Abstract
The SCF (Skp1-Cul1-F-box) complex contributes to a variety of cellular events including meiotic cell cycle control, but its function during meiosis is not understood well. Here we describe a novel function of SCF/Skp1 in meiotic recombination and subsequent chromosome segregation. The skp1 temperature-sensitive mutant exhibited abnormal distribution of spindle microtubules in meiosis II, which turned out to originate from abnormal bending of the spindle in meiosis I. Bent spindles were reported in mitosis of this mutant, but it remained unknown how SCF could affect spindle morphology. We found that the meiotic bent spindle in skp1 cells was due to a hypertension generated by chromosome entanglement. The spindle bending was suppressed by inhibiting double strand break (DSB) formation, indicating that the entanglement was generated by the meiotic recombination machinery. Consistently, Rhp51/Rad51-Rad22/Rad52 foci persisted until meiosis I in skp1 cells, proving accumulation of recombination intermediates. Intriguingly bent spindles were also observed in the mutant of Fbh1, an F-box protein containing the DNA helicase domain, which is involved in meiotic recombination. Genetic evidence suggested its cooperation with SCF/Skp1. Thus, SCF/Skp1 together with Fbh1 is likely to function in the resolution of meiotic recombination intermediates, thereby ensuring proper chromosome segregation.
Introduction
SCF (Skp1-Cullin 1-F-box) complexes constitute a ubiquitin ligase family, members of which are involved in ubiquitylation and degradation of a number of cellular factors including cell cycle regulators and transcription factors. SCF complexes are composed of an unaltered core of Cullin 1, the RING finger protein Rbx1/Roc1/Hrt1, and Skp1, which can bind a number of different F-box proteins. F-box proteins act as receptors for specific substrates for ubiquitylation. The fission yeast Schizosaccharomyces pombe has eighteen F-box proteins, according to the S. pombe gene database (GeneDB; www.genedb.org/genedb/pombe/), which may regulate various cellular processes. It has been shown that F-box proteins Pop1 and Pop2 are required for the maintenance of genome ploidy through the timely destruction of Rum1 and Cdc18, which are a CKI (cyclin-dependent kinase inhibitor) and a DNA replication factor, [1], [2], [3], [4]. Another F-box protein Pof1 regulates the cadmium response by targeting the transcription factor Zip1, which controls expression of cadmium-induced genes [5]. Pof3, an F-box protein also, is involved in the maintenance of genomic integrity together with its binding partner Mcl1 [6], [7]. Phenotypes of the pof3-deletion (pof3Δ) mutant displays defects in genome integrity such as shortened telomeres, and the Rad3-dependent DNA damage checkpoint is indispensable for the survival of pof3Δ cells [7]. Pof3 is involved in the downregulation of the GATA-type transcription factor Ams2, which activates transcription of the core histone genes during S phase [8]. Further, Pof6 is required for cell separation [9], with its binding partner Sip1 [10]. Fbh1, an F-box protein containing a DNA helicase domain, is involved in the processing of chromosomal recombination intermediates [11].
Mutants of Skp1 are known to exhibit defective phenotypes in the cell cycle control. The skp1-a7 temperature-sensitive mutant arrests in G2 phase of the mitotic cell cycle due to activation of the Rad3-dependent DNA damage checkpoint [12]. The rad3Δ skp1-a7 double mutant suppressed the G2 arrest at the high temperature. This indicates that the DNA damage checkpoint is ectopically activated in the skp1-a7 mutant [12]. Interestingly, the skp1-a7 mutant exhibits abnormally-bent spindles and fails in nuclear division at anaphase [13]. This implies that Skp1 may regulate organization of spindle microtubules and/or elasticity of the nuclear envelope. The rad3Δ skp1-a7 double mutant, however, no longer shows the bent-spindle phenotype in anaphase [13]. The F-box protein responsible for this bent-spindle phenotype has not been identified.
Thus, fission yeast Skp1 regulates a wide variety of cellular events in the mitotic cell cycle, cooperating with many F-box proteins, but little is known about its function in meiosis. To investigate the possible commitment of Skp1 to meiosis, we observed the behavior of the skp1 mutant in meiosis and subsequent sporulation. We noticed that abnormal spores were generated in the skp1-a7 mutant. We also realized that skp1-a7 cells frequently displayed abnormal X-shaped spindles in meiosis II and failed in nuclear division. Further analyses revealed that these phenotypes originate from the bent spindle generated in meiosis I, which is similar to the one previously observed in mitosis. We show that phenotypic abnormalities observed in skp1 mutant cells are attributable to defects in Fbh1-mediated meiotic recombination.
Results
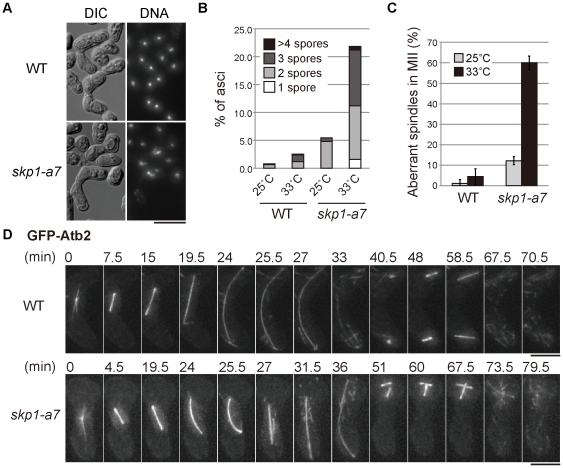
To investigate the meiotic function of SCF/Skp1 in fission yeast, meiosis was induced in the skp1-a7 mutant [12]. Throughout this study we performed observation of meiotic progression at the semi-restrictive temperature for the mutant (33°C), not at the restrictive temperature (36°C), because meiosis is intrinsically sensitive to high temperature and does not proceed at 36°C. After conjugation, wild-type zygotes underwent meiosis I (MI) and meiosis II (MII) and generated four spores in an ascus (Fig. 1A). In contrast, skp1-a7 zygotes frequently produced an abnormal number of chromosome masses (less than four) and showed a high rate of two- or three-spored ascus formation (Fig. 1A,B). To pinpoint which stage of the cell cycle was impaired by the loss of functional Skp1, microtubules were visualized using GFP-Atb2 (green fluorescent protein-fused α2-tubulin). 14–15 hours after the induction of meiosis, we frequently found zygotes with abnormal spindle structure in MII. Most of wild-type zygotes (WT) showed a pair of spindles at MII, whilst spindles in about 60% of skp1-a7 zygotes appeared to be abnormal, represented typically by a ‘cross’ of two spindles as shown in Fig. 1C.
Figure 1. The skp1-a7 mutant shows the bent spindle in MI.
(A) Meiosis and sporulation were induced for wild-type (WT) and the skp1-a7 strains at 33°C, the semi-restrictive temperature. Images of DIC and DNA stained with DAPI are shown. (B) The number of spores in WT and skp1-a7 asci were counted. Percentages of aberrant asci only are shown in the graph. Most of WT cells generated four spores, whereas skp1-a7 cells produced abnormal asci with 1∼3 and ≥5 spores. Meiosis was induced at the indicated temperature. n = 250. (C) Percentages of aberrant ‘crossing’ spindles in MII. Meiosis was induced at the indicated temperature. n≥30. (D) Time-lapse images of WT and skp1-a7 cells expressing GFP-Atb2 (α2-tubulin) from the MI onset until anaphase/telophase of MII. In the skp1-a7 cell, the spindle abnormally bent at 25.5 min before full elongation, and collapsed at 27 min. Note that the WT spindle never bent before reaching the cell cortex. The crossing spindles in MII are shown at 51∼67.5 min. Bars, 5 µm.
Time-lapse imaging of WT and skp1-a7 meiotic cells was performed to elucidate the reason for the spindle abnormality. In WT zygotes, a spindle elongated in straight in anaphase I until both ends of the spindle reached to the cell tips (24 min, top; Fig. 1D). Then the spindle bent slightly in accordance with the shape of the zygote and disassembled upon MI exit (33 min). After the completion of MI, an MII spindle started to form in each of the two nuclei (40.5 min) and elongated. In the skp1-a7 mutant, the spindle morphology was almost normal in the early stage of MI (bottom; Fig. 1D). The spindle, however, did not fully elongate even nearly 20 minutes after the MI onset (19.5 min, Fig. 1D). Interestingly, the spindle started to bend without touching the cell cortex (24–25.5 min), and then collapsed (the V-shape spindle at 27 min), implying that an unusual intolerable tension was imposed to the spindle. After the collapse, two bundles were merged into one and resumed to grow to a comparable length to the WT spindle (31.5 and 36 min). This rebuilt spindle cannot be bipolar: two spindle poles are located only to one side of the broken spindle, and the other side (the broken ends of microtubules) has no SPBs. This unequal segregation of SPBs then caused the co-existence of two spindles in one nucleus in MII (48 min). We conclude that the collapse of an abnormally-bent spindle is the cause for the ‘crossing spindle’ observed in skp1-a7 cells.
The bent spindle was observed previously during mitosis of the skp1-a7 mutant [12], but the reason remained unclear. We thus sought for the reason to generate a bent spindle. One possibility might be that the spindle is hyperstabilized in the skp1-a7 mutant. Alternatively, the architecture of the nuclear envelope might be altered. However, we could not obtain supporting data for these hypotheses (data not shown). We then hypothesized that the abnormal tension might be caused by some event(s) specifically seen before or during MI, because the MII spindle of the skp1-a7 mutant was not bent (51, 60 and 67.5 min, Fig. 1D). MI is characterized by the unique chromosome organization: homologous chromosomes derived from the parents are paired and form chiasmata, and then they are segregated (reductional division), in contrast to the separation of the sister-chromatids in mitosis and MII (equational division; reviewed in [14], [15]).
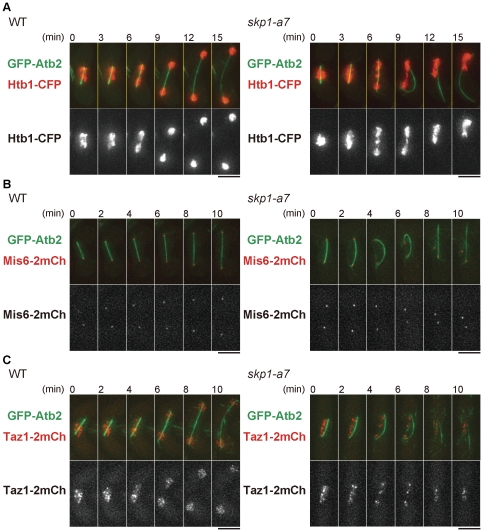
The behavior of chromosomes in the skp1-a7 mutant was monitored using Htb1-CFP, histone H2B fused with cyan fluorescent protein, together with GFP-Atb2 to visualize microtubules. In MI of WT cells, Htb1-CFP segregated in equal amounts (Fig. 2A). In the skp1-a7 mutant, however, an unsegregated bulge of Htb1-CFP was seen before the spindle started to bend (6 min, skp1-a7). Then the spindle bent, as if it were a bow with a series of ‘bridged’ chromosomes as a bowstring. When the spindle collapsed, most of the chromosome mass gathered to one side of the zygote (12 min), resulted in missegregation of the chromosomes. We then tracked the behavior of kinetochores by visulalizing Mis6 [16] tagged with two copies of mCherry. In WT, Mis6-2mCherry foci segregated equally in MI anaphase (anaphase I) (WT, Fig. 2B). In the skp1-a7 mutant, Mis6-2mCherry foci split equally and reached to SPBs in anaphase I as in WT (0 min, skp1-a7), but the two foci moved back close to each other when the spindle bent and collapsed (6–10 min). This indicates that the kinetochore-microtubule attachment and kinetochore segregation in MI once occurred normally in the skp1-a7 mutant, by the time the spindle collapsed. Telomeres, the ends of chromosomes, were next visualized using the telomere-binding protein Taz1-2mCherry [17]. In WT, Taz1-2mCherry foci segregated equally as anaphase I proceeded (Fig. 2C). In skp1-a7, however, some Taz1-2mCherry foci remained in the middle of the nucleus when the spindle bent (4, 6 min). Taking these observations together, the centromeric region of chromosomes appeared to separate normally but the arm region was not fully segregated at the anaphase I onset in the skp1-a7 mutant. This was a likely cause of the chromosomal entanglement observed when the spindle bent. To confirm that the defects in arm separation and the emergence of the bent spindle are linked, we removed chromosome cohesion by deleting the rec8 gene. Rec8 protein is a meiotic cohesin, which adheres sister and homologous chromosomes until metaphase I [18]. Upon the anaphase I onset, Rec8 is cleaved by separase and homologous chromosomes are segregated. Cells with rec8 disrupted (rec8Δ) lose chromosomal cohesion in meiosis. Importantly, in the double mutant of skp1-a7 rec8Δ, the bent-spindle phenotype was rarely observed (Fig. 3A). This strongly indicates that chromosomes cannot be completely resolved in skp1-a7 cells. As a consequence, the spindle might induce an intolerable tension that causes the sudden collapse.
Figure 2. Chromosome segregation defects in skp1-a7 cells at MI.
Time-lapse images of WT and skp1-a7 zygotes at MI with indicated fluorescent proteins filmed at 33°C. (A) GFP-Atb2 for microtubules (green) and Htb1-CFP for histone H2B (red). In skp1-a7, the chromosome bridge is seen at 6∼9 min, followed by an appearance of the bent spindle evident at 9 min. The spindle then collapsed and the rebuilt spindle is seen at 12∼15 min. (B) GFP-Atb2 (green) with Mis6-2mCh for kinetochores (red). (C) GFP-Atb2 (green) with Taz1-2mCh for telomeres (red). Bars, 5 µm.
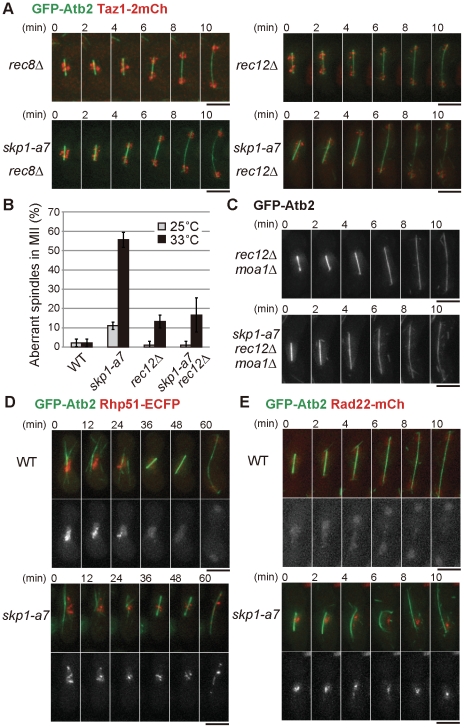
Figure 3. The chromosome entanglement is made through meiotic recombination.
