Abstract
Solitary Fibrous Tumor (SFT) is a mesenchymal neoplasm composed of CD34-positive fibroblastic cells. The pathogenesis driving this neoplasm remains unclear, with no recurrent genetic aberrations described to date. Previous reports suggest a role for IGF2 overexpression in the pathogenesis of these tumors, implicated in triggering hypoglycemia in some patients. The expression profiling of 23 SFTs was investigated using an Affymetrix U133A platform. The transcriptional signature was compared to a set of 34 soft tissue sarcomas spanning 7 subtypes. Potential candidate genes were then further investigated for activating mutations or loss of imprinting (LOI). SFT had a distinct expression signature and clustered in a tight genomic cluster, separate from all other sarcoma subtypes. A number of overexpressed receptor tyrosine kinase (RTK) genes were identified in SFT, including DDR1, ERBB2 and FGFR1, however no mutations were identified by cDNA sequencing. Overexpression of IGF2 was uniformly detected in SFT, regardless of anatomic location and was related to LOI. In contrast, IGF1 and JUN overexpression was seen in pleural, but not meningeal location. SFT shows a distinctive expression signature, with overexpression of DDR1, ERBB2, and FGFR1. Despite of lack of activating mutations in these RTKs, therapy with specific inhibitors targeting these kinases might be considered in advanced/metastatic cases. Our results confirm LOI in several tumors expressing high levels of IGF2, which may explain the observed paraneoplastic hypoglycemia.
Keywords: solitary fibrous tumor, expression profiling, IGF2, DDR1
INTRODUCTION
Solitary Fibrous Tumor (SFT) is a mesenchymal neoplasm with distinctive histopathological features and consistent CD34 expression. The constituent cells show a fibroblastic phenotype, being arranged in a patternless growth of alternating cellularity and collagenous stroma. Hemangiopericytoma (HPC), previously regarded as a distinct entity, displays a more uniform cellularity and a prominent staghorn vascular network. Due to its considerable morphological overlap and similar CD34 reactivity, hemangiopericytoma has been reclassified as a histologic variant of SFT [1]. SFT may develop virtually in any region of the body and generally follow a benign clinical course [2–3]. However, 15–20% of patients progress with either local recurrence or distant metastases [2–3]. Poor prognosis has been linked to tumor size greater than 10 cm and increased mitotic activity [2]. Fibrosarcomatous transformation of SFT is defined as presence of increased mitotic activity and increased cellularity, with or without tumor necrosis. This more frankly malignant component of SFT is associated with a higher risk for recurrence and metastasis. The clinical management for primary tumors is surgery alone. However, there are no effective therapies for metastatic or advanced disease.
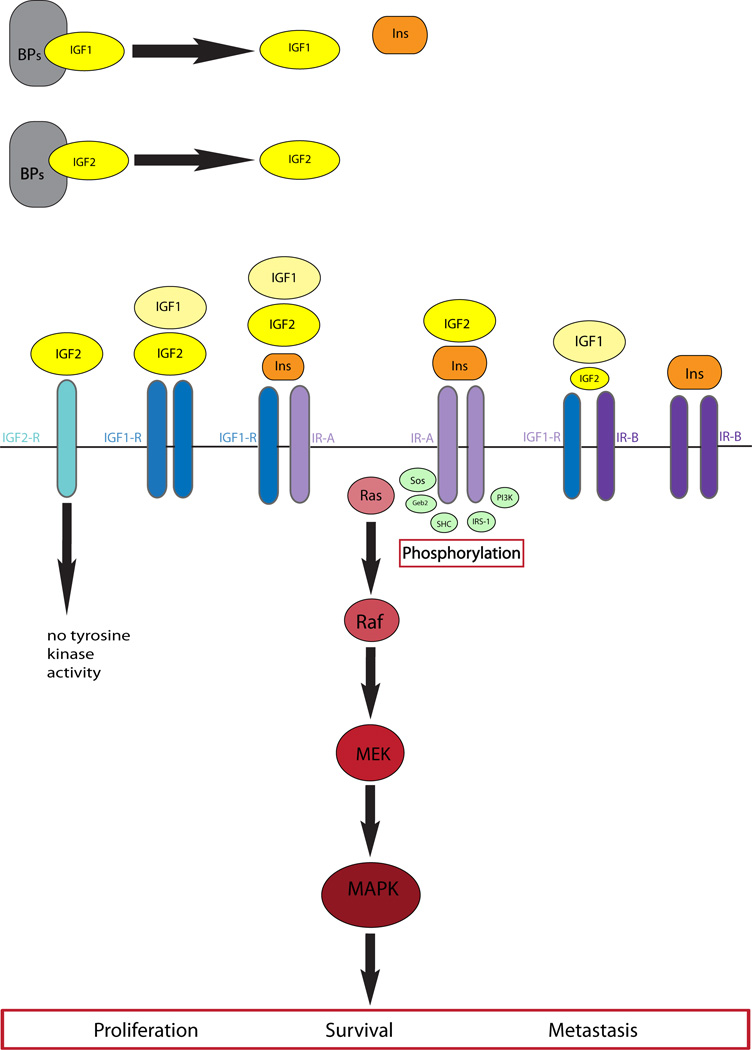
A small subset (<5%) of patients with SFT present with hypoglycemia, known as Doege-Potter syndrome, which has been associated with large tumor size or aggressive clinical behavior and is resolved by surgical resection of the lesion [4]. Paraneoplastic hypoglycemia has been reported in a large variety of other malignancies, including mesenchymal, epithelial and hematopoietic origin [5]. The common mechanism is the excessive release of a prohormone form of Insulin-like growth factor II, IGF2, also referred as “big IGF-II”. IGF2 is a protein hormone that shares similarities with insulin and its gene is subject to imprinting on the paternal allele in most adult tissues. Although IGF2 exerts its biologic function by binding to IGF1R, it was recently shown that SFTs lack IGF1R expression and thus IGF2 signaling occurs via Insulin Receptor-A (IR-A) pathway [6].
