Abstract
Crystallization of membrane proteins remains a significant challenge. For proteins resistant to the traditional approach of directly crystallizing from detergents, lipidic phase crystallization can be a powerful tool. Bicelles are an excellent medium for crystallizing membrane proteins in a lipidic environment. They can be described as bilayer discs formed by the mixture of a long-chain phospholipid and an amphiphile in an aqueous medium. Membrane proteins can be readily reconstituted into bicelles, where they are maintained in a native-like bilayer environment. Importantly, membrane proteins have been shown to be fully functional in bicelles under physiological conditions. Protein-bicelle mixtures can be manipulated with almost the same ease as detergent-solubilized membrane proteins, making bicelles compatible with standard equipment including high-throughput crystallization robots. A number of membrane proteins have now been successfully crystallized using the bicelle method, including bacteriorhodopsin, β2 adrenergic receptor, voltage-dependent anion channel, xanthorhodopsin and rhomboid protease. Because of the success with a variety of membrane proteins and the ease of implementation, bicelles should be part of every membrane protein crystallographer's arsenal.
Keywords: membrane protein crystallization, bicelle, lipidic crystallization
1. Introduction
The pace of membrane protein structure determination is rising at a rapid rate thanks to recent technological advances covered in this issue. The most popular and successful approach for membrane protein crystallization so far has been detergent-based crystallization [1,2]. Membrane proteins are generally isolated from the cell using detergents and it is convenient to place the purified detergent-solubilized protein directly into the same crystallization trials used for soluble proteins. Because detergents are not a perfect mimic of a natural bilayer, however, many membrane proteins are not stable enough in detergent to produce high quality crystals [3]. Moreover, detergent solubilization of membrane proteins often leads to loss of structural lipids, resulting in impaired protein integrity [4]. Detergent micelles also mask much of the surface area available for crystal contacts. Consequently, most detergent-based crystals have Type II packing formed by contacts only between the hydrophilic surfaces of membrane proteins [3]. Accordingly, proteins that lack well-ordered soluble domains can be difficult to crystallize.
One strategy to improve the chances of obtaining high-quality crystals is to augment crystal contacts by expanding the soluble domains. This can be carried out using epitope-specific antibodies that provide a large stable polar domain for crystal formation. Originally developed by Hunte and Michel to facilitate crystallization of cytochrome c oxidase [5,6] and cytochrome bc1 complex [7], the antibody method involves raising Fab or Fv fragments from mice. It has also been extensively utilized by the MacKinnon lab to crystallize KcsA [8], KvAP [9] and ClC channels [10]. More recently, the structures of proton pump AdiC [11], SecYE translocon [12], nitric oxide reductase [13] and anion selective cys-loop receptor [14] were obtained using Fab fragments. Antibodies including the recently identified camelid single chain antibody fragments (nanobodies), have also played a significant role in stabilizing G protein-coