Abstract
Studies on the intracellular trafficking of influenza virus ribonucleoproteins are currently limited by the lack of a method enabling their visualization during infection in single cells. This is largely due to the difficulty of encoding fluorescent fusion proteins within the viral genome. To circumvent this limitation, we used the split-green fluorescent protein (split-GFP) system (S. Cabantous, T. C. Terwilliger, and G. S. Waldo, Nat. Biotechnol. 23:102–107, 2005) to produce a quasi-wild-type recombinant A/WSN/33/influenza virus which allows expression of individually fluorescent PB2 polymerase subunits in infected cells. The viral PB2 proteins were fused to the 16 C-terminal amino acids of the GFP, whereas the large transcomplementing GFP fragment was supplied by transient or stable expression in cultured cells that were permissive to infection. This system was used to characterize the intranuclear dynamics of PB2 by fluorescence correlation spectroscopy and to visualize the trafficking of viral ribonucleoproteins (vRNPs) by dynamic light microscopy in live infected cells. Following nuclear export, vRNPs showed a transient pericentriolar accumulation and intermittent rapid (∼1 μm/s), directional movements in the cytoplasm, dependent on both microtubules and actin filaments. Our data establish the potential of split-GFP-based recombinant viruses for the tracking of viral proteins during a quasi-wild-type infection. This new virus, or adaptations of it, will be of use in elucidating many aspects of influenza virus host cell interactions as well as in screening for new antiviral compounds. Furthermore, the existence of cell lines stably expressing the complementing GFP fragment will facilitate applications to many other viral and nonviral systems.
INTRODUCTION
Influenza virus continues to pose a serious threat to worldwide public health due to its rapid and unpredictable evolution. This has been highlighted by the regular occurrence of human cases of infection with highly pathogenic H5N1 avian influenza viruses since 2003 (46) and by the emergence of a new H1N1 influenza virus of swine origin in 2009 (33). Even during typical epidemic years, despite the availability of influenza vaccines, approximately 250,000 to 500,000 people die worldwide due to severe complications associated with influenza virus infections (World Health Organization [http://www.who.int/topics/influenza/en/]). A better understanding of the fundamental biology of the virus, notably at the level of intraspecies transmissibility and virulence, is required to enable society to better address the problem.
The influenza virus genome consists of eight segments of negative-sense RNA (viral RNA [vRNA]), each of which is encapsidated with multiple copies of the nucleoprotein (NP) and one trimeric polymerase (Pol) complex (subunits PB1, PB2, and PA) to form ribonucleoprotein particles (RNPs). Upon endocytosis of the virion and low-pH-dependent fusion of the viral envelope with the endosomal membrane, viral RNPs (vRNPs) are released into the cytoplasm and transported to the nucleus, where transcription and replication of the viral genome occur (28). Newly synthesized vRNPs may serve as templates for new rounds of transcription/replication or are exported from the nucleus and transported to the sites of viral assembly, where, by largely unknown mechanisms, the correct complement of eight segments is incorporated into progeny virions which bud from the cell membrane.
The RNA-dependent RNA polymerase, functioning in the context of the RNP, is the key viral enzyme responsible for virus replication and transcription. It is thus an attractive target for the development of new antivirals, which are needed to complement vaccination and overcome limitations in existing antivirals, notably resistance (12). Interspecies transmission of influenza virus requires that viral proteins have to adapt to function optimally in the particular environment of the new host. It is now clear that mutational adaptation of the polymerase is essential for successful interspecies transmission (27). A contributory factor to this is likely the need for the polymerase to adapt to the various host cell proteins required to facilitate its function. Indeed, an increasing number of host factors that directly or indirectly interact with the polymerase have been identified (15, 27, 39). However, despite recent advances in understanding polymerase function, including the emergence of high-resolution structures of polymerase subunit domains (37), many aspects of viral replication remain obscure. These include, for instance, a detailed characterization of the nuclear microenvironments where transcription and replication occur and the mechanisms which underlie RNP nuclear export, cytoplasmic trafficking, and packaging into virions.
Fluorescence microscopy provides unprecedented opportunities to analyze the molecular and cellular dynamics associated with the entire viral replication cycle, going beyond what is possible, for instance, by immunofluorescence (IF) imaging of fixed cells. Dynamic imaging of viral proteins in live cells can be achieved in theory by genetically fusing a fluorescent reporter to the virus-encoded protein of interest, ensuring minimal perturbation of either virus or cell behavior. Such live-cell imaging was recently performed on transiently coexpressed influenza virus proteins (13, 20) and on a plasmid-based “minireplicon” system which consists of the three polymerase subunits, NP, and synthetic viral RNAs (1). However, the minireplicon systems do not perfectly reflect a real infection, and, in particular, they do not reflect the evolution of the concentration and distribution of viral proteins as a function of time postinfection. Encoding fluorescent tags or fusions within the genome segments is, in the case of influenza virus, complicated by the limited coding capacity of each segment, combined with the need to maintain correct packaging of all the segments; too large perturbations lead to genetic instability and/or impaired viral functions (6). Several previous attempts that involved partial or complete replacement of a viral protein by green fluorescent protein (GFP) (e.g., neuraminidase [40], hemagglutinin [22], or NS1 [21]) thus resulted in either replication-deficient virions and/or severe impairment of viral protein function. Another approach involved incorporation of a tetracysteine (TC) biarsenical labeling tag into NS1 (18) but has the drawback that it requires a potentially perturbing chemical labeling step after cells are infected. To circumvent these limitations, we applied the split-GFP technique to the PB2 gene, which allowed us to fluorescently label PB2 although only a short additional sequence was added to the PB2 gene. Indeed, the split-GFP method is based on the spontaneous and essentially irreversible assembly of a highly stable fluorophore by recombination of a 16-amino-acid C-terminal GFP-peptide (residues 215 to 230 [GFP11]) fused to the protein of interest with a larger complementary GFP fragment (GFP1-10) that is supplied in trans (4, 16, 19, 32). The split-GFP system was recently used to track bacterial secretion effectors upon translocation in host cells (42). Here, we rescued a recombinant replication-competent nonattenuated influenza virus expressing a tagged PB2-GFP11 polymerase subunit. This virus allowed visualization of PB2 and viral RNPs by live microscopy throughout the infectious cycle and characterization of the intranuclear dynamics of PB2 by fluorescence correlation spectroscopy (FCS), for the first time in live infected cells. To our knowledge, this is the first application of the split-GFP system to a virus-encoded protein, demonstrating its potential for the tracking of viral proteins during a quasi-wild-type (wt) infection. This new tool, or adaptations of it, will have numerous applications in influenza virus biology.
MATERIALS AND METHODS
Cells.
293T and A549 cells were grown in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). MDCK cells were grown in modified Eagle's medium (MEM) supplemented with 5% FBS. The A549-GFP1-10 and MDCK-GFP1-10 cells were generated by lentiviral transduction, as described by Beignon et al. (2). Vero cells were grown in MEM supplemented with 10% FBS.
Plasmids.
The eight pPolI plasmids containing the sequences corresponding to the genomic segments of influenza virus A/WSN/33 (WSN) and the four recombinant pcDNA3.1 plasmids for the expression of WSN-PB1, -PB2, -PA, and -NP proteins (10) were kindly provided by G. Brownlee (Sir William Dunn School of Pathology, Oxford, United Kingdom). The pPolI-WSN-PB2 plasmid was modified using standard PCR and cloning procedures in order to fuse the PB2 open reading frame (ORF) to the following sequences: (i) a linker sequence encoding three alanine residues, (ii) the GFP11 coding sequence (RDHMVLHEYVNAAGIT), (iii) a stop codon combined with an NheI restriction site (TAGCATGC), and (iv) the 5′ terminal 143 nucleotides of the PB2 segment. The resulting pPolI-WSN-PB2-GFP11 construct was verified by sequencing positive clones using a BigDye Terminator sequencing kit and an automated sequencer (Perkin Elmer). The sequence encoding the WSN-PB2-GFP11 fusion protein was subcloned into the pcDNA3.1 plasmid. The pPR7-FluA-Renilla luciferase plasmid was obtained by subcloning the Renilla luciferase coding sequence in positive orientation between the two BbsI restriction sites in plasmid pPR7 (5). The sequences of the oligonucleotides used for amplification and sequencing can be provided upon request. The pCMV-GFP1-10 (where CMV is cytomegalovirus) and pCMV-GFP11 plasmids were purchased from Sandia Biotech; pCI plasmid was from Promega. Plasmids encoding fluorescently tagged proteins (PB1-mCherry, PB1-monomeric cyan fluorescent protein [mCFP], enhanced GFP [EGFP]-PB2, PA-mCFP, PA-mCherry, γ-tubulin–DsRed1, and mCherry-MAP4) were described elsewhere (11, 13, 17, 36). A maltose binding domain-GFP11 (MBD-GFP11) construct was obtained by placing the MBD coding sequence (13) into the multiple cloning site of pCMV-GFP11 backbone upstream to GFP11. mRuby-lifeact was designed in the group of Wedlich-Soldner (36); an mCherry-Rab11 construct was made by subcloning the Rab11 coding sequence into the multiple cloning site of pmCherry-C1 vector (Clontech). Golgi cisternae were visualized with CellLight Golgi-RFP (red fluorescent protein) BacMam 2.0 reagent (Invitrogen). EGFP-Rab1A plasmid was a kind gift of Jaakko Saraste (Bergen, Norway). The pCMV-firefly luciferase plasmid was kindly provided by Yves Jacob (Institut Pasteur, France).