The limited progress in understanding the pathogenesis of this disease is also a reflection of the lack of recurrent genetic abnormalities. Several cytogenetic reports failed to identify a consistent phenotype, revealing gains or loses in several chromosomes, as well as structural rearrangements in 4q13, 9p22–9p23, 12q24, 12q13–12q15 [7–8]. Furthermore, no consistent receptor tyrosine kinase overexpression has been identified in this tumor, to suggest sensitivity to targeted therapy with small molecule inhibitors. The main objective of this study was to identify therapeutic targets and candidate genes driving sarcomagenesis, by mining genome-wide transcriptional profiling.
METHODS
Patient selection and clinicopathologic features
Thirty-eight consecutive patients who were treated surgically at MSKCC with pathologic diagnosis confirmed of SFT and frozen tissue available were identified from the institutional prospectively collected sarcoma database and included in the molecular analysis. Patient demographics, treatment data and follow-up information were obtained by chart review. Pathologic diagnosis was confirmed using standard hematoxylin and eosin staining and immunoreactivity for CD34 (Ventana Medical Systems, Inc, Tucson, Arizona; pre-diluted) on formalin fixed paraffin embedded tissue. In cases originating in the abdominal cavity or metastatic to different organs, a CD117 was performed to exclude the alternative diagnosis of a gastrointestinal stromal tumor. Adequate quality RNA for transcriptional profiling was obtained in 23 samples from 22 patients, which represented the study group for this analysis. There were 12 females and 10 males, with a median age of 52 years (range 31–74). The primary tumor location included: soft tissue in 8 patients (retroperitoneum, 4, trunk, 3, and extremity, 1); meninges in 7; pleura in 6 and visceral (bladder) in one patient. None of the samples included for analysis were from a sinonasal primary. Ten of the primary tumors were larger than 10 cm, while three tumors were smaller than 5 cm. Six tumors were classified as benign, including 3 from soft tissue location, 2 pleural and 1 meningeal The remaining 14 cases were deemed as malignant SFT, based on a mitotic count of >4 MF/10HPFs, plus/minus areas of necrosis. Among these 14 patients all except two patients developed local or distant recurrence as follows: 5 patients recurred locally, 6 patients developed distant metastases, and 3 developed both. At last follow-up 6 were dead of disease, 3 were alive with disease and 5 were alive with no evidence of disease. The two patients classified as having malignant SFT who did not develop progression had their primary tumors in the soft tissue, however follow-up information was available for only a brief period of time (under 2 years). The metastatic sites included: lung in 7 patients; bone and liver in 2; and kidney and soft tissue in one. The samples tested on the array were taken from primary tumors in 9 (38%), local recurrences in 4 (17%) and distant metastases from 10 (45%). Information of the serum glucose levels was obtained by review of the charts and lab tests performed before surgical resection. Evidence of pre-treatment hypoglycemia was diagnosed in 2 (9%) patients with large (>10 cm) soft tissue SFTs, who eventually died of their disease. The study was approved by the Institutional Review Board (IR# 02-060).