(A) Chromosome segregation at MI in WT and skp1-a7 cells was monitored in the rec8Δ and rec12Δ background. Note that the spindle was not bent in the skp1-a7 rec8Δ and skp1-a7 rec12Δ. GFP-Atb2 (green) and Taz1-2mCherry (red) are shown. (B) Suppression of the crossing-spindle phenotype in skp1-a7 cells by rec12Δ. Frequency of ‘crossing’ spindles in MII at 25°C and 33°C. n≥30. Error bars, SEM. (C) The spindle was straight in skp1-a7 rec12Δ moa1Δ zygotes. Note that in rec12Δ moa1Δ cells, sister-chromatids are joint until metaphase I and then the sister-chromatids are segregated because of the kinetochore bipolarity. (D) Rhp51-ECFP foci persisted until MI in the skp1-a7 mutant. GFP-Atb2 (green) and Rhp51-ECFP (red) are shown. (E) Rad22-mCherry foci also persisted until MI in the skp1-a7 mutant. Bars, 5 µm.
The chromosomal non-disjuntion can be possibly due to meiotic recombination defects. To test this, we removed Rec12 from the skp1-a7 mutant. Rec12 is a fission yeast ortholog of the Spo11 endonuclease, which induces double-strand breaks (DSBs) in chromosomes during meiotic prophase [19], [20]. In rec12Δ cells, homologous chromosomes are not recombined and hence do not form chiasmata [20]. The bent-spindle phenotype of skp1-a7 cells was suppressed by introducing rec12Δ (Fig. 3A,B), indicating that the recombination of homologous chromosomes is involved in production of the chromosome non-disjunction observed in the skp1-a7 mutant. The suppression was similarly seen in rec12Δ moa1Δ skp1-a7 cells (Fig. 3C), in which the chromosome segregation is performed in an equational manner due to lack of kinetochore monopolarity [21]. This indicates that the non-disjunction does not occur among sister chromatids, supporting the possibility of the recombination-dependent entanglement.
We then visualized the DNA repair protein Rhp51 (the fission yeast ortholog of Rad51/RecA [22], [23]). Rhp51 binds to single- and double-strand DNA and promotes annealing and exchange of strands via its recombinase activity, and nuclear Rhp51 foci are markers of recombination intermediates that contain single-strand DNA (ssDNA) [24]. Rhp51-ECFP formed intense foci in the WT nucleus of meiotic prophase, which diminished as the cells entered MI (WT, Fig. 3D). By contrast, in skp1-a7 zygotes, the Rhp51-ECFP foci persisted even during MI, when the cells displayed chromosome non-disjunction (skp1-a7, Fig. 3D). Rad22, another repair protein that interacts with Rhp51, also forms foci at DSB sites [25], [26]. skp1-a7 zygotes exhibited prolonged localization of Rad22-mCherry foci even in MI (Fig. 3E). Persistence of Rhp51 and Rad22 foci indicates that the recombination complex, comprising the invading ssDNA and the recombination factors, was not properly processed. The persistence of the complex may cause the significant non-disjunction of chromosome arms.
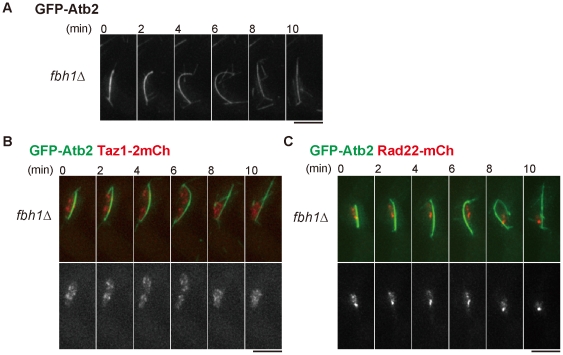
SCF functions as a complex of Skp1, Cullin 1 and F-box proteins, which are thought to determine the specificity of binding proteins or degradation substrates [27]. We next sought for an F-box protein responsible for the meiotic function of SCF/Skp1 among eighteen F-box proteins reported to date (GeneDB; www.genedb.org/genedb/pombe/). Among deletion mutants of these F-box protein, we screened all viable strains for the one that reproduced the bent spindle seen in the skp1-a7 mutant. None showed a bent-spindle in MI, except for fbh1Δ (Fig. 4A). Fbh1/Fdh1 is a unique F-box protein, which has an UvrD/REP helicase domain at the C-terminus, in addition to the conserved F-box motif at the N-terminus [11], [28]. Fbh1 is known to process the recombination intermediates in mitosis [11] and meiosis [29]. The frequency of the appearance of a bent spindle at MI in the fbh1Δ mutant reached almost 40% (see Fig. 5C), which was comparable to the percentage in the skp1-a7 mutant. Similarly to skp1-a7, fbh1Δ cells showed defects in chromosome disjunction (indicated by Taz1-2mCherry, Fig. 4B). fbh1Δ cells also exhibited persistence of Rad22-mCherry foci during MI, like skp1-a7 cells (Fig. 4C). Thus, the fbh1Δ mutant displayed nearly the same phenotype as the skp1-a7 mutant during meiosis.
Figure 4. The fbh1 deletion mutant shows similar phenotypes to the skp1-a7 mutant.
(A) The bent spindle was frequently observed in fbh1Δ cells at MI. Time-lapse imaging for GFP-Atb2 is shown. (B) The fbh1Δ mutant exhibited non-disjunction of chromosome arms visualized by Taz1-2mCherry (red) with GFP-Atb2 (green). (C) The fbh1Δ mutant showed persistent Rad22-mCherry foci until MI. Bars, 5 µm.