Production of recombinant viruses by reverse genetics.
The method used for the production of recombinant influenza viruses by reverse genetics was adapted from previously described procedures (10, 34). Briefly, the eight pPolI-WSN and four pcDNA3.1-WSN plasmids (0.5 μg of each) were cotransfected into a subconfluent monolayer of cocultivated 293T and MDCK cells (4 × 105 and 3 × 105 cells, respectively, seeded in a 35-mm dish), using 10 μl of Fugene 6 transfection reagent (Roche). After 24 h of incubation at 37°C, the supernatant was removed, and cells were washed twice with DMEM and incubated at 37°C in DMEM containing tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin at a final concentration of 0.5 μg/ml for 48 h. The efficiency of reverse genetics was evaluated by titrating the supernatant on MDCK cells in plaque assays. Subsequent amplification of the recombinant WSN-PB2-GFP11 virus and its wild-type counterpart, WSN-wt was performed on MDCK cells at a multiplicity of infection (MOI) of 0.0001 for 3 days at 35°C in DMEM containing TPCK-trypsin at a concentration of 1 μg/ml. Viral stocks were titrated by plaque assays on MDCK and MDCK-GFP1-10 cells as described previously (23). Viral RNA was extracted and subjected to reverse transcription and amplification using specific oligonucleotides anchored on the 3′ and 5′ terminal 22 to 38 nucleotides of the PB2 segment, as described earlier (35). The products of amplification were purified using a QIAquick gel extraction kit (Qiagen) and were sequenced using a BigDye Terminator sequencing kit and an automated sequencer (Perkin Elmer). The sequences of the oligonucleotides used for amplification and sequencing can be provided upon request.
Ribonucleoprotein activity assays.
A mixture of plasmids pPR7-FluA-Renilla luciferase (100 ng) and pCMV-firefly luciferase (5 ng) was transfected into 3 × 105 A549 or A540-GFP1-10 cells, using the Fugene-HD transfection reagent (Roche). At 24 h posttransfection, cells were infected with the WSN-wt or WSN-PB2-GFP11 virus at an MOI of 5 PFU/cell. At 6, 12, or 24 h postinfection (hpi), cells were washed twice with phosphate-buffered saline (PBS) supplemented with Ca2+ and Mg2+ and lysed in 100 μl of lysis buffer provided with the Renilla Luciferase Assay System kit (Promega). Renilla and firefly luciferase activities were measured on 40 μl of cell extracts, using the Renilla luciferase substrate provided with the above-mentioned kit, the firefly luciferase substrate kindly provided by Y. Jacob (Institut Pasteur, France), and a Centro luminometer (Berthold).
Infection.
For viral growth assays, confluent monolayers of MDCK or MDCK-GFP1-10 cells were infected at an MOI of 0.001 PFU/cell. Following 1 h of adsorption at 35°C, cells were washed three times and then incubated with DMEM supplemented with 1 μg/ml of TPCK-trypsin (Worthington). Viral titers in the supernatants collected at different time points were measured by plaque assays on MDCK cells as described previously (23). For imaging, subconfluent 293T or Vero cells plated on polylysine-coated coverslips (BD Biosciences), LabTekII chambered cover glasses (growing surface per well, 1.7 cm2) (Nunc), or on eight-well Ibidi μ-Slides (Ibidi GmbH, Germany) were transfected with the pCMV-GFP1-10 or the pCI plasmid (1 μg) using the Fugene-HD transfection reagent (Roche). At 24 or 48 h posttransfection, the cells were incubated for 1 h with the WSN-wt or WSN-PB2-GFP11 recombinant virus at an MOI of 3 to 5 PFU/cell or mock infected. For indirect immunofluorescence assays, the inoculum was replaced with DMEM supplemented with 2% FBS. For imaging and FCS acquisitions, the inoculum was replaced by a CO2-independent medium without phenol red (Invitrogen) supplemented with 20% FBS, 2 mM glutamine, 100 μg/ml penicillin, and 100 U/ml streptomycin.
Indirect immunofluorescence assay.
293T or Vero cells were fixed with PBS–4% paraformaldehyde for 20 min, permeabilized with PBS–0.1% Triton X-100 for 10 min, treated with 3% normal rabbit serum for 60 min, and then incubated with either a mixture of anti-PB2 monoclonal antibodies (MAbs) (31) (a kind gift from J. Ortin, Centro Nacional de Biotecnologia, Madrid, Spain) diluted 1:5 or with the anti-vRNP MAb clone 3/1 (a kind gift of Robert G. Webster, St. Jude Children's Research Hospital, Memphis, TN) diluted 1:200. Subsequently, cells were incubated with AlexaFluor 555-coupled anti-mouse IgG secondary antibody diluted 1:500 or AlexaFluor 647-coupled anti-mouse secondary antibody diluted 1:200 (both, Molecular Probes) and stained with 4′,6′-diamidino-2-phenylindole (DAPI).
Purification and analysis of WSN-PB2-GFP11 virions.
MDCK-GFP1-10 cells at about 80% confluence were infected with the WSN-PB2-GFP11 or the WSN-wt virus at an MOI of 0.05 PFU/cell. Two days after infection, the culture medium was collected for virus purification. The medium was centrifuged at 4,400 rpm in a 50-ml Falcon tube using an Eppendorf table-top centrifuge for 10 min to remove cells and cell debris. The supernatant was centrifuged through 4 ml of 20% sucrose cushion (SW28 rotor [Beckman]; 4°C at 25,000 rpm for 2 h), and the pellet was resuspended in PBS. The virus particles were further separated from macromolecular complexes or aggregates on a 15 to 60% sucrose gradient (SW60 rotor; 4°C at 20,700 rpm for 55 min). Fractions from the gradient were collected, and aliquots were analyzed for the presence of GFP1-10 and NP by Western blotting using monoclonal antibody JL8 against GFP (Clontech) and rabbit polyclonal antibody directed against A/PR/8/34 virions (42, 43). The remaining fractions were inactivated by glutaraldehyde fixation, negatively stained with phosphotungstic acid, and analyzed by transmission electron microscopy.
Light microscopy.
Microscopy was performed using three systems: a TCS SP2 AOBS (acousto-optic beam splitter) confocal laser scanning microscope (Leica Microsystems, Germany), an Andor Revolution Nipkow disk microscope (Andor Technologies) equipped with a CSU22 confocal scan head (Yokogawa) and a DV885 electron-multiplying charge-coupled-device (EMCCD) camera (Andor Technologies), and an LSM 7 DUO system equipped with a confocal line scan head and CCD line detector (Carl Zeiss, Germany). On the SP2 AOBS system, excitation and emission were set as follows: at 488 and 500 to 550 nm for GFP; 405 and 420 to 470 nm for DAPI; 561 and 590 to 670 nm for mCherry, AlexaFluor 555, and tagRFP; and 633 nm and 643 to 742 nm for AlexaFluor 647. On the Nipkow disk and LSM 7 DUO, standard dichroic and filter combinations for GFP and RFP fluorescence were used for GFPcomp (the fluorophore formed by complementation of nonfluorescent GFP1-10 and GFP11 fragments) and mCherry signals. In all multicolor imaging, signals were acquired sequentially, except for in Movie S3 in the supplemental material, where the GFP and mCherry signals were acquired simultaneously upon separation with standard combinations of dichroic filters. Image quantification and colocalization analyses were performed with ImageJ, version 1.44c, software (http://rsb.info.nih.gov/ij/). The Pearson correlation coefficient (varies from −1 to 1) for experimental images was calculated by ImageJ, version 1.45a. The occurrence of false colocalization was excluded by calculating the Pearson coefficient for “scrambled” images according to the Costes method using the colocalization test plug-in of ImageJ. Images were corrected for possible cross talk between the channels; for presentation purpose, brightness and contrast of raw images were optimized.
Fluorescence recovery after photobleaching (FRAP) experiments were performed on the SP2 AOBS system using the manufacturer's software. The perinuclear area enriched with PB2-GFPcomp was photobleached in a circular region of interest (ROI) of ∼2-μm radius with 458-, 476-, and 488-nm lines of an argon laser at maximal intensity and at maximal zoom for 1.6 s. Pre- and postbleach frames were acquired with excitation at 488 nm with 2-s intervals during 150 to 200 s with 40× or 63× oil immersion objectives at an illumination intensity of <1/80 of that used for photobleaching. The shifts of the field of view due to stage instability were corrected using the StackReg plug-in of ImageJ. Signal was corrected for background and acquisition photobleaching and quantified in manually drawn polygonal regions of interest corresponding to the shape of the perinuclear PB2-GFPcomp-enriched areas. FRAP curves were fitted to a single-exponential model with MicroCal Origin, version 8.0, software.