Transcriptional Profiling for Mining Candidate Genes and Data Analysis
Adequate tumor tissue for molecular studies was selected on H&E stained sections from frozen tissue tumor blocks. Areas of viable tumor highlighted on the slides were matched and macrodissected from the cryomolds for further nucleic acid extraction. Twenty-three samples with adequate quality RNA were studied on the U133A Affymetrix platform (22,000 transcripts), as previously described [9]. The transcriptional profile was compared to the expression of a well-characterized set of 34 soft tissue sarcomas (STS), spanning the most common histologic types, including: synovial sarcoma, clear cell sarcoma, gastrointestinal stromal tumor, myxoid liposarcoma, dedifferentiated liposarcoma, leiomyosarcoma, malignant fibrous histiocytoma, and fibrosarcoma [10]. These STS samples used in our original publication [10] were initially studied on the Affymetrix U95 chip platform, and then subsequently re-analyzed on the newer chip version, U133A, using the same RNA aliquot. CEL files and normalized value in MIAME format from the SFT and STT samples analyzed on the U133A chip are publically available, at below website: http://cbio.mskcc.org/Public/SFT
Hierarchical clustering and differentially expressed genes were obtained using Partek® Genomics Suite™ software. The default normalization used was the Robust Multi-chip Average (RMA), Log probeset using base: 2 and ProbesetSummarization:Median Polish. The clustering was performed according to ‘Euclidean metric’ and ‘Average Linkage method’ for Rows and Columns, using a Hierarchical clustering method.
Additionally, we have imported and analyzed the cDNA array data set of 13 SFT samples from West and colleagues [11], which is publically available from the Stanford Microarray Database. Significance Analysis of Microarray (SAM) was performed on this data set as described by Tusher et al. [12]. In contrast with the Affymetrix oligonucleotide platform, the cDNA array technology is based on measuring expression between competing test and reference cDNAs. This data-set was used for comparison and validation of the differentially expressed genes obtained from our study, since it investigated a different group of patients, using a different platform and statistical software.
Sequencing of Target Genes and Mutation Detection
Genomic DNA was extracted from frozen tissue in all 38 patients. Matching normal DNA was extracted from frozen tissue in cases with detectable mutations. Candidate genes found to be overexpressed in SFT samples compared to other sarcoma types were selected and were fully sequenced. Putative exonic regions for the entire human genome (NCBI Human Genome Build 36.1) were broken into target regions of 500 bp or less, and specific primers were designed using Primer 3. PCR reactions were carried out in 384 well plates, in a Duncan DT-24 water bath thermal cycler, with 10 ng of DNA as template, using a touchdown PCR protocol with HotStart Taq (Kapa Biosystems, Cape Town, South Africa), as previously described [13]. Mutations were detected using an automated detection pipeline at the MSKCC Bioinformatics Core. Bi-directional reads and mapping tables (to link read names to sample identifiers, gene names, read direction, and amplicon) were subjected to a QC filter which excludes reads that have an average phred score of < 10 for bases 100–200 [13]. All putative mutations were confirmed by a second PCR and direct sequencing, in parallel with amplification and sequencing of matched normal tissue DNA.
Additionally, PCR and DNA sequencing for hot spots mutations were performed for PDGFRB exon 18 and ERBB2 exons 18–23, using standard procedures.
Examination of IGF2 imprinting status
IGF2 is a paternally imprinted gene, whose overexpression was previously associated with loss of imprinting. An ApaI site, located in IGF2 exon 9 (3’UTR), is a common site for polymorphism [14]. The restriction enzyme site is found in both genomic DNA and at the cDNA level, and was previously associated with IGF2 LOI [15]. Genomic DNA was extracted from cases with highest IGF2 mRNA expression. Cases harboring ApaI site polymorphism between paternal and maternal alleles at genomic level were considered informative and further studied at cDNA level. The following IGF2 exon 9 primers were used: Fwd, 5’-CTTGGACTTTGAGTCAAATTGG-3’ and Rev, 5’-GGTCGTGCCAATTACATTTCA-3. RNA from informative cases was subjected to RT-PCR after being DNA-sed. The presence of ApaI IGF2 exon 9 polymorphism at cDNA level was investigated to determine the status of LOI.
