Western blotting with anti-M1 and anti-HA monoclonal antibodies. Proteins in the cell lysates and VLPs are shown in the upper and lower panels, respectively. The intensity of bands for M1 and HA was quantified by using ImageJ software (NIH). The percentage of incorporation of M1 and HA into VLPs was calculated (the value for the wild-type VLP was set as 100%) by using the following formula: {ratio of the amount of protein in the mutant VLP to the total amount of mutant protein (VLP + cell lysate)/ratio of the amount of protein in the wild-type VLP to the total amount of wild-type protein (VLP + cell lysate)} × 100. The averages of three independent experiments are shown at the bottom of the gel images.
Fig 8.
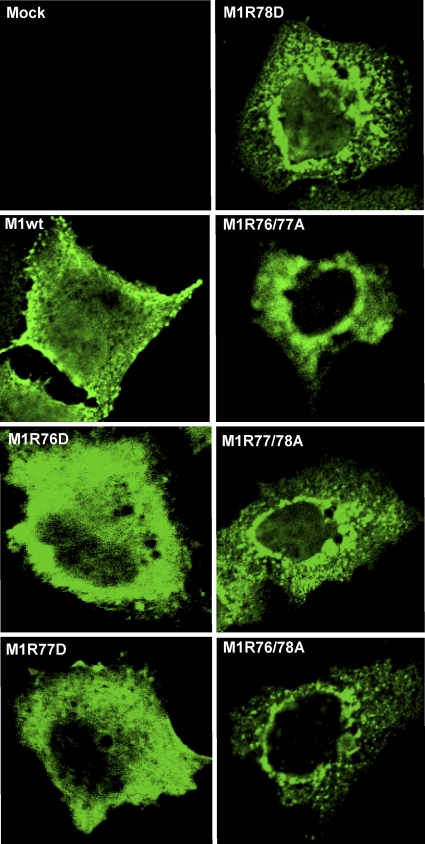
Intracellular distribution of mutant proteins in plasmid-transfected cells. A549 cells were transfected with an expression plasmid for either the wild-type or mutated M1 along with expression plasmids for HA and NA. At 24 h posttransfection, the cells were fixed and stained with an anti-M1 monoclonal antibody. Slides were observed under a confocal laser scanning microscope with an objective with ×40 magnification, and the images were processed as described in Materials and Methods.
Since M1R76D, M1R77D, M1R78D, M1R76/77A, M1R77/78A, and M1R76/78A were not efficiently incorporated into VLPs, we examined their intracellular distribution in cells transfected with the expression plasmid sets that were used for the VLP formation assays by use of indirect immunofluorescence assays (Fig. 8). At 12 h posttransfection, wild-type M1 localized in the cytoplasm and the nucleus and partially at the cell periphery. In cells transfected with the expression plasmids for M1R76D and M1R77D, there was partial accumulation of M1 close to the nucleus, whereas most of the M1 was diffusely distributed throughout the cytoplasm. The other M1 mutants (i.e., M1R78D, M1R76/77A, M1R77/78A, and M1R76/78A) tended to accumulate at the periphery of the nucleus. These data indicate that the M1 mutants that did not support virus recovery are aberrant in intracellular trafficking.
DISCUSSION
In the present study we examined the role in virus replication of the arginine residues at positions 76 to 78 of influenza A virus M1. By using site-directed mutagenesis and reverse genetics, we showed that these residues play an important role in virus replication. Single, double, or triple substitution of the arginines with either alanine or aspartic acid resulted in either the failure of virus recovery or the generation of viruses with slower growth kinetics. The mutant M1s that did not support virus recovery were defective in intracellular trafficking and failed to be incorporated into VLPs. Furthermore, our results show that some of these M1 mutants exhibited abnormal virus budding. Our study thus demonstrates the importance of these amino acids for M1 intracellular trafficking, virus assembly, and virus budding.
Although M1 mutants possessing the single arginine-to-alanine substitution were recovered, the double alanine substitutions did not support virus recovery (Table 1). Moreover, the single substitution with aspartic acid at any position abrogated virus recovery, whereas substitutions with lysine supported virus recovery (Table 1). Growth curve analysis demonstrated that the M1 mutants WSN-M1R77A and WSN-M1R78A had slower growth kinetics compared to those of the wild-type virus (Fig. 1D), although the overall levels of protein synthesis in the virus-infected cells were comparable (Fig. 2A). These results suggest that the positive charge of the basic amino acid residues in this region is necessary for optimal virus replication at the posttranslational stages. In fact, on the basis of crystallographic data, Sha and Luo (43) proposed that amino acids at positions 77 to 79 would fall in the linker region that connects the N-terminal and middle domains of M1 and might be critical in maintaining the spatial orientation of helical structures. Changing the charges of any of these residues would affect the overall charge in the region, resulting in perturbation of the spatial arrangement of the N-terminal and middle domains. These findings are further supported by the conserved nature of the basic amino acids among influenza A and B viruses (Fig. 1B). Sha and Luo (43) also reported that the amino acids at positions 77 and 78 could be oriented in such a way that they could interact with the phospholipids of the plasma membrane. Altering the charges of these M1 amino acids could prevent such an interaction between M1 and the plasma membrane and thus interfere with the incorporation of M1 into VLPs. The results of our VLP assay (Fig. 7) support this contention.
Immunofluorescence analysis of virus-infected cells revealed that M1R77A and M1R78A accumulated in patches at the cell periphery during the late stage of infection (Fig. 3 and 4). Excessive accumulation of M1 and other viral proteins in patches at the cell periphery was observed only in WSN-M1R77A- and WSN-M1R78A-infected cells. There are two possible explanations for the slower growth kinetics of the M1 mutants: (i) despite of the presence of all of the virion components at the budding site, mutated M1-mediated virion assembly may be inefficient, or (ii) the budding of virions from the M1 mutant-infected cells may be abnormal. The electron microscopy data are consistent with the latter possibility in that they showed the budding of these M1 mutants to be aberrant (Fig. 5). This defective budding could also be due to defective interactions between M1 and host cellular factors. Recently, it was demonstrated that influenza A virus budding takes place in an endosomal sorting complex required for transport (ESCRT)-independent pathway and is mediated by one of the small GTPase family proteins Rab11 (3, 10). It is tempting to speculate that the mutant M1s may have lost the ability to interact with Rab11 or another host cellular protein that is required for efficient virus budding.
In conclusion, our study identifies the importance of the conserved amino acids in influenza A virus M1 for virus replication, specifically the intracellular trafficking of M1, virus assembly, and virus budding. The novel mutants generated here may serve as a tool to better understand these processes. Further research is warranted to identify the host factors that might have lost the ability to interact with M1 due to these mutations and the role they play in influenza virus assembly and budding. A better understanding of these interactions may help in the design of strategies for influenza virus control and prevention.
ACKNOWLEDGMENTS
We thank Susan Watson for editing the manuscript.
This study was supported by ERATO (Japan Science and Technology Agency); by a grant-in-aid for Specially Promoted Research from the Ministries of Education, Culture, Sports, Science, and Technology; by grants-in-aid from Health, Labor, and Welfare of Japan; by a Contract Research Fund for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases; and by National Institute of Allergy and Infectious Disease Public Health Service research grants.
The authors declare that they have no conflicts of interest.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Arzt S, et al. 2001. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology 279:439–446 [DOI] [PubMed] [Google Scholar]
- 2. Baudin F, Petit I, Weissenhorn W, Ruigrok RW. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102–108 [DOI] [PubMed] [Google Scholar]
- 3. Bruce EA, Digard P, Stuart AD. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 84:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bui M, Whittaker G, Helenius A. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bui M, Wills EG, Helenius A, Whittaker GR. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74:1781–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. 2005. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 79:1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen BJ, Leser GP, Morita E, Lamb RA. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhiman N, et al. 2010. Evidence for amino acid changes in a 57-base-pair region of the highly conserved matrix gene of pandemic (H1N1) 2009 influenza A virus. J. Clin. Microbiol. 48:3817–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eierhoff T, Ludwig S, Ehrhardt C. 2009. The influenza A virus matrix protein as a marker to monitor initial virus internalisation. Biol. Chem. 390:509–515 [DOI] [PubMed] [Google Scholar]
- 10. Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. 2011. RAB11A is essential for influenza genome transport to the plasma membrane. J. Virol. 85:6117–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elster C, Larsen K, Gagnon J, Ruigrok RW, Baudin F. 1997. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 78(Pt 7):1589–1596 [DOI] [PubMed] [Google Scholar]
- 12. Gomez-Puertas P, Albo C, Perez-Pastrana E, Vivo A, Portela A. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538–11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gregoriades A. 1980. Interaction of influenza M protein with viral lipid and phosphatidylcholine vesicles. J. Virol. 36:470–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris A, Forouhar F, Qiu S, Sha B, Luo M. 2001. The crystal structure of the influenza matrix protein M1 at neutral pH: M1–M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 289:34–44 [DOI] [PubMed] [Google Scholar]
- 15. Helenius A. 1992. Unpacking the incoming influenza virus. Cell 69:577–578 [DOI] [PubMed] [Google Scholar]
- 16. Herz C, Stavnezer E, Krug R, Gurney T., Jr 1981. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell 26:391–400 [DOI] [PubMed] [Google Scholar]
- 17. Huang X, Liu T, Muller J, Levandowski RA, Ye Z. 2001. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology 287:405–416 [DOI] [PubMed] [Google Scholar]
- 18. Kobasa D, Rodgers ME, Wells K, Kawaoka Y. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 71:6706–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamb RA, Lai CJ, Choppin PW. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. U. S. A. 78:4170–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamb RA, Zebedee SL, Richardson CD. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627–633 [DOI] [PubMed] [Google Scholar]
- 21. Latham T, Galarza JM. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z, et al. 2009. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J. Virol. 83:4153–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu T, Ye Z. 2004. Introduction of a temperature-sensitive phenotype into influenza A/WSN/33 virus by altering the basic amino acid domain of influenza virus matrix protein. J. Virol. 78:9585–9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X, et al. 2009. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell Microbiol. 11:730–741 [DOI] [PubMed] [Google Scholar]
- 25. Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117–130 [DOI] [PubMed] [Google Scholar]
- 26. Martin K, Helenius A. 1991. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 65:232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mena I, Vivo A, Perez E, Portela A. 1996. Rescue of a synthetic chloramphenicol acetyltransferase RNA into influenza virus-like particles obtained from recombinant plasmids. J. Virol. 70:5016–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neumann G, et al. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neumann G, Watanabe T, Kawaoka Y. 2000. Plasmid-driven formation of influenza virus-like particles. J. Virol. 74:547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 32. Noda T, et al. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439:490–492 [DOI] [PubMed] [Google Scholar]
- 33. O'Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ozawa M, et al. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 81:30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patterson S, Gross J, Oxford JS. 1988. The intracellular distribution of influenza virus matrix protein and nucleoprotein in infected cells and their relationship to haemagglutinin in the plasma membrane. J. Gen. Virol. 69(Pt 8):1859–1872 [DOI] [PubMed] [Google Scholar]
- 36. Perez DR, Donis RO. 1998. The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology 249:52–61 [DOI] [PubMed] [Google Scholar]
- 37. Pinto LH, Holsinger LJ, Lamb RA. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528 [DOI] [PubMed] [Google Scholar]
- 38. Rees PJ, Dimmock NJ. 1981. Electrophoretic separation of influenza virus ribonucleoproteins. J. Gen. Virol. 53:125–132 [DOI] [PubMed] [Google Scholar]
- 39. Rey O, Nayak DP. 1992. Nuclear retention of M1 protein in a temperature-sensitive mutant of influenza (A/WSN/33) virus does not affect nuclear export of viral ribonucleoproteins. J. Virol. 66:5815–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robb NC, Smith M, Vreede FT, Fodor E. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398–1407 [DOI] [PubMed] [Google Scholar]
- 41. Robertson BH, Bennett JC, Compans RW. 1982. Selective dansylation of M protein within intact influenza virions. J. Virol. 44:871–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sarkar G, Sommer SS. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8:404–407 [PubMed] [Google Scholar]
- 43. Sha B, Luo M. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 4:239–244 [DOI] [PubMed] [Google Scholar]
- 44. Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. 2011. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS Lett. 585:41–46 [DOI] [PubMed] [Google Scholar]
- 45. Sugrue RJ, Hay AJ. 1991. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology 180:617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wakefield L, Brownlee GG. 1989. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 17:8569–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang D, et al. 2010. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J. Virol. 84:4673–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe K, Handa H, Mizumoto K, Nagata K. 1996. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J. Virol. 70:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Whittaker G, Bui M, Helenius A. 1996. Nuclear trafficking of influenza virus ribonuleoproteins in heterokaryons. J. Virol. 70:2743–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Whittaker G, Bui M, Helenius A. 1996. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 6:67–71 [DOI] [PubMed] [Google Scholar]
- 51. Whittaker G, Kemler I, Helenius A. 1995. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J. Virol. 69:439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 73:7467–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ye Z, Robinson D, Wagner RR. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 69:1964–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ye ZP, Baylor NW, Wagner RR. 1989. Transcription-inhibition and RNA-binding domains of influenza A virus matrix protein mapped with anti-idiotypic antibodies and synthetic peptides. J. Virol. 63:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye ZP, Pal R, Fox JW, Wagner RR. 1987. Functional and antigenic domains of the matrix (M1) protein of influenza A virus. J. Virol. 61:239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zvonarjev AY, Ghendon YZ. 1980. Influence of membrane (M) protein on influenza A virus virion transcriptase activity in vitro and its susceptibility to rimantadine. J. Virol. 33:583–586 [DOI] [PMC free article] [PubMed] [Google Scholar]