For particle tracking, time-lapse series were acquired on the LSM 7 DUO microscope at 1.8 to 4 frames per second (fps); Vero cells transiently expressing GFP1-10 and either mCherry-MAP4 or mRuby-lifeact were infected with WSN-PB2-GFP11. A total of 101 cells from five independent infections were analyzed; time series of 50 to 100 frames/cell were used for single-particle tracking analysis. Depolymerization of microtubules and actin filaments was controlled visually using the fluorescent protein markers (mCherry-MAP4 and mRuby-lifeact, respectively). Imaris, version 7.2.3 (Bitplane), software was used to detect PB2-GFPcomp particles, to build the tracks by the auto-processive motion algorithm, and to analyze the obtained data; only tracks with more than three frames were used; motions with speeds of <0.1 μm/s were filtered out.
FCS setup and data analysis.
The Leica TCS SP2 AOBS system with avalanche photodiode detectors (APDs) and a corrected water immersion Apochromat 63× objective lens (numerical aperture [NA], 1.2) was used for FCS experiments (described in detail in reference 9). FCS data were acquired from 4 to 7 h postinfection as the reliability of FCS analysis at later time points was compromised by the high concentration of PB2-GFPcomp species. FCS data were acquired with the FCS module and analyzed as described previously (13). GFPcomp and mCherry fluorophores were excited at 488 and 561 nm. The autocorrelation curves were globally fitted with a one-species model assuming anomalous intracellular diffusion of the labeled subunits (13) using Origin, version 8, software. From these fits, the parameter Neff, the mean number of fluorescent particles within the detection volume, was estimated. The parameter τdiff, the characteristic diffusion time of the fluorescent species, was estimated from the fits of the autocorrelation curves. Diffusion time is a characteristic time which a fluorescent molecule on average resides in the detection volume. τdiff depends both on the diffusion coefficient of the molecule and on the size of the detection volume (38). Thus, for more correct description of the diffusion properties of PB2, we used the relative diffusion time given by the equation τdif_rel = τdif/τdifGFP, where τdiffGFP is the diffusion time measured for uncoupled EGFP or MBD-GFPcomp in the nuclei in the control well for each experiment. From FCS data, two parameters were extracted: (i) the characteristic translational diffusion time (τdif_rel) and (ii) the relative brightness, calculated by division of the count rate in the APD detector by the number of the particles in the FCS detection volume (N).
RESULTS
Tagging the influenza polymerase PB2 subunit with split-GFP in a replication-competent virus.
To achieve the split-GFP tagging in the context of a replication-competent A/WSN/33 (WSN) influenza virus, the PB2 genomic segment was engineered as shown in Fig. 1A. Nucleotides corresponding to GFP residues 215 to 230 (GFP11) were fused to the 5′ end of the PB2 ORF. In addition, a duplication of the last 109 nucleotides (nt) encoding PB2 was inserted between the stop codon and the 5′ noncoding region to ensure the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the segment as previously described (6). A WSN-PB2-GFP11 virus and its wild-type (WSN-wt) counterpart were produced by reverse genetics (10) and amplified in MDCK cells in parallel. Both viruses grew to high titers (107 to 108 PFU/ml). Viral RNA was extracted and subjected to reverse transcription-PCR (RT-PCR) using primers designed to amplify the whole PB2 segment. Direct sequencing of the PCR products confirmed that the WSN-PB2-GFP11 virus retained the GFP11 coding sequence and the PB2 ORF duplication during three serial passages at a low MOI on MDCK cells (data not shown).
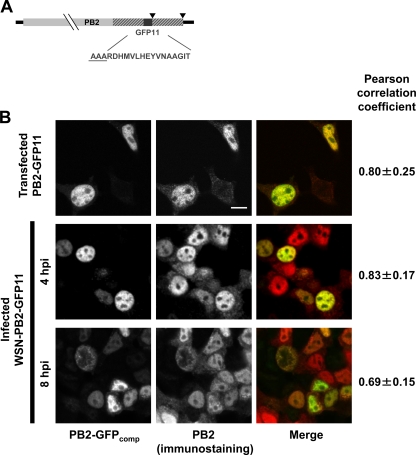
Fig 1.
(A) Schematic representation of the PB2-GFP11 genomic segment. The PB2 ORF (light gray box) is fused to the sequence encoding the GFP11 domain (dark gray box) and is flanked by the 3′ and 5′ noncoding regions (black lines). The duplicated PB2 ORF sequences are represented as hatched boxes. The stop codons are indicated by black arrowheads. The GFP11 amino acid sequence is indicated. The three-alanine linker at the junction between PB2 and GFP11 is underlined. (B) Colocalization of the PB2-GFPcomp signal with PB2-specific immunofluorescence signal. 293T cells were transfected with the GFP1-10 expression plasmid, subsequently infected with the WSN-PB2-GFP11 virus at an MOI of 3, and fixed at indicated times or cotransfected with the PB2-GFP11 and GFP1-10 expression plasmids and fixed at 48 h posttransfection. Immunostaining was performed as described in Materials and Methods, using anti-PB2 monoclonal antibodies. On the merged images, PB2-GFPcomp and immunostaining signals are pseudo-colored green and red, respectively. Scale bar, 10 μm. Pearson correlation coefficient, as determined on GFP1-10 expressing cells, is indicated on the right (mean ± SD; 3 to 8 cells per condition). The cells which were not transfected with GFP1-10 plasmid and did not develop GFP fluorescence were not included in the colocalization analysis.
The WSN-PB2-GFP11 or WSN-wt virus was used to infect 293T cells previously transfected with the pCMV-based plasmid expressing the GFP1-10 fragment of the split-GFP (described in Materials and Methods) or with a control plasmid. Bright fluorescence was detected only in the GFP1-10-transfected and WSN-PB2-GFP11-infected cells and not in mock-transfected or WSN-wt-infected control cells (data not shown). Thus, efficient complementation of PB2-GFP11 with GFP1-10 occurred and produced a specific fluorescence signal. Cells were fixed at 4 or 8 h postinfection and stained with an anti-PB2 antibody. The PB2 immunofluorescence (IF) signal and the fluorescent signal of complemented GFP (GFPcomp) showed a high degree of colocalization, as indicated by the Pearson coefficient (Fig. 1B, middle and bottom panels). At 4 h postinfection (hpi), some PB2 IF signal was observed in the cytoplasm while the GFPcomp fluorescence appeared only in the nucleus. This difference was also observed in cells cotransfected with the PB2-GFP11 and GFP1-10 expression plasmids (Fig. 1B, top panels). This may be due to the fact that the anti-PB2 antibody, directed against a denatured N-terminal fragment of PB2 (31), binds to nascent PB2 proteins in the cytoplasm, whereas GFPcomp fluorescence occurs only upon synthesis of full-length PB2 protein and complementation with GFP1-10, which possibly occurs only after rapid nuclear import. At 8 h postinfection, both the PB2 IF and GFPcomp fluorescence signals were present in the cytoplasm (Fig. 1B, bottom panels), in agreement with the fact that vRNPs undergo nucleocytoplasmic export at late stages of the viral cycle. To estimate the efficiency of the reaction GFP1-10 + GFP11 → GFPcomp, we transfected 293T cells with an excess of GFP1-10 plasmid and, 1 day later, with mCherry-GFP11 plasmid. The concentrations of green fluorophore (representing GFP11 tag combined with GFP1-10) and red fluorophore (representing mCherry) were subsequently quantified by FCS (data not shown). From 5 to 11 h after the second transfection, green fluorescence appeared in a subpopulation of the cells; 28% of these cells also showed red fluorescence; cells showing red fluorescence only were not detected. The presence of green-only cells suggests that complementation of GFPcomp was faster than maturation of mCherry. At 20 h posttransfection and later, all green fluorescent cells also showed red fluorescence, and the green/red concentration ratio was 1.0 ± 0.5 (mean value ± standard deviation [SD]; n = 18), indicating that at steady state most of the GFP11-tagged mCherry molecules were complemented with GFP1-10.
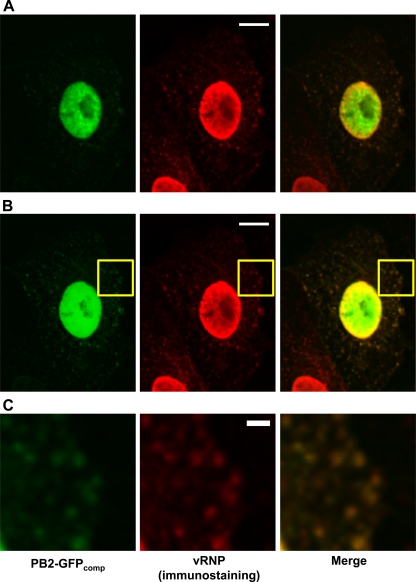
Cells transiently expressing GFP1-10 and infected with WSN-PB2-GFP11 were fixed 6 h 30 min postinfection and immunostained with the MAb 3/1 antibody which specifically recognizes influenza virus vRNPs (7). In the cytoplasm, PB2-GFPcomp particles showed a strong colocalization with vRNP staining (Pearson coefficient of 0.82 ± 0.09, mean ± SD; n = 12) (Fig. 2B and C). In the nucleus, colocalization of PB2-GFPcomp with vRNPs was only partial and occurred mostly at the periphery of the nucleus, where vRNP staining was the strongest. In contrast, PB2-GFPcomp showed no preferential accumulation near the nuclear envelope (Fig. 2A), thus indicating that, in the nucleus, only a subpopulation of the viral polymerase is associated with mature, ready-for-export vRNP complexes.
Fig 2.
Colocalization of the PB2-GFPcomp signal with vRNP-specific immunofluorescence signal. Vero cells were transfected with the GFP1-10 expression plasmid, subsequently infected with the WSN-PB2-GFP11 virus at an MOI of 3, and fixed at 6 h 30 min postinfection. Immunostaining was performed as described in Materials and Methods, using the MAb 3/1. On the merged images, PB2-GFPcomp and immunostaining signals are pseudo-colored green and red, respectively. (A) The microscope settings were optimized for the acquisition of nuclear signals. Scale bar, 10 μm. (B) The microscope settings were optimized for the acquisition of cytoplasmic signals. Scale bar, 10 μm. (C) Zoomed-in image of the area defined by a yellow rectangle in panel B. Scale bar, 2 μm.
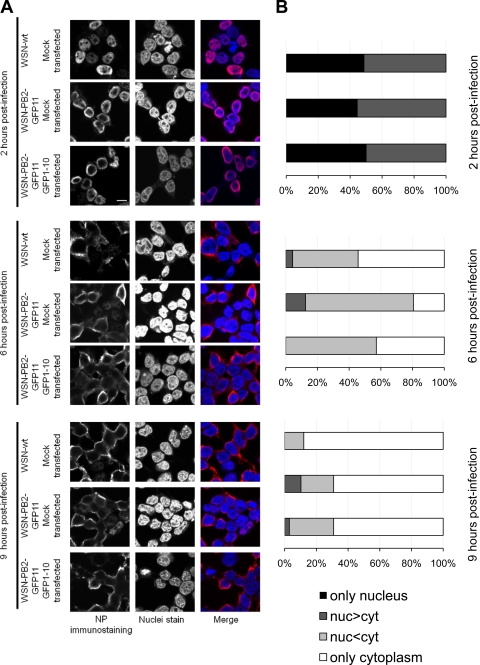
In order to assess whether viral replication occurred at similar rates in the presence of PB2-wt, PB2-GFP11, or PB2-GFPcomp, the nuclear and cytoplasmic accumulation of the viral nucleoprotein (NP) was monitored by indirect immunofluorescence upon infection with WSN-wt or WSN-PB2-GFP11 virus in control or GFP1-10-expressing cells. In agreement with published data (25), all or most NP was detected in the nucleus at 2 h postinfection, marked NP relocalization to the cytoplasm was observed at 6 h postinfection, and most cells showed an exclusively cytoplasmic NP staining at 9 h postinfection (Fig. 3A and B). No difference was observed between the WSN-wt, WSN-PB2-GFP11, and WSN-PB2-GFP11 plus GFP1-10 samples, except for a minor proportion of cells showing predominant nuclear NP staining at 9 h postinfection for the WSN-PB2-GFP11 virus (with or without GFP1-10) but not for the WSN-wt virus (Fig. 3B, dark gray bars). This difference might reflect a slight slowdown of vRNP nuclear export for the tagged virus, which is, however, unlikely to impair viral replication as the WSN-PB2-GFP11 virus grows just as efficiently as its WSN-wt counterpart.
Fig 3.
Intracellular distribution of NP in 293T cells infected with the WSN-wt or WSN-PB2-GFP11 virus. Mock-transfected 293T cells were infected with the WSN-wt or WSN-PB2-GFP11 virus at an MOI of 3. 293T cells transfected with the GFP1-10 expression plasmid were infected with the WSN-PB2-GFP11 virus at an MOI of 3. At 2, 6, and 9 h postinfection, immunofluorescence assays were performed as described in Materials and Methods, using an anti-NP monoclonal antibody. (A) Representative images. In the merged images, the NP-specific staining and DAPI nucleic acid staining were pseudo-colored red and blue, respectively. Scale bar, 10 μm. (B) Quantification of NP nuclear and cytoplasmic distribution. The cells were classified according to the mean intensities of NP-specific immunostaining signal (corrected for nonspecific staining) in their nuclei and cytoplasms. The proportions of cells showing only nuclear, more nuclear than cytoplasmic (nuc>cyt), less nuclear than cytoplasmic (nuc<cyt), or only cytoplasmic NP-specific signals are indicated. A total of 36 to 90 cells per condition were analyzed. In the GFP1-10 plus WSN-PB2-GFP11 condition, only cells showing GFPcomp fluorescent signal were selected for analysis.
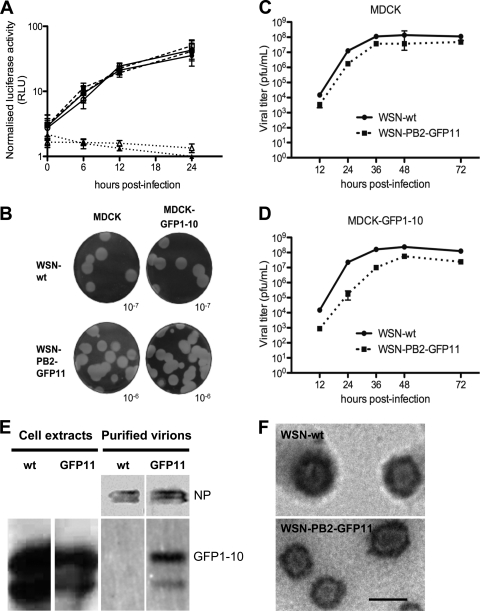
A virus-inducible reporter gene was used to compare the viral polymerase activity in the presence of a PB2-wt or PB2-GFP11 subunit in control or GFP1-10-expressing cells. To this end, A549 cells and A549-GFP1-10 cells that stably expressed GFP1-10 were transfected with a plasmid encoding a virus-like Renilla luciferase reporter RNA, together with a firefly luciferase expression plasmid. At 24 h posttransfection, the cells were infected with the WSN-wt or WSN-PB2-GFP11 virus at an MOI of 5 PFU/cell. As assessed by measuring the relative luciferase activities in cell extracts at various time points, the efficiency of transcription/replication of the virus-like reporter RNA was very similar for the WSN-wt and the WSN-PB2-GFP11 viruses in the presence or absence of the GFP1-10 protein (Fig. 4A). The PB2-GFP11 and PB2-GFPcomp proteins thus behave like the wild-type PB2 protein in this assay. In agreement with these observations, fluorescence cross-correlation spectroscopy (data not shown) indicated that transiently expressed PB2-GFPcomp interacted with the PB1 polymerase subunit in the nucleus, as we previously had observed for EGFP-PB2 (13). The growth properties of the WSN-wt and WSN-PB2-GFP11 viruses were compared on MDCK cells and MDCK-GFP1-10 cells that stably expressed the GFP1-10 protein. When plaque assays were performed, the plaques formed on MDCK and on MDCK-GFP1-10 cells were very similar in number and size for the WSN-wt and the WSN-PB2-GFP11 viruses (Fig. 4B). Both cell lines were infected in parallel at an MOI of 0.001, and viral titers in the culture supernatants were determined at different times postinfection by plaque assays on MDCK cells. As shown in Fig. 4C and D, a moderate slowdown in the growth kinetics of the WSN-PB2-GFP11 virus was observed from 12 to 36 h postinfection. This slowdown was more pronounced on MDCK-GFP1-10 cells (Fig. 4D), with the difference in titers achieved by the WSN-PB2-GFP11 virus compared to the WSN-wt virus being 2 and 1 log at 24 h and 36 h postinfection, respectively. However, maximum titers of >107 PFU/ml were observed with both viruses at 48 h postinfection on MDCK as well as on MDCK-GFP1-10 cells. Overall, our data suggest that the presence of the GFP11 tag at the C terminus of PB2 only moderately alters viral growth and that the GFP11-tagged virus is a relevant tool for live imaging of influenza virus-infected cells.
Fig 4.
Phenotypic comparison of the WSN-wt and WSN-PB2-GFP11 viruses. (A) A plasmid allowing the expression of a pseudo-viral Renilla luciferase reporter RNA was cotransfected with a firefly luciferase expression plasmid in A549 cells (open symbols) or A549-GFP1-10 cells (closed symbols). At 24 h posttransfection, cells were either mock infected (dotted lines), infected with the WSN-wt virus (solid lines), or infected with the WSN-PB2-GFP11 virus (dashed lines) at an MOI of 5 PFU/cell. The Renilla luciferase activities measured in cell extracts at the indicated times postinfection were normalized with respect to firefly luciferase activities and are expressed as the mean ± SD of quadruplicates. RLU, relative light units. (B) Plaque phenotype of the WSN-wt and WSN-PB2-GFP11 viruses assayed on MDCK and MDCK-GFP1-10 cells. Cell monolayers were stained with crystal violet after 72 h of incubation at 35°C. The dilutions of the viral stock used for infection are indicated. (C and D) Growth curves of the WSN-wt (solid lines) and WSN-PB2-GFP11 viruses (dashed lines) on MDCK or MDCK-GFP1-10 cells. Cell monolayers were infected at an MOI of 0.001 and incubated for 72 h at 35°C. At the indicated time points, the supernatants were harvested, and virus titers were determined by plaque assays on MDCK cells. (E) Western blot analysis of WSN-wt and WSN-PB2-GFP11 purified virions produced from MDCK cells stably expressing GFP1-10. (F) Electron microscopy images of the WSN-wt and WSN-PB2-GFP11 virions, which correspond to lanes 3 and 4 in panel E, respectively. Scale bar, 100 nm.
Since the possibility existed that a portion of PB2-GFP11 did not complement with GFP1-10 in the infected cells and that only these GFP1-10-free vRNPs but not PB2-GFPcomp-containing vRNPs are incorporated into budding virions, we tested the progeny WSN-PB2-GFP11 virions for the presence of GFP1-10. The supernatants of MDCK-GFP1-10 cells infected with WSN-PB2-GFP11 and, as a control, with the WSN-wt virus, were used to purify virus particles by ultracentrifugation and sucrose gradient density centrifugation. Fractions containing the virions were identified by Western blotting with antibodies against NP (Fig. 4E) and by visualization of the virions with electron microscopy (Fig. 4F). GFP1-10 was detected by Western blotting in the fractions containing the virus encoding the GFP11 tag but not in the fractions containing the wild-type virus (Fig. 4E). This confirms that progeny vRNPs containing PB2-GFPcomp are correctly transported to the plasma membrane and incorporated into the virions and strongly indicates that the GFPcomp tag does not disturb the higher-order assembly of vRNPs.
Live imaging of the PB2 polymerase subunit in influenza virus-infected cells.
293T cells transiently expressing the GFP1-10 domain were infected at a high MOI with the WSN-PB2-GFP11 virus and live imaged. To characterize the PB2 distribution in infected cells throughout the infection cycle, selected fields of view were repeatedly imaged with 10- to 15-min intervals for 12 to 18 h following infection (Fig. 5A; see also Movie S1 in the supplemental material). Until 6 to 7 h postinfection, the fluorescence intensity gradually increased in the nucleus (representative data are shown in Fig. 5A and C, black dashed-dotted line, and in Movie S1), reflecting nuclear import and accumulation of PB2, in agreement with previous observations (35). The PB2-GFPcomp signal was distributed nonhomogeneously throughout the nucleus, except in the nucleoli, which consistently showed low fluorescence. From 7 h postinfection, the fluorescence intensity in the nuclei decreased, most likely due to the nuclear export of vRNPs, but it remained significant until >16 h postinfection (Fig. 5A and C, black dashed-dotted line). In the cytoplasm, fluorescence became detectable at 3 to 4 h postinfection, increased until roughly 10 h postinfection, and then remained apparently stable up to 12 h postinfection (Fig. 5A and C, black dashed line). In the presence of leptomycin B, a specific inhibitor of the CRM1 nuclear export pathway used by progeny vRNPs (8, 29, 45), the PB2-GFPcomp signal remained almost undetectable in the cytoplasm during the whole period of observation (Fig. 5C, red dashed line, and D and E), while the nuclear signal continued to grow until 14 h postinfection (Fig. 5C, red dashed-dotted line). Based on these observations and the abundant colocalization of cytoplasmic PB2-GFPcomp particles with vRNPs (Fig. 2), the sensitivity to the CRM1 pathway inhibitor is a strong indication that the GFP-fluorescent particles that are detected in the cytoplasm at late time points indeed represent PB2-GFPcomp exported from the nucleus and incorporated into vRNPs and hence may serve to track vRNPs in the living cells.
Fig 5.
Imaging of the PB2-GFPcomp protein in live 293T cells infected with the WSN-PB2-GFP11 virus. (A) Selected frames from Movie S1A in the supplemental material showing individual PB2-GFPcomp-positive 293T cells at the times postinfection indicated (min). 293T cells transiently expressing GFP1-10 were infected at an MOI of 3 with the WSN-PB2-GFP11 virus, and images (PB2-GFPcomp signal) of live cells throughout the infectious cycle were acquired with a Leica TCS SP2 AOBS microscope, as described in Materials and Methods. Single confocal slices are shown. Brightness and contrast were adjusted for the whole series of images. Arrowheads point to the cell used for quantification shown on Fig. 5C. Scale bar, 10 μm. (B) 293T cells transfected with GFP1-10 and infected with WSN-PB2-GFP11 virus, imaged (PB2-GFPcomp signal) 9 h (left) and 1 day (right) postinfection. Accumulation of PB2-GFPcomp at the plasma membrane is indicated by arrowheads. Scale bars, 10 μm. (C) Quantification of PB2-GFPcomp content over time in representative infected cells treated with 20 nM leptomycin B (red lines) and mock treated (black lines; the corresponding cell is marked with arrowheads in Fig. 5A). PB2-GFPcomp mean fluorescence intensities corrected for background in an ROI in the nucleus (dash-dotted line) and in an ROI in the cytoplasm (dashed line) are represented. Positions and shapes of the ROIs were manually adjusted in each frame to cover significant portions of the appropriate compartments of the cell. au, arbitrary units. (D) Effect of leptomycin B treatment on the intracellular distribution of the PB2-GFPcomp protein in live 293T cells transiently expressing GFP1-10 and infected at an MOI of 3 with the WSN-PB2-GFP11 virus. After viral adsorption, cells were incubated with 15 nM leptomycin B or with the corresponding dilution of dimethyl sulfoxide. Representative images of PB2-GFPcomp-expressing cells acquired at 6 h postinfection are shown. Brightness and contrast were adjusted for presentation purposes. Scale bar, 10 μm. (E) Effect of leptomycin B treatment on the intracellular distribution of the PB2-GFPcomp protein in live infected cells. Values are the ratio of nuclear to cytoplasmic intensities of PB2-GFPcomp fluorescence in the absence (black bars) or presence (gray bars) of leptomycin B. The data are shown as mean values ± SD calculated from >10 cells/condition.
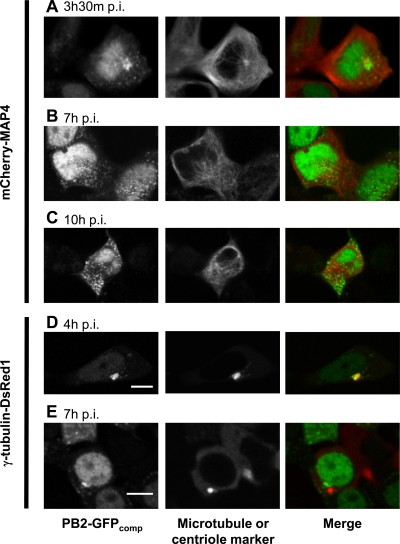
Live imaging was then performed on Vero or 293T cells that transiently expressed, in addition to the GFP1-10 domain, either a recombinant microtubule marker, MAP4, or a centriole marker, γ-tubulin, fused to mCherry and DsRed1 fluorescent proteins, (Fig. 6; see also Movie S2 in the supplemental material). At 3.5 to 4 h postinfection with the WSN-PB2-GFP11 virus, perinuclear bright spots of PB2-GFPcomp localized at the microtubule organizing center (MTOC) or in its vicinity (Fig. 6A and D). No significant colocalization of the pericentriolar PB2-GFPcomp with the endoplasmic reticulum ([ER] labeled with ER-Tracker TR dye), mitochondria (labeled with CellLight Mitochondria-RFP), the Golgi network (labeled with CellLight Golgi-RFP), or the ER-Golgi intermediate compartment (ERGIC; labeled with the mCherry-Rab1A protein) was observed (data not shown). At later times postinfection, PB2-GFPcomp spots were distributed throughout the cytoplasm without any marked accumulation near the MTOC or in the perinuclear area in general (Fig. 6B, C, and E); at 9 h postinfection or later, PB2-GFPcomp accumulated near the plasma membrane, presumably representing regions of viral assembly and budding (Fig. 5B). The observed changes in PB2 intracellular distribution over the infection cycle and, in particular, its transient pericentriolar accumulation agree with those recently reported for vRNPs in fixed cells (1, 7, 14, 26).
Fig 6.
Covisualization of the PB2-GFPcomp protein with microtubule network markers in live infected cells. 293T cells transiently expressing GFP1-10 and mCherry-MAP4 (A to C) or γ-tubulin-DsRed1 (D and E) fusion proteins were infected at an MOI of 3 with the WSN-PB2-GFP11 virus. The PB2-GFPcomp and mCherry-MAP4/γ-tubulin–DsRed1 fluorescence signals in live cells were acquired with a Leica TCS SP2 AOBS microscope, as described in Materials and Methods. Merged images are shown in which the PB2-GFPcomp signal is pseudo-colored green and Cherry-MAP4 or γ-tubulin–DsRed1 signal is pseudo-colored red. Scale bar, 10 μm.
To evaluate the turnover rate of the pericentriolar PB2-GFPcomp body, FRAP experiments were performed on infected cells (Fig. 7). After photobleaching of the perinuclear PB2-GFPcomp-enriched area, a substantial fraction of the initial fluorescence was recovered (time constant, 32 ± 13 s; apparent mobile fraction, 44% ± 17%, mean ± SD), indicating the dynamic state of this perinuclear PB2-GFPcomp-enriched area and suggesting its relevance for the cytoplasmic trafficking of newly synthesized PB2, most probably within vRNPs. The discrepancy between the mobile fraction values found here and those reported for vRNPs reconstituted in a minireplicon system (44% and 11%, respectively) (1) might be due to differences between cellular environments in the infection and transfection settings as well as to differences in the FRAP experimental setup.
Fig 7.
FRAP analysis of perinuclear PB2-GFPcomp particles in live infected cells. 293T cells transiently expressing GFP1-10 were infected at an MOI of 3 with the WSN-PB2-GFP11 virus. Representative data set of a FRAP experiment (squares) and the fitted curve (solid line). The signal was background subtracted, normalized to the prebleach level, and corrected for photobleaching during acquisition. The prebleach signal level is shown as a dashed line.
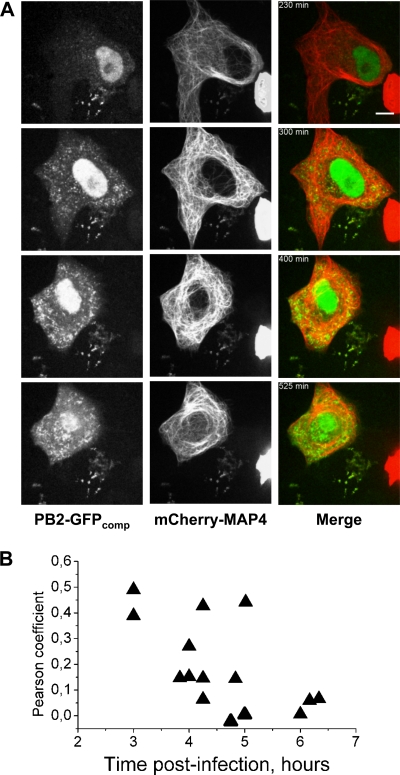
Microtubule-perturbing agents, such as nocodazole, were shown to moderately affect trafficking of minireplicon-derived vRNPs (1) and progeny virus titers (25). Here, when Vero cells expressing the mCherry-MAP4 microtubule marker and GFP1-10 were infected with WSN-PB2-GFP11 and imaged over extended periods of time, the majority of PB2-GFPcomp spots spreading across the cytoplasm did not colocalize markedly with the peripheral microtubule network (Fig. 8A; see also Movie S2 in the supplemental material). This was confirmed by colocalization analysis, which revealed Pearson coefficient values of 0.4 to 0.5 at 3 h postinfection but values close to 0 at 6 h postinfection (Fig. 8B). The PB2-GFPcomp particles were then tested for colocalization with Rab11, a GTPase residing in recycling endosome which was recently shown to be essential for the trafficking of influenza virus vRNPs (1, 7, 25). A549-GFP1-10 cells transiently expressing mCherry-Rab11 were infected with the WSN-PB2-GFP11 virus and imaged at a high frame rate (4 fps) at 7 h postinfection. Most of PB2-GFPcomp particles in the cytoplasm colocalized with mCherry-Rab11 (see Movie S3), indicating that progeny vRNPs containing PB2-GFPcomp are associated with the recycling endosome vesicles which are used for the trafficking of vRNPs.
Fig 8.
Time-lapse imaging of live infected cells and colocalization analysis for PB2-GFPcomp and microtubules. (A) A representative Vero cell transiently expressing GFP1-10 and mCherry-MAP4 and infected with the WSN-PB2-GFP11 virus. Selected frames from a time-lapse series (see Movie S2 in the supplemental material) acquired during ∼700 min with 5-min intervals using an Andor Revolution Nipkow disk microscope, as described in Materials and Methods, are shown. In merged images, the PB2-GFPcomp and Cherry-MAP4 signals are pseudo-colored green and red, respectively. Scale bar, 10 μm. (B) Colocalization between PB2-GFPcomp and mCherry-MAP4 in the cytoplasm of infected cells, as characterized by the Pearson coefficient, throughout the course of infection. Each triangle represents the Pearson coefficient measured in an individual cell (data from four independent infections are shown).
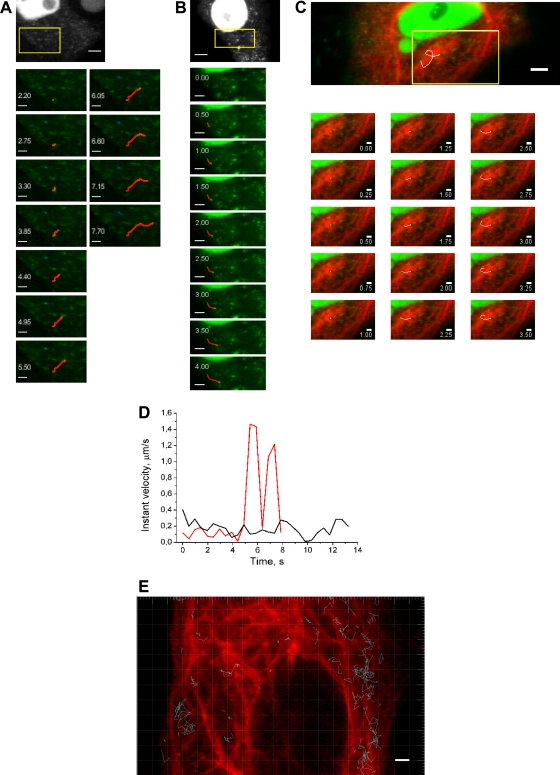
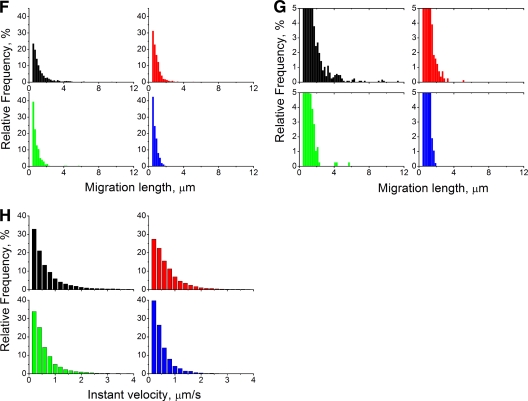
To obtain further insights into the dynamics of PB2-GFPcomp-containing particles present in the cytoplasm of infected cells, single-particle tracking analysis was performed. For most of the observation time, PB2-GFPcomp particles moved slowly (<0.25 μm/s) and changed direction randomly (Fig. 9A, blue line, and D, black curve). Some particles occasionally showed rapid (>1 μm/s) processive motions typical of filament transport (Fig. 9A, red line, and D, red curve; see also Move S4A in the supplemental material). Although some of the fast-motion tracks coincided with microtubules, a substantial proportion of the tracks did not appear to overlap with the microtubules (Fig. 9C and E). Treatment with nocodazole caused a moderate redistribution of the particle migration lengths and, in particular, a suppression of the longest (>4 μm) migrations (Fig. 9F and G, red histogram compared to black one), without visibly affecting the global distribution of instant velocities (Fig. 9H). Indeed, rapid directional motions were still occasionally observed in the presence of nocodazole (Fig. 9B, red line; see also Movie S4B). Cytochalasin D (an actin filament-depolymerizing agent [24]) treatment caused a redistribution of the migration lengths in favor of shorter migrations, with suppression of the longest migrations as well (Fig. 9F and G, green; see also Movie S4C). Finally, combined treatment with nocodazole and cytochalasin D led to a pronounced change of the migration length distribution: no migrations of >2 μm were observed, and most of them were shorter than 1 μm (Fig. 9F and G, blue; see also Movie S4D), i.e., representing practically immobile particles. Furthermore, the combined treatment caused pronounced global redistribution of the instant velocities, with nearly all values being <1 μm/s. Overall, our observations strongly suggest that both microtubules and actin filaments are involved in the trafficking of vRNPs in infected cells.
Fig 9.
Single-particle tracking of PB2-GFPcomp particles in live Vero cells infected with the WSN-PB2-GFP11 virus. Vero cells transiently expressing GFP1-10 and mCherry-MAP4 or mRuby-lifeact were infected at an MOI of 3 with the WSN-PB2-GFP11 virus and incubated with either 30 μM nocodazole, 2.5 μM cytochalasin D, a combination of 30 μM nocodazole with cytochalasin D, or with dimethyl sulfoxide (mock treatment). The image series was acquired at 6 to 9 h postinfection and no later than 1 h after the addition of cytochalasin D. Single-particle tracking analysis was performed on GFP-positive particles detected in the cytoplasm. (A to C) Selected frames from the time-lapse series, corresponding to the areas defined by the yellow rectangles on the full-frame images: mock treatment (A), treatment with nocodazole (B), and mock treatment, overlaid on the image of microtubules (mCherry-MAP4; red) acquired immediately before time series recording (C). Time (in seconds) is indicated. Representative tracks of rapid and directional motions of PB2-GFPcomp are shown as red lines (A and B), and a track of a randomly moving particle is shown as a blue line (A). A track apparently overlapping with microtubules is shown as a white line (C). Scale bars, 5 μm (full-frame images) and 2 μm (magnified images). (D) Variation of PB2-GFPcomp particle instant velocity over time. Data from two representative tracks with (red line) and without (black line) a transient rapid motion are shown. (E) Tracks of PB2-GFPcomp particles (blue lines) overlaid on the image of the microtubule network (mCherry-MAP4 signal acquired at zero time point, pseudo-colored red) in a representative cell. Scale bar, 2 μm. (F to H) Distribution of the migration lengths (distance between the initial and the final positions) (F and G) and distribution of the instant velocities (H) of PB2-GFPcomp particles. Data from ≈900 to 2,200 tracks per condition are presented. Red, 30 μM nocodazole-treated cells; green, 2.5 μM cytochalasin D-treated cells; blue, nocodazole and cytochalasin D-treated cells; black, mock-treated cells (dimethyl sulfoxide). In panel G, the y scale was stretched in order to highlight the distribution for small values.