RESULTS
Solitary fibrous tumors show a distinct genomic profile compared to other sarcoma types
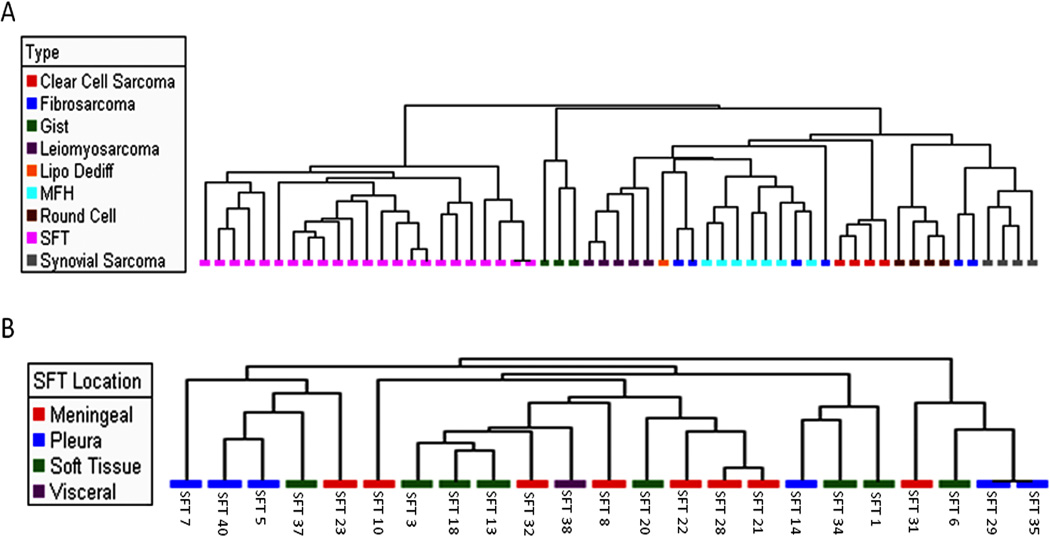
By unsupervised clustering 1,662 genes were differentially expressed between SFT and STS (1201 gene overexpressed, 461 underexpressed in the SFT group). In addition we performed clustering using a restricted gene list of 150 common human kinases present on the U133A chip, curated by GO mining, resulting in a similar pattern of a tight SFT genomic cluster, separate from all other sarcoma types (Fig.1). As expected, CD34 expression was significantly up-regulated in SFT (p=0.01). Differentially expressed genes between SFT and STS were obtained using the t-test, using an FDR of 0.05 and >2 fold change (FC). Among the upregulated tyrosine kinases in the SFT group, the top four genes included: FGFR1 (fibroblast growth factor receptor-1; FC, 4.3), ERBB2 (human epidermal growth factor receptor 2; FC, 3.6), DDR1 (Discoidin domain receptor family, member 1; FC, 2.7), and INSR (insulin receptor; FC, 2.1). Among non-kinase genes, significant up-regulation of the ADLH1A (Aldehyde dehydrogenase 1 family, member A1; FC, 24.4), IGF2 (insulin like growth factor 2; FC, 13.2; top 9th ranked gene), CHI3L1 (Chitinase 3-like 1, cartilage glycoprotein-39; FC, 33.8), GRIA2 (Glutamate receptor, ionotropic, AMPA 2; FC, 23.8), FGF2 (fibroblast growth factor 2, FC 3.6) and ApoD (Apolipoprotein D, FC 3.2) were seen in the SFTs. SFT showed a number of collagen genes that were upregulated compared to other sarcoma types, including: COL4A1, COL16A1, COL11A1, COL6A3, COL14A1, and COL17A1.
Fig 1.
Hierarchical unsupervised clustering was performed using Partek® Genomics Suite™ across all genes present on the Affymetrix U133A chip: A) a tight SFT genomic cluster separate from all other sarcoma types was observed; B) primary location did not appear to influence clustering. Note that samples: SFT28 and SFT32 were obtained from the same patient.
Using the published Stanford data from the 13 SFT tested on the cDNA arrays [11] and analyzed by SAM, a similar profile of upregulated kinases were obtained, including: FGFR1 (FC, 3.1), ERBB2 (FC, 2.0), and DDR1 (FC, 3.4). Furthermore, ALDH1A1 and ApoD were two of the top up-regulated genes on the cDNA array. In order to investigate the transcriptional heterogeneity among the SFT family, we performed in a second step an unsupervised clustering of the 23 SFT samples. No distinct subgroups were detected based on anatomic location (soft tissue, meningeal, pleural), or other clinicopathologic factors. Furthermore, by unsupervised clustering, the soft tissue SFT restricted to primary tumor samples (6 of the total of 8 samples tested) did not group together and were dispersed among 3 different subclusters. Similarly, the pleural SFT samples originating from primary location did not group together in a genomic cluster. Thus anatomic location does not seem to have a significant impact on the transcriptional signature of SFTs, regardless of the origin of sample tested on the array (primary versus recurrence).
In a supervised analysis comparing meningeal versus pleural location, 14 genes were differentially expressed with an FDR of 0.05 and FC>2. Among them, IGF1 (insulin like growth factor-1; FC, 8.8) and JUN (jun oncogene; FC, 6.5) were overexpressed in the pleural SFT, while PTGDS (prostaglandin D2 synthase; FC, 13.9) in meningeal SFT. A similar comparison between the meningeal versus soft tissue SFT, showed that ApoD (F.C., 5.6) was upregulated in the soft tissue location, while IGF1 (F.C., 5.6) and IGFBP7 (F.C., 3.3) were upregulated in the pleural SFT compared to the soft tissue location.
Overexpression of FGFR1, DDR1 and ERRB2 Receptor Tyrosine Kinases in SFT is not due to activating mutations
Full-length cDNA sequencing of FGFR1 and DDR1 genes was performed in 15 and 8 SFT samples, respectively, selected based on their highest mRNA expression. In addition, the entire kinase domain of ERBB2 (exons 18–23) was sequenced in 10 SFT tumors with highest ERBB2 mRNA levels [16]. No activating mutations were detected. A DDR1 exon 13 N502S substitution was detected in one pleural SFT tumor, which has been previously reported in acute myeloid leukemia [17]. However, the matched normal tissue of this patient revealed an identical substitution, confirming this being a polymorphism.
Additionally, we tested all the 39 SFT samples for the presence of a putative mutation in the kinase domain of PDGFRB (Asp850Val), previously reported by Rossi et al. in a malignant pleural SFT. [18]. However none of our tumors showed mutations in this locus. Other PDGFRB exons were not further investigated, since the PDGFRB mRNA levels were consistently low in all of 23 SFT samples.
IGF2 Loss of Imprinting was detected in SFTs with high IGF2 expression
Investigation of 15 SFTs showing highest IGF2 mRNA expression revealed 6 informative cases, with IGF2 exon 9 ApaI polymorphism at genomic level. In 5 of them adequate RNA was available for further RT-PCR analysis. In 3 of the 5 cases an ApaI polymorphism was detected by PCR and sequencing at both genomic and cDNA level. This result is in keeping with expression of both maternal and paternal IGF2 alleles and is consistent with IGF2 LOI. Interestingly, one of these 3 patients had low serum levels of glucose before the surgical removal of the mass.
DISCUSSION
SFT is a rare fibrous neoplasm, which was initially described in the pleural cavity as “localized or fibrous mesothelioma”, and thought to arise mainly in relationship with serosal surface. The widespread anatomic distribution of SFT was only recently appreciated, reflecting an increasing endorsement for a unifying concept of SFT and hemangiopericytoma as a single histological entity. The motivation behind this genome-wide investigation of SFT was the lack of consistent genetic abnormalities or known therapeutic targets that can be explored in patients with metastatic or advanced disease. Using the Affymetrix oligonucleotide platform, SFTs were characterized by a homogenous transcriptional signature, clustering in a distinct genomic group from all other sarcoma types.
Interestingly, among the SFT tumors no genomic subgroups emerged based on anatomic site. This finding is particularly relevant for meningeal tumors, which was considered to have a potentially distinct biology from pleural or soft tissue sites. Furthermore, a disconnect between the terminology applied in the meningeal vs other locations of SFT still persists. In the latest WHO classification of CNS tumors, meningeal SFT and HPC are discussed separately. As such, SFT of the meninges (histologic WHO grade I) is considered a benign fibrous lesion, associated with diffuse and strong CD34 reactivity. In contrast, meningeal HPC is described as a tumor with diffuse and evenly dispersed cellularity and variable expression for CD34. Based on mitotic activity and/or presence of necrosis, meningeal HPC is further subdivided into two histologic grades (WHO grade II and III). In our point of view, the artificial separation between SFT and HPC is merely a reflection of histologic grading (degree of cellularity and mitotic activity) rather than two distinct neoplasms. Our expression data further emphasizes this viewpoint, with meningeal tumors having similar profile with SFT/HPC located at other sites.
As the diversity of fibroblastic differentiation has emerged recently, with different expression signatures of fibroblasts from different anatomic location, one might expect that these differences may translate into transcriptional variations of SFT arising at different sites. The fetal lung fibroblast signature included several genes which were implicated in lung development, including DDR1, FLT1, FGF7, FOXF1, BMP4 and HGF [19]. In contrast the soft tissue fibroblast signature, from arm and abdomen location, included genes involved in Wnt signaling (WISP2, DAAM2). Although some of these genes were identified as upregulated in SFT, no correlation was noted between the topographic normal fibroblastic differentiation and the corresponding gene signature of various anatomic sites of the SFT. However, in a supervised analysis, certain differences in expression of IGF1, JUN and PTGDS were identified between meningeal and pleural SFT.
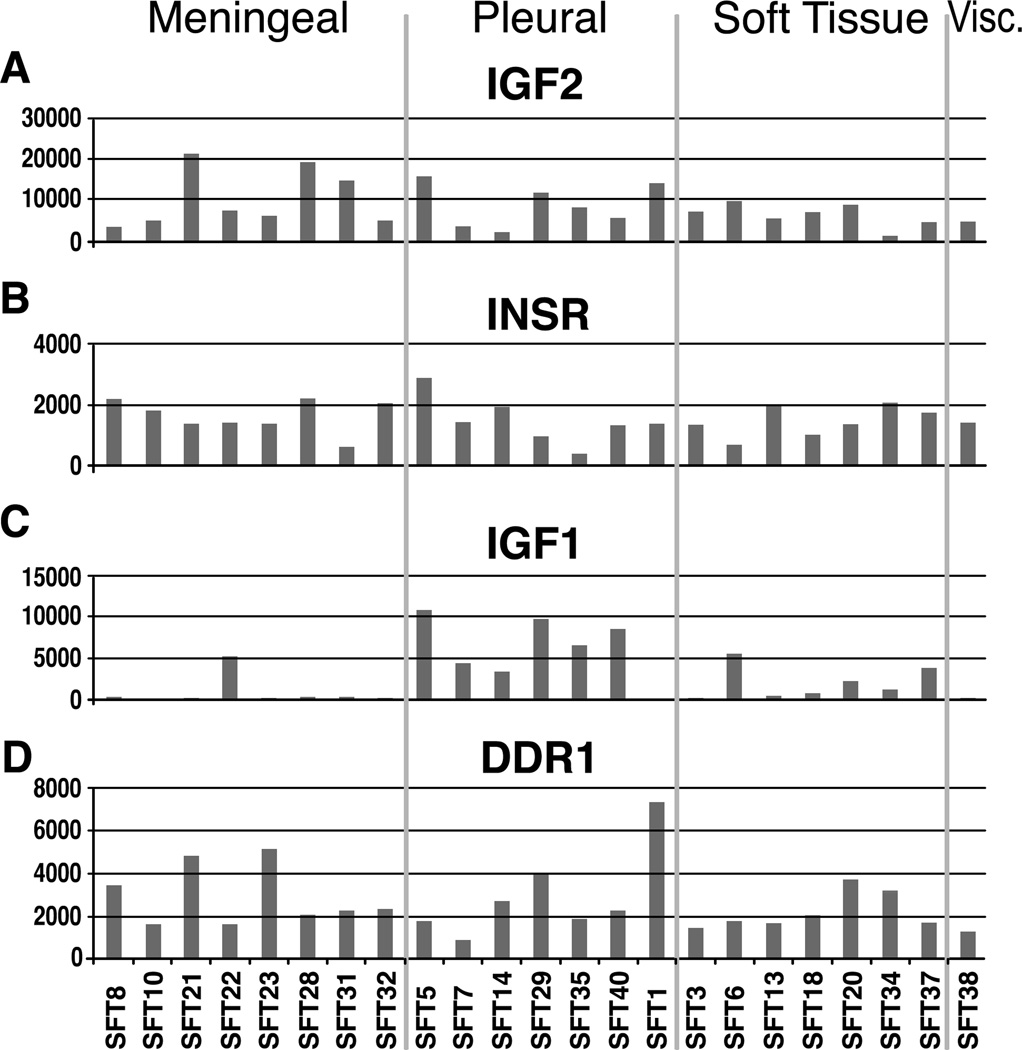
In contrast, IGF2 expression was consistently upregulated in SFT of all anatomic sites (Fig 2). As suggested by Li et al. [6], our results support that the IGF2-mediated downstream signaling pathway occurs through insulin receptor (IR), rather than IGF1R (Fig 3). Overexpression of IR paralleled that of IGF2 in most SFTs (Fig 2), while IGF1R expression was consistently negligible. SFT also showed high levels of IGFBP3 and IGFBP6, irrespective of anatomic site. There are two IR isoforms (IR-A and IR-B) resulting from the alternative splicing of exon 11. IGF2 binds only to IR-A, but not IR-B isoform, and as such a predominant IR-A transcript has been demonstrated in SFT [6]. One putative mechanism of IGF2 up-regulation is through loss of imprinting (LOI), which has been previously implicated in a single SFT, as well as in other malignancies [15, 20]. Our results provide further support of this mechanism, as 3 tumors with highest IGF2 expression showed evidence of LOI. One of these patients developed hypoglycemia, diagnostic of Doege-Potter syndrome.
Fig. 2.
Distribution of mRNA expression based on different anatomic locations of SFT: A-C: including genes involved in IGF pathway, such as IGF2, INSR and IGF1; and D. DDR1 expression.
Fig. 3.
Diagram of IGF/INSR/IGF1R signaling pathway.
SFTs showed a distinct signature of overexpressed tyrosine kinases compared to other sarcoma types. Not surprisingly for a tumor of fibroblastic lineage, FGFR1 (fibroblast growth factor receptor-1) was the top upregulated receptor tyrosine kinase (RTK). Its ligand FGF2 was also found to be overexpressed in SFT compared to other sarcoma types. Mutations in the FGFR1 have been previously described in the type I Pfeiffer craniosynostosis syndrome, which triggers FGF2 enhanced ligand binding [21]. In cancer, overexpression of FGFR1 is triggered by different mechanisms of activation, including mutations (i.e. glioblastoma multiforme, melanoma), gene amplification (breast, oral cancer) or chromosomal translocation (leukemia) [22–25]. Despite high levels of expression, no mutations were identified in FGFR1.
DDR1 (Discoidin Domain Receptor 1) is a unique receptor tyrosine kinase activated by binding to its ligand collagen. DDR1 is constitutively expressed in normal tissues, such as lung, kidney, brain and in dermal fibroblasts [26–27]. The overexpression of DDR1 was detected in several different human cancers, suggesting a function in tumor progression. However some investigators argue against classifying DDR1 as a bona fide transforming oncogene due to remarkably slow activation of its tyrosine kinase[26–27]. DDR1 overexpression was also detected in the fibroblastic foci of idiopathic pulmonary fibrosis, where it may contribute to fibroblast activation and survival [23]. As shown by Chang et al. genes implicated in extracellular matrix synthesis, such as DDR1, were upregulated in the fetal lung fibroblasts, compared to fetal or adult cutaneous fibroblasts [19]. In SFT, DDR1 expression was uniformly increased, regardless of anatomic location (Fig. 2 D). Furthermore, mutations in DDR1, located in exon 13 N502S, have been identified recently in 3 patients with acute myeloid leukemia [17]. Although the same substitution was identified in one of the SFT lesions, this was considered a polymorphism, since was present in the normal tissue as well. Despite the lack of DDR1 activating mutations in SFT, the significance of its upregulation in SFTs should be further investigated, due to its implications in fibroblast function and differentiation.
In summary, our results further underscore the importance of the IGF2-INSR pathway in SFT oncogenesis and confirm that LOI contributes to high expression of IGF2. The homogeneous expression profile of SFT independent of anatomic location reinforces clinicopathologic studies that unify pleural and extrapleural SFT as a single biologic entity.
Acknowledgements
We would like to thank: Nicholas Socci and the Bioinformatics Core for assistance with the expression data analysis, Agnes Viale and the staff of the Genomic Core lab, Adriana Heguy and the Sequencing Core; Nicole Moraco for reviewing clinical charts, and Milagros Soto for editorial assistance. Also would like to thank Dr. Marc Rosenblum for his insight over the meningeal SFT/HPC terminology and valuable discussions of the manuscript.
Grant support: P01CA47179 (SS, RGM, GKS, CRA), CA148260 (RGM), NCI-ASCO Cancer Foundation Clinical Investigator Team Leadership Supplemental Award (RGM), Cycle for Survival (RGM, GKS), the Shuman Fund for GIST Research (RGM, CRA), Alan Rosenthal Fund for research in sarcoma (MLK).
Footnotes
The raw data of the expression arrays is made available http://cbio.mskcc.org/Public/SFT
Conflict of interest: none
REFERENCES
- 1.Guillou L, Fletcher JA, Fletcher CDM, Mandahl N. Extrapleural solitary fibrous tumour and haemangiopericytoma. In World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. edn) Lyon: IARC Press; 2002. [Google Scholar]
- 2.Gold JS, Antonescu CR, Hajdu C, Ferrone CR, Hussain M, Lewis JJ, et al. Clinicopathologic correlates of solitary fibrous tumors. Cancer. 2002;94:1057–1068. [PubMed] [Google Scholar]
- 3.Park MS, Araujo DM. New insights into the hemangiopericytoma/solitary fibrous tumor spectrum of tumors. Curr Opin Oncol. 2009;21:327–331. doi: 10.1097/CCO.0b013e32832c9532. [DOI] [PubMed] [Google Scholar]
- 4.Zafar H, Takimoto CH, Weiss G. Doege-Potter syndrome: hypoglycemia associated with malignant solitary fibrous tumor. Med Oncol. 2003;20:403–408. doi: 10.1385/MO:20:4:403. [DOI] [PubMed] [Google Scholar]