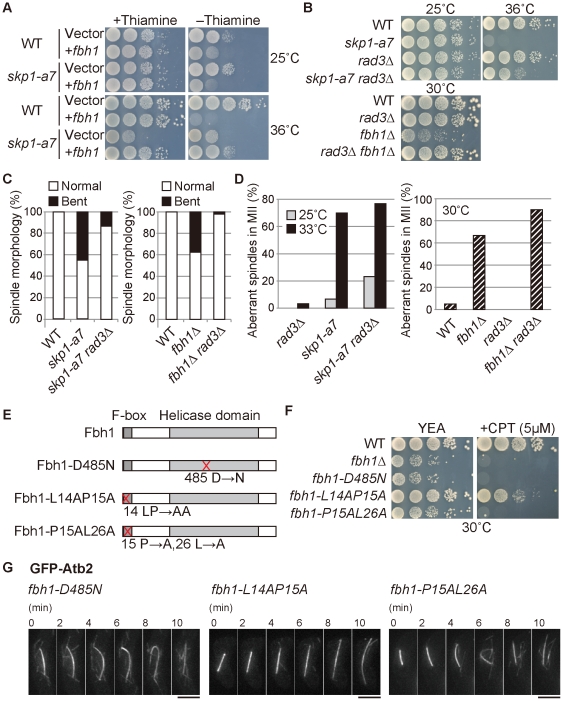
Figure 5. Skp1 together with Fbh1 resolves meiotic recombination intermediates.
(A) Temperature sensitivity of the skp1-a7 mutant was partially suppressed by overexpression of Fbh1 from the plasmid vector pREP1. Ten-fold serial dilution of the indicated strains was spotted on minimal media with or without thiamine and then incubated at 25°C and 36°C. Overexpression is induced on the medium without thiamine (−thiamine). (B) Growth defects of skp1-a7 and fbh1Δ mutants were suppressed by rad3Δ, removal of the Rad3-dependent DNA damage checkpoint. (C, D) Frequency of the normal and bent spindle in mitosis (C) and the aberrant ‘crossing’ spindles in MII (D) in the indicated strains. n≥30. (E) Schematic diagram for Fbh1 WT and Fbh1-D485N (the helicase-dead form), Fbh1-L14A P15A and Fbh1-P15A L26A (the F-box-deficient form) mutant proteins. (F) Sensitivity to 5 µM CPT (camptothecin) was tested for the indicated strains. (G) fbh1-D485N and fbh1-P15A L26A mutants exhibited the bent-spindle phenotype, whereas fbh1-L14A P15A did not. GFP-Atb2 in MI of the indicated strains is shown. Bars, 5 µm.
To further investigate if the phenotypes of skp1-a7 cells could be explained by a lack of Fbh1 as the responsible F-box protein, the genetic interaction of skp1 and fbh1 was then tested. Elevated expression of Fbh1 partially suppressed the growth defects of skp1-a7 at the restrictive temperature (Fig. 5A). As reported previously [12], the temperature sensitivity of the skp1-a7 mutant was suppressed by removal of Rad3, the ATR kinase of the DNA damage checkpoint machinery (Fig. 5B). Deletion of the rad3 + gene also suppressed the growth defects of the fbh1Δ mutant (Fig. 5B). These results prove the relevance of the skp1-a7 and fbh1Δ phenotypes.
In mitosis, the bent-spindle phenotype was no longer observed in either the skp1-a7 rad3Δ or the fbh1Δ rad3Δ mutants (Fig. 5C) [13]. This could mean that the Rad3-dependent DNA damage checkpoint was ectopically activated in skp1-a7 and fbh1Δ cells, which caused the defects [12]. Interestingly, however, removal of Rad3 did not suppress the ‘crossing’ spindles in MII (Fig. 5D). This suggests that the bent-spindle in MI was caused by persistent recombination intermediates generated in the process of meiotic recombination, but independently of the Rad3-dependent checkpoint machinery. To further investigate if SCF/Skp1 functions in resolution of recombination intermediates in cooperation with the DNA helicase activity of Fbh1, we created the fbh1-P15A L26A [30] and fbh1-L14A P15A [28] mutants carrying the dysfunctional F-box domain, which is required for the interaction to Skp1, and the fbh1-D485N mutant defective in the helicase activity [28] (Fig. 5E). The fbh1-P15A L26A mutant showed sensitivity to genotoxins during vegetative growth as the fbh1 disruptant did (Fig. 5F) [30], whilst the fbh1-L14A P15A mutant showed only minor sensitivity (Fig. 5F) [28]. It is possible that the F-box function in the latter mutant might not be fully deteriorated. Reflecting the genotoxin sensitivity, the fbh1-P15A L26A mutant showed the bent spindle, whereas the fbh1-L14A P15A mutant did not (Fig. 5G). These results together indicate that the bent spindle emerges when the function of the F-box in Fbh1 is fully inhibited. The fbh1-D485N mutant also frequently displayed the bent spindle, confirming the necessity of helicase activity (Fig. 5G). Taken together, we conclude that not only the helicase activity of Fbh1, but also its binding to Skp1 is required for the resolution of the chromosomal entanglement. Thus, SCF/Skp1, together with the Fbh1 DNA helicase, is responsible to resolve the recombination intermediates in meiosis.
Discussion
This study has illuminated the mechanism how the bent spindle is generated in the skp1 mutant, clarifying the function of SCF/Skp1 in fission yeast meiosis. The bent spindle in skp1-a7 is seen during anaphase of mitosis and MI. The spindle starts to bend before reaching the cell cortex. The nucleus containing the bent spindle neither elongate nor divide, indicating that abnormal tension is generated against microtubule extension in anaphase. It has been speculated that this is due to some defects in microtubule organisation, and/or in elasticity of the nuclear envelope. Our results shown here, however, demonstrate that the bent spindle is generated mainly by chromosomal entanglement, rather than the defects in the spindle or nuclear envelope, at least in MI. Telomeres failed to segregate, whilst kinetochores did segregate, supporting this notion. The bent spindle was no longer seen in the double mutants of skp1-a7 rec8Δ, indicating that the abnormal spindle tension caused by skp1-a7 mutation is due to the chromosomal junction. Moreover, the bent-spindle phenotype was also suppressed by removal of Rec12, strongly indicating that the entanglement was generated through meiotic recombination. Indeed, Rhp51 and Rad22 foci, which localize to DSB sites during meiotic recombination in prophase, persisted even in MI, supporting the notion that the DSB is not fully repaired in the skp1-a7 mutant. An accumulation of Rad22 and Rhp51 foci indicates that the ssDNA-containing recombination intermediates may not be resolved. Alternatively, DNA repair might be blocked in the skp1-a7 mutant at an earlier stage before proceeding to an intermediate that can be resolved. These two possibilities are not mutually exclusive. At least in budding yeast, the DNA damage response blocks induction of gene expression necessary for the resolution of recombination intermediates [31], [32]. Thus, a failure to repair some breaks could cause a regulatory block to intermediate resolution at other breaks. This might explain why the bent spindle in the skp1-a7 mutant was suppressed by rad3Δ in the mitotic cell cycle [13]. In contrast, the aberrant spindle of skp1-a7 cells in MII was not suppressed by rad3Δ (Fig. 5D), indicating that in addition to the Rad3-dependent DNA damage checkpoint, some other mechanism specifically equipped for meiotic recombination may operate to block the function of repair machinery. It may also be possible that the Rad3-dependent checkpoint is not responsible for the bent spindle in meiosis.
We screened for an F-box mutant that could generate bent spindles as in the skp1-a7 mutant, and identified Fbh1. Recently it is reported that Fbh1 is involved in the resolution of meiotic recombination [29]. This is, therefore, consistent with our results, and we further propose that SCF/Skp1 together with the F-box protein Fbh1, is involved in the repair of DSBs generated by Rec12 for meiotic recombination. We speculate that the recombination intermediates remain in the skp1-a7 and fbh1Δ mutants even in MI, which results in the entanglement of chromosomes and generation of abnormal tension against the spindle. There are two DNA helicases implicated in the processing of recombination intermediates in yeast, namely Srs2 and Rqh1/RecQ [33], [34]. In S. pombe, srs2Δ cells do not show significant defects in meiosis [35]. The rqh1Δ mutant did not show the bent spindle in MI, in contrast to the fbh1Δ mutant (our unpublished results). This is consistent with the previous study reporting that Rqh1 does not play a major role in DSB formation and repair [35]. As indicated in the previous report [11], Srs2 or Rqh1 may repress accumulation of spontaneously arising recombination intermediates during the mitotic cell cycle, and Fbh1 is required for the resolution of both mitotic and meiotic recombination intermediates. Hence three DNA helicases in fission yeast play distinct roles in DNA recombination during both mitosis and meiosis.
We also found that the F-box mutant fbh1-P15A L26A showed the bent spindle phenotype as fbh1Δ and skp1-a7 did, indicating that Skp1 and Fbh1 act together to resolve the meiotic recombination intermediates. It remains unclear, however, what biological benefit Fbh1 accepts by acting together with SCF/Skp1. It is previously reported that the F-box of Fbh1 is required for the localization of Fbh1 itself and Skp1 to the sites damaged by genotoxins [30]. We now envision that some proteins involved in recombination repair may need to be degraded through SCF-dependent proteolysis. It would be interesting to investigate the protein stability and ubiquitilation of a number of recombination repair factors, to identify the crucial substrates of SCF/Skp1-Fbh1, which would give us the new molecular insight as to how recombination intermediates are resolved in mitosis and meiosis.
Materials and Methods
Yeast Strains, Plasmids and Media
S. pombe strains used in this study are listed in Table 1. Standard methods for yeast genetics were used [36]. Briefly, for vegetative growth, a rich medium YEA, a synthetic medium SD, and a minimal medium MM with a nitrogen source were used. For induction of meiosis, h 90 heterothallic strains were plated onto the sporulation agar (SPA) or SSA and incubated at the indicated temperature (25°C–33°C) for 23∼26 hours to count the number of nuclei and/or spores, or mainly 12∼17 hours for subsequent live-cell imaging. For overexpression of the fbh1 gene, the plasmid pREP1 [37] was used as the vector. Overexpression was induced on the minimal media without thiamine. The rhp51-ECFP strain is a gift from Y. Tsutsui and H. Iwasaki [38]. The GFP-Atb2 strain used in this study is previously described [39]. For construction of strains expressing other fluorescent protein tags from the native promoter and gene disruptants, the conventional methods for the PCR-based gene targeting were used [39], [40].