Dynamics of the PB2 polymerase subunit in the nuclei of live influenza virus-infected cells.
Fluorescence correlation spectroscopy (FCS) was used to characterize the dynamics of the PB2 protein in the nucleus of 293T cells transiently expressing the GFP1-10 domain and either transfected with the PB2-GFP11 expression plasmid or infected at a high MOI with the WSN-PB2-GFP11 virus. 293T cells transiently coexpressing MBD-GFP11 were used as a control. The diffusion time (τdif) of the PB2-GFPcomp protein, when transiently expressed in the absence of other viral proteins, was about three times longer than that of MBD-GFPcomp (Fig. 10B, bottom black bar). This difference suggested that PB2 interacts with slowly diffusing nuclear partners and is in agreement with previously published observations from our group using the mCherry-PB2 fusion protein (13) and from others (20). Interestingly, in infected cells, the relative diffusion time of PB2-GFPcomp was five times longer than that of MBD-GFPcomp (Fig. 10A, red curve compared to the black one, and B, middle black bar), suggesting that in the course of infection, at least a portion of PB2 molecules either interacts with a distinct, overall slower-diffusing set of partner(s) and/or forms larger aggregates than when it is transiently expressed on its own.
Fig 10.
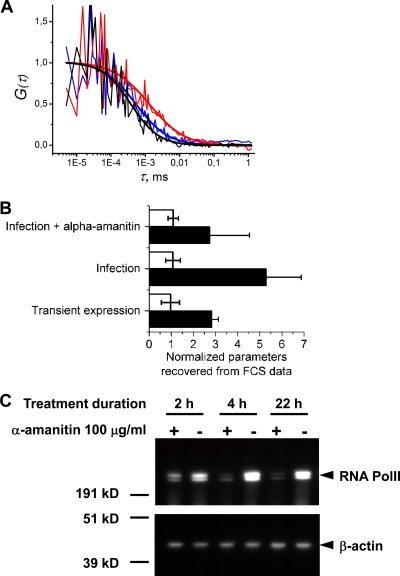
FCS analysis of PB2-GFPcomp protein dynamics in the nuclei of live infected cells. 293T cells transiently expressing GFP1-10 were infected at an MOI of 3 with the WSN-PB2-GFP11 virus, incubated in the absence or presence of 100 μg/ml α-amanitin from 2 h postinfection, and submitted to FCS measurements from 4 to 6.5 h postinfection as described in Materials and Methods. 293T cells cotransfected with the GFP1-10 and MBD-GFP11 expression vectors were used as a control. (A) Representative raw individual autocorrelation curves and the curves obtained by global fitting of 12 to 20 individual curves/condition are shown for cells expressing the MBD-GFP11 protein (black curves) and cells infected with WSN-PB2-GFP11 without (red curves) or with (blue curves) α-amanitin treatment. (B) The characteristic diffusion time (black bars) and molecular brightness (white bars) recovered from FCS data analysis. The values measured for PB2-GFP11 transient expression, WSN-PB2-GFP11 infection, and WSN-PB2-GFP11 infection in the presence of α-amanitin (originally in ms and in counts per second per particle, respectively) were normalized with respect to the corresponding values for the transiently expressed MBD-GFP11 protein. In each transfection/infection experiment, 8 to 20 individual autocorrelation curves were acquired for each condition. The diffusion time for each condition was determined from global fitting of all curves acquired under that condition. The data are shown as mean values ± SD of two to four independent transfection/infection experiments. (C) Western blot analysis of RNA Pol II steady-state levels in α-amanitin-treated cells. 293T cells were incubated in the absence (−) or presence (+) of 100 μg/ml of α-amanitin for the indicated time. Whole-cell lysates were prepared, loaded on a denaturing polyacrylamide gel, and analyzed by Western blotting using monoclonal antibodies specific for the human RNA Pol II large subunit and ß-actin, as described in Materials and Methods. Protein molecular mass markers (kD) are indicated.
To discriminate between the two latter possibilities, we reasoned that if the 5-fold increase of PB2-GFPcomp τdif compared to MBD-GFPcomp τdif was due exclusively to the formation of aggregates, it would reflect a 125-fold increase in size of PB2-GFPcomp aggregates compared to the MBD-GFPcomp monomer as the translational diffusion time is proportional to the cubic root of the particle hydrodynamic volume. The relative brightness levels of MBD-GFPcomp and PB2-GFPcomp, either expressed alone or in the context or the viral infection, were determined. No significant difference was observed (Fig. 10B, white bars), which argues against the presence of large aggregates of PB2 or aggregates of trimeric polymerase complexes and thus suggests that interactions with slowly diffusing partners is the major cause for the decreased mobility of the PB2 protein. A likely cause for the slowing down of the PB2 diffusion rate is the association of viral polymerase with cellular factors, together with its incorporation within vRNPs. As a functionally important association between the viral polymerase and the cellular RNA polymerase II (Pol II) large subunit has been described (9, 41, 44), FCS measurements were repeated on cells treated with α-amanitin, known to specifically induce Pol II degradation (30). The efficacy of α-amanitin treatment was controlled by Western blot analysis of total cell lysates using an anti-Pol II antibody (Fig. 10C). When α-amanitin was added immediately after infection of GFP1-10-expressing 293T cells with the WSN-PB2-GFP11 virus, no PB2-GFPcomp fluorescence was detected (data not shown), in agreement with previous reports that α-amanitin inhibits influenza virus replication (41). When α-amanitin was added at 2 h postinfection and removed at 4.5 to 5 h postinfection, the diffusion time of PB2-GFPcomp was apparently reduced compared to that in untreated cells (Fig. 10A, blue curve compared to the red one, and B, top black bar), whereas the diffusion time of transiently expressed EGFP or MBD-GFPcomp remained unchanged (data not shown). Our observations were in agreement with the α-amanitin-induced 1.6-fold increase in mobility of transiently expressed PB1-PB2-PA complexes previously reported (20) and confirmed in an infectious setting that influenza virus polymerase dynamics is affected by the steady-state levels of Pol II.
DISCUSSION
Studies on the intracellular trafficking of influenza virus RNPs and polymerase subunits are currently limited by the absence of a method enabling their direct visualization during infection in single cells. The split-GFP-based recombinant virus described here enables direct and extended real-time observation of the nuclear accumulation of the influenza virus PB2 polymerase subunit and subsequent cytoplasmic trafficking of newly synthesized vRNPs in the context of an infection. Growth curves, polymerase activity, the high level of agreement of our results with those obtained by immunofluorescence of fixed cells, and, finally, the demonstration that PB2-GFPcomp-containing RNPs are incorporated into progeny infectious virions demonstrate that the WSN-PB2-GFP11 virus behaves essentially the same as the wild type. It is thus an excellent tool for the study of vRNP trafficking and other late events of the influenza virus life cycle such as vRNP packaging. Although we applied the split-GFP method to label PB2, the same approach in principle could be used to study other influenza virus proteins in living cells. An alternative to our approach is the biarsenical-tetracysteine (TC) labeling system which has already been used to label the NS1 protein of an infectious influenza virus (18). However, applications of the TC labeling method are limited by toxicity and a high background fluorescence due to nonspecific labeling of cellular proteins, which do not occur with the split-GFP labeling method. Another limitation is that the TC-tagged molecules synthesized after exposure to the biarsenical dye remain undetectable. On the other hand, TC labeling is particularly useful for instant visualization of viral protein molecules accumulated at the time of labeling, whereas the split-GFP labeling method is less appropriate for this purpose because reconstitution of the GFP by transcomplementation is not instantaneous. The split-GFP method could potentially be combined with the biarsenical-tetracysteine labeling to allow simultaneous visualization of two or more viral proteins in single live cells. An alternative method to visualize vRNPs in live infected cells was reported while the manuscript was in preparation, involving the delivery of a fluorescently labeled anti-vRNP antibody to the cytoplasm using a transfection reagent (26). This method presents certain advantages (different viral proteins can be visualized with a wide choice of fluorescent labels without the need for genetic engineering of the virus) but also substantial drawbacks, such as a strong nonspecific signal yielded by unbound antibodies and a limited efficiency of delivery of the antibodies to some subcellular compartments, such as the nucleus. High intracellular levels of the large GFP1-10 fragment are required for the split-GFP method, which initially limited its application to highly transfectable cell lines. However, we were able to develop A549 and MDCK cell lines stably expressing GFP1-10, which overcomes this limitation and opens the way for easier and broader applications of the split-GFP system to both viral and nonviral systems.