- 5.Zapf J. Insulinlike growth factor binding proteins and tumor hypoglycemia. Trends Endocrinol Metab. 1995;6:37–42. doi: 10.1016/1043-2760(94)00144-s. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Chang Q, Rubin BP, Fletcher CD, Morgan TW, Mentzer SJ, et al. Insulin receptor activation in solitary fibrous tumours. J Pathol. 2007;211:550–554. doi: 10.1002/path.2136. [DOI] [PubMed] [Google Scholar]
- 7.Debiec-Rychter M, de Wever I, Hagemeijer A, Sciot R. Is 4q13 a recurring breakpoint in solitary fibrous tumors? Cancer Genet Cytogenet. 2001;131:69–73. doi: 10.1016/s0165-4608(01)00489-7. [DOI] [PubMed] [Google Scholar]
- 8.Mandahl N, Orndal C, Heim S, Willen H, Rydholm A, Bauer HC, et al. Aberrations of chromosome segment 12q13-15 characterize a subgroup of hemangiopericytomas. Cancer. 1993;71:3009–3013. doi: 10.1002/1097-0142(19930515)71:10<3009::aid-cncr2820711020>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 9.Antonescu CR, Viale A, Sarran L, Tschernyavsky SJ, Gonen M, Segal NH, et al. Gene expression in gastrointestinal stromal tumors is distinguished by KIT genotype and anatomic site. Clin Cancer Res. 2004;10:3282–3290. doi: 10.1158/1078-0432.CCR-03-0715. [DOI] [PubMed] [Google Scholar]
- 10.Segal NH, Pavlidis P, Antonescu CR, Maki RG, Noble WS, DeSantis D, et al. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West RB, Nuyten DS, Subramanian S, Nielsen TO, Corless CL, Rubin BP, et al. Determination of stromal signatures in breast carcinoma. PLoS Biol. 2005;3:e187. doi: 10.1371/journal.pbio.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonescu CR, Yoshida A, Guo T, Chang NE, Zhang L, Agaram NP, et al. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009;69:7175–7179. doi: 10.1158/0008-5472.CAN-09-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tadokoro K, Fujii H, Inoue T, Yamada M. Polymerase chain reaction (PCR) for detection of ApaI polymorphism at the insulin like growth factor II gene (IGF2) Nucleic Acids Res. 1991;19:6967. doi: 10.1093/nar/19.24.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawson EA, Zhang X, Crocker JT, Wang WL, Klibanski A. Hypoglycemia from IGF2 overexpression associated with activation of fetal promoters and loss of imprinting in a metastatic hemangiopericytoma. J Clin Endocrinol Metab. 2009;94:2226–2231. doi: 10.1210/jc.2009-0153. [DOI] [PubMed] [Google Scholar]
- 16.Landau M, Ben-Tal N. Dynamic equilibrium between multiple active and inactive conformations explains regulation and oncogenic mutations in ErbB receptors. Biochim Biophys Acta. 2008;1785:12–31. doi: 10.1016/j.bbcan.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Loriaux MM, Levine RL, Tyner JW, Frohling S, Scholl C, Stoffregen EP, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–4796. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi G, Schirosi L, Giovanardi F, Sartori G, Paci M, Cavazza A. Pleural malignant solitary fibrous tumor with sarcomatous overgrowth showing PDGFRbeta mutation. Chest. 2006;130:581–583. doi: 10.1378/chest.130.2.581. [DOI] [PubMed] [Google Scholar]
- 19.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dejeux E, Olaso R, Dousset B, Audebourg A, Gut IG, Terris B, et al. Hypermethylation of the IGF2 differentially methylated region 2 is a specific event in insulinomas leading to loss-of-imprinting and overexpression. Endocr Relat Cancer. 2009;16:939–952. doi: 10.1677/ERC-08-0331. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahimi OA, Zhang F, Eliseenkova AV, Linhardt RJ, Mohammadi M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum Mol Genet. 2004;13:69–78. doi: 10.1093/hmg/ddh011. [DOI] [PubMed] [Google Scholar]
- 22.Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, et al. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freier K, Schwaenen C, Sticht C, Flechtenmacher C, Muhling J, Hofele C, et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC) Oral Oncol. 2007;43:60–66. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Acevedo VD, Ittmann M, Spencer DM. Paths of FGFR-driven tumorigenesis. Cell Cycle. 2009;8:580–588. doi: 10.4161/cc.8.4.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel W. Discoidin domain receptors: structural relations and functional implications. FASEB J. 1999;13 Suppl:S77–S82. doi: 10.1096/fasebj.13.9001.s77. [DOI] [PubMed] [Google Scholar]
- 27.Matsuyama W, Watanabe M, Shirahama Y, Mitsuyama H, Higashimoto I, Osame M, et al. Discoidin domain receptor 1 contributes to the survival of lung fibroblast in idiopathic pulmonary fibrosis. Am J Pathol. 2006;168:866–877. doi: 10.2353/ajpath.2006.050801. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]