Table 1. The S. pombe strains used in this study.
| Strain | Genotype | Figures |
| JY3 | h 90 | 1A |
| SO1650 | h 90 skp1-a7 | 1A |
| SO657 | h 90 GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 1B, 1C, 1D, 3B, 5B, 5C, 5D |
| SO656 | h 90 skp1-a7 GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 1B, 1C, 1D, 3B, 5B, 5C, 5D |
| SO974 | h 90 htb1-CFP-kan GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 2A |
| SO976 | h 90 skp1-a7 htb1-CFP-kan GFP-atb2-kan ade6-M216 leu1-32 | 2A |
| SO1006 | h 90 mis6-2mCherry-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 2B |
| SO1004 | h 90 skp1-a7 mis6-2mCherry-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 2B |
| SO1043 | h 90 taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 2C |
| SO1041 | h 90 skp1-a7 taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 2C |
| SO1345 | h 90 rec12::ura4+ taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3A |
| SO1343 | h 90 skp1-a7 rec12::ura4+ taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3A |
| SO1129 | h 90 rec8::ura4+ taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3A |
| SO1127 | h 90 skp1-a7 rec8::ura4+ taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3A |
| SO1056 | h 90 rec12::hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3B |
| SO1054 | h 90 skp1-a7 rec12::hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3B |
| SO1367 | h 90 rec12::hph moa1::bsd GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3C |
| SO1365 | h 90 skp1-a7 rec12::hph moa1::bsd GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3C |
| SO1529 | h 90 rhp51-ECFP-ura4+-rhp51+ GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3D |
| SO1527 | h 90 skp1-a7 rhp51-ECFP-ura4+-rhp51+ GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3D |
| SO1514 | h 90 rad22-mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3E |
| SO1512 | h 90 skp1-a7 rad22-mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 3E |
| SO1393 | h 90 fbh1::bsd GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 4A, 5C, 5D |
| SO1569 | h 90 fbh1::bsd taz1-2mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 4B |
| SO1658 | h 90 fbh1::bsd rad22-mCh-hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 4C |
| SO1593 | h 90 ade6-M216 leu1-32 ura4-D18 pREP1 | 5A |
| SO1595 | h 90 ade6-M216 leu1-32 ura4-D18 pREP1-fbh1 | 5A |
| SO1605 | h 90 skp1-a7 ade6-M216 leu1-32 ura4-D18 pREP1 | 5A |
| SO1553 | h 90 skp1-a7 ade6-M216 leu1-32 ura4-D18 pREP1-fbh1 | 5A |
| SO920 | h 90 rad3::hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 5B, 5D |
| SO922 | h 90 skp1-a7 rad3::hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 5B, 5C, 5D |
| JY878 | h 90 ade6-M216 leu1-32 ura4-D18 | 5B, 5F |
| SO918 | h 90 rad3::hph ade6-M216 leu1-32 ura4-D18 | 5B |
| SO1316 | h 90 fbh1::bsd ade6-M216 leu1-32 ura4-D18 | 5B, 5F |
| SO1652 | h 90 fbh1::bsd rad3::hph ade6-M216 leu1-32 ura4-D18 | 5B |
| SO1644 | h 90 fbh1::bsd rad3::hph GFP-atb2-kan ade6-M216 leu1-32 ura4-D18 | 5C, 5D |
| SO1813 | h 90 fbh1-D485N ade6-M216 leu1-32 ura4-D18 | 5F |
| SO1877 | h 90 fbh1-L14AP15A ade6-M216 leu1-32 ura4-D18 | 5F |
| SO1891 | h 90 fbh1-P15AL26A ade6-M216 leu1-32 ura4-D18 | 5F |
| SO1853 | h 90 fbh1-D485N ade6-M216 leu1-32 ura4-D18 pREP81-GFP-atb2 | 5G |
| SO1886 | h 90 fbh1-L14AP15A ade6-M216 leu1-32 ura4-D18 pREP81-GFP-atb2 | 5G |
| SO1898 | h 90 fbh1-P15AL26A ade6-M216 leu1-32 ura4-D18 pREP81-GFP-atb2 | 5G |
The original strain for rhp51-ECFP is a gift from Y. Tsutsui and H. Iwasaki.
Microscopy
Living cells were observed in Edinburgh minimal medium without a nitrogen source for meiosis. Images in Fig. 1A were taken using Zeiss AxioplanII microscope. Other images were taken using a microscope (IX71, Olympus) with a DeltaVision-SoftWoRx system (Applied Precision) at 25°C, 30°C or 33°C. Z sectioning was done with 0.4 µm intervals, and images were taken every 1∼3 min. Images were then deconvolved, and a Z-stack projection was created.