Split-GFP labeling of PB2 generated a strong and specific fluorescent signal and did not perturb viral infection. Combined with fluorescence correlation spectroscopy, it allowed us to confirm, in an infectious setting, that the intranuclear dynamics of the viral polymerase is affected by the steady-state levels of cellular RNA polymerase II (20). This approach should prove useful to further explore interactions of the viral polymerase with nuclear proteins. Live imaging of cells infected with a split-GFP-based virus demonstrated, at 3 to 7 h postinfection, a pericentriolar accumulation of viral RNPs followed by a wide distribution throughout the cytoplasm and an accumulation at the plasma membrane. These observations were in agreement with previous observations of fixed cells (3, 14) and with recent studies suggesting that exported RNPs initially concentrate at the pericentriolar recycling endosome through interactions with Rab11 (1, 7, 26). In the cytoplasm of cells infected with the split-GFP-based virus, vRNP-containing particles generally moved nondirectionally with an average speed of <0.25 μm/s. Rapid directional movements were sporadically observed, some of which coincided with microtubules. The observed dynamics of PB2-GFPcomp particles qualitatively agrees with the observations by Momose et al. of in vivo immunolabeled vRNPs showing intermittent, rapid, directional motion events in addition to continuous slow diffusion-like motions (26). The discrepancies between the distribution of the velocities reported by Momose et al. and those reported in the present study are most probably due to the use of different algorithms for particle tracking analysis, different instrumental settings, and different cell lines. When microtubules were disrupted by nocodazole, we no longer observed the longest migrations of PB2-GFPcomp particles (>4 μm); but sporadic rapid directional motions could still be detected, and the overall distribution of the instant velocities remained almost unchanged. Our observations are in agreement with the proposed involvement of microtubules in the transport of vRNPs (1, 26). However, microtubule disruption results in only a moderate decrease of the production of viral particles (1), suggesting that other mechanisms of transport are also important. We therefore studied the effect of actin depolymerization and observed, for the first time, that this also significantly affects the motion of progeny vRNPs. Our particle tracking data strongly suggest that both microtubules and actin filaments are involved in the trafficking of vRNPs in the infected cells.
In conclusion, we have shown that split-GFP-based recombinant viruses will be useful tools to elucidate the mechanisms underlying influenza virus vRNP trafficking and packaging as well as the dynamics of the viral proteins and their interactions with host proteins in live infected cells. Furthermore, these viruses could also be the basis for the development of new, high-throughput assays for inhibitors of influenza virus replication or genome-wide small interfering RNA (siRNA) screens for host factors important in the later stages of viral infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jan Ellenberg, Catherine Chapuis, Nathalie Daigle, Sebastien Huet, and Peter Lenart (EMBL, Heidelberg, Germany), Laurent Blanchoin (CEA, Grenoble, France), Jaakke Saraste (University of Bergen, Norway), and Yves Jacob (Institut Pasteur, Paris, France) for advice, discussions, and plasmids; Pierre Charneau and Philippe Souque (Institut Pasteur, Paris, France) for advice and help with the production of recombinant lentiviruses; George Brownlee (Oxford University) for reverse genetics plasmids; Juan Ortin and Lorena Ver (CSIC, Madrid, Spain) for anti-PB2 monoclonal antibodies, the IF protocol, and plasmids; Roland Wedlich-Soldner for the lifeact plasmid; Robert G. Webster and Scott Krauss (St. Jude's Children Research Hospital, Memphis, TN) for MAb 3/1; Thomas Lunardi (EMBL, Grenoble, France) and Mehdi Cherrar (Institut Pasteur) for technical help; Alexei Grichine for advice on instrumentation at the Microscopie Photonique—Imagerie Cellulaire facility of the Institute Albert Bonniot (Grenoble, France); and Emmanuelle Perret and Jean-Yves Tinevez for advice on instrumentation at the Plateforme d'Imagerie Dynamique facility at Institut Pasteur (Paris, France).
S.A. was supported by a Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme (grant agreement PIEF-GA-2009-236959), and S.A. and O.S. were supported by EIPOD fellowships (EMBL). This work was supported in part by the EU FP7-funded FLU-PHARM project (FP7-INFLUENZA-2010 grant 259751).
Footnotes
Published ahead of print 23 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Amorim MJ, et al. 2011. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 85:4143–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beignon AS, et al. 2009. Lentiviral vector-based prime/boost vaccination against AIDS: pilot study shows protection against Simian immunodeficiency virus SIVmac251 challenge in macaques. J. Virol. 83:10963–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruce EA, Digard P, Stuart AD. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 84:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabantous S, Terwilliger TC, Waldo GS. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23:102–107 [DOI] [PubMed] [Google Scholar]
- 5. Crescenzo-Chaigne B, Naffakh N, van der Werf S. 1999. Comparative analysis of the ability of the polymerase complexes of influenza viruses type A, B and C to assemble into functional RNPs that allow expression and replication of heterotypic model RNA templates in vivo. Virology 265:342–353 [DOI] [PubMed] [Google Scholar]
- 6. Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S, Naffakh N. 2005. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341:34–46 [DOI] [PubMed] [Google Scholar]
- 7. Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. 2011. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85:6117–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elton D, et al. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75:408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329–345 [DOI] [PubMed] [Google Scholar]
- 10. Fodor E, et al. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerlich D, Beaudouin J, Gebhard M, Ellenberg J, Eils R. 2001. Four-dimensional imaging and quantitative reconstruction to analyse complex spatiotemporal processes in live cells. Nat. Cell Biol. 3:852–855 [DOI] [PubMed] [Google Scholar]
- 12. Hayden F. 2009. Developing new antiviral agents for influenza treatment: what does the future hold? Clin. Infect. Dis. 48(Suppl 1):S3–S13 [DOI] [PubMed] [Google Scholar]
- 13. Huet S, et al. 2010. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 84:1254–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jo S, et al. 2010. Involvement of vesicular trafficking system in membrane targeting of the progeny influenza virus genome. Microbes Infect. 12:1079–1084 [DOI] [PubMed] [Google Scholar]
- 15. Jorba N, et al. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077–2088 [DOI] [PubMed] [Google Scholar]
- 16. Kent KP, Childs W, Boxer SG. 2008. Deconstructing green fluorescent protein. J. Am. Chem. Soc. 130:9664–9665 [DOI] [PubMed] [Google Scholar]
- 17. Keppler A, Ellenberg J. 2009. Chromophore-assisted laser inactivation of alpha- and gamma-tubulin SNAP-tag fusion proteins inside living cells. ACS Chem. Biol. 4:127–138 [DOI] [PubMed] [Google Scholar]
- 18. Li Y, et al. 2010. Genetically engineered, biarsenically labeled influenza virus allows visualization of viral NS1 protein in living cells. J. Virol. 84:7204–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Listwan P, Terwilliger TC, Waldo GS. 2009. Automated, high-throughput platform for protein solubility screening using a split-GFP system. J. Struct. Funct Genomics 10:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loucaides EM, et al. 2009. Nuclear dynamics of influenza A virus ribonucleoproteins revealed by live-cell imaging studies. Virology 394:154–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manicassamy B, et al. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U. S. A. 107:11531–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinez-Sobrido L, et al. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miranda AF, Godman GC, Tanenbaum SW. 1974. Action of cytochalasin D on cells of established lines. II. Cortex and microfilaments. J. Cell Biol. 62:406–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Momose F, Kikuchi Y, Komase K, Morikawa Y. 2007. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect. 9:1422–1433 [DOI] [PubMed] [Google Scholar]
- 26. Momose F, et al. 2011. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One 6:e21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 62:403–424 [DOI] [PubMed] [Google Scholar]
- 28. Neumann G, Brownlee GG, Fodor E, Kawaoka Y. 2004. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 283:121–143 [DOI] [PubMed] [Google Scholar]
- 29. Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nguyen VT, et al. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24:2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ochoa M, et al. 1995. Epitope mapping of cross-reactive monoclonal-antibodies specific for the influenza-a virus PA and PB2 polypeptides. Virus Res. 37:305–315 [DOI] [PubMed] [Google Scholar]
- 32. Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 33. Peiris JS, Tu WW, Yen HL. 2009. A novel H1N1 virus causes the first pandemic of the 21st century. Eur. J. Immunol. 39:2946–2954 [DOI] [PubMed] [Google Scholar]
- 34. Pleschka S, et al. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 70:4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rameix-Welti MA, Tomoiu A, Dos Santos Afonso E, van der Werf S, Naffakh N. 2009. Avian influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J. Virol. 83:1320–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Riedl J, et al. 2010. Lifeact mice for studying F-actin dynamics. Nat. Methods 7:168–169 [DOI] [PubMed] [Google Scholar]
- 37. Ruigrok RW, Crepin T, Hart DJ, Cusack S. 2010. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 20:104–113 [DOI] [PubMed] [Google Scholar]
- 38. Schwille P, Meyer-Almes FJ, Rigler R. 1997. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 72:1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shapira SD, et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shinya K, Fujii Y, Ito H, Ito T, Kawaoka Y. 2004. Characterization of a neuraminidase-deficient influenza a virus as a potential gene delivery vector and a live vaccine. J. Virol. 78:3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Spooner LL, Barry RD. 1977. Participation of DNA-dependent RNA polymerase II in replication of influenza viruses. Nature 268:650–652 [DOI] [PubMed] [Google Scholar]
- 42. Van Engelenburg SB, Palmer AE. 2010. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods 7:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vignuzzi M, Gerbaud S, van der Werf S, Escriou N. 2001. Naked RNA immunization with replicons derived from poliovirus and Semliki Forest virus genomes for the generation of a cytotoxic T cell response against the influenza A virus nucleoprotein. J. Gen. Virol. 82:1737–1747 [DOI] [PubMed] [Google Scholar]
- 44. Vreede FT, Chan AY, Sharps J, Fodor E. 2010. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396:125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watanabe K, et al. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 77:31–42 [DOI] [PubMed] [Google Scholar]
- 46. Yen HL, Webster RG. 2009. Pandemic influenza as a current threat. Curr. Top. Microbiol. Immunol. 333:3–24 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.