Acknowledgments
We thank Y. Tsutsui and H. Iwasaki for advice, discussion over unpublished data and the rhp51-ECFP strain.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: MS and MY are supported by Grants-in-Aid from Japan Society for the Promotion of Science (JSPS) and also by a PRESTO program of JST (MS) and a Global COE program (MY) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. TT is supported by Cancer Research UK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kominami K, Ochotorena I, Toda T. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells. 1998;3:721–735. doi: 10.1046/j.1365-2443.1998.00225.x. [DOI] [PubMed] [Google Scholar]
- 2.Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- 3.Wolf DA, McKeon F, Jackson PK. F-box/WD-repeat proteins Pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of Cdc18p. Curr Biol. 1999;9:373–376. doi: 10.1016/s0960-9822(99)80165-1. [DOI] [PubMed] [Google Scholar]
- 4.Jallepalli PV, Tien D, Kelly TJ. sud1 + targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc Natl Acad Sci U S A. 1998;95:8159–8164. doi: 10.1073/pnas.95.14.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison C, Katayama S, Dhut S, Chen D, Jones N, et al. SCF(Pof1)-ubiquitin and its target Zip1 transcription factor mediate cadmium response in fission yeast. EMBO J. 2005;24:599–610. doi: 10.1038/sj.emboj.7600536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamnun YM, Katayama S, Toda T. Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem Biophys Res Commun. 2006;350:125–130. doi: 10.1016/j.bbrc.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Katayama S, Kitamura K, Lehmann A, Nikaido O, Toda T. Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol Biol Cell. 2002;13:211–224. doi: 10.1091/mbc.01-07-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takayama Y, Mamnun YM, Trickey M, Dhut S, Masuda F, et al. Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell. 2010;18:385–396. doi: 10.1016/j.devcel.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermand D, Bamps S, Tafforeau L, Vandenhaute J, Makela TP. Skp1 and the F-box protein Pof6 are essential for cell separation in fission yeast. J Biol Chem. 2003;278:9671–9677. doi: 10.1074/jbc.M211358200. [DOI] [PubMed] [Google Scholar]
- 10.Jourdain I, Spielewoy N, Thompson J, Dhut S, Yates JR, et al. Identification of a conserved F-box protein 6 interactor essential for endocytosis and cytokinesis in fission yeast. Biochem J. 2009;420:169–177. doi: 10.1042/BJ20081659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, et al. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann A, Katayama S, Harrison C, Dhut S, Kitamura K, et al. Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells. 2004;9:367–382. doi: 10.1111/j.1356-9597.2004.00730.x. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann A, Toda T. Fission yeast Skp1 is required for spindle morphology and nuclear membrane segregation at anaphase. FEBS Lett. 2004;566:77–82. doi: 10.1016/j.febslet.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Sakuno T, Watanabe Y. Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions. Chromosome Res. 2009;17:239–249. doi: 10.1007/s10577-008-9013-y. [DOI] [PubMed] [Google Scholar]
- 15.Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 16.Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 17.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Smith GR. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharif WD, Glick GG, Davidson MK, Wahls WP. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome. 2002;1:1. doi: 10.1186/1475-9268-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Muris DF, Vreeken K, Carr AM, Broughton BC, Lehmann AR, et al. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 1993;21:4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol. 2008;6:e88. doi: 10.1371/journal.pbio.0060088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Octobre G, Lorenz A, Loidl J, Kohli J. The Rad52 homologs Rad22 and Rti1 of Schizosaccharomyces pombe are not essential for meiotic interhomolog recombination, but are required for meiotic intrachromosomal recombination and mating-type-related DNA repair. Genetics. 2008;178:2399–2412. doi: 10.1534/genetics.107.085696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du LL, Nakamura TM, Moser BA, Russell P. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol Cell Biol. 2003;23:6150–6158. doi: 10.1128/MCB.23.17.6150-6158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 28.Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Lorenz A, Osman F, Whitby MC. A failure of meiotic chromosome segregation in a fbh1Δ mutant correlates with persistent Rad51-DNA associations. Nucleic Acids Res. 2011;39:1718–1731. doi: 10.1093/nar/gkq977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaguchi C, Morishita T, Shinagawa H, Hishida T. Essential and distinct roles of the F-box and helicase domains of Fbh1 in DNA damage repair. BMC Mol Biol. 2008;9:27. doi: 10.1186/1471-2199-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 32.Sunnerhagen P. Cytoplasmatic post-transcriptional regulation and intracellular signalling. Mol Genet Genomics. 2007;277:341–355. doi: 10.1007/s00438-007-0221-5. [DOI] [PubMed] [Google Scholar]
- 33.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbour L, Xiao W. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat Res. 2003;532:137–155. doi: 10.1016/j.mrfmmm.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Cromie GA, Hyppa RW, Smith GR. The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics. 2008;179:1157–1167. doi: 10.1534/genetics.108.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 37.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 38.Akamatsu Y, Tsutsui Y, Morishita T, Siddique MS, Kurokawa Y, et al. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J. 2007;26:1352–1362. doi: 10.1038/sj.emboj.7601582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato M, Toya M, Toda T. Visualization of fluorescence-tagged proteins in fission yeast: the analysis of mitotic spindle dynamics using GFP-tubulin under the native promoter. Methods Mol Biol. 2009;545:185–203. doi: 10.1007/978-1-60327-993-2_11. [DOI] [PubMed] [Google Scholar]
- 40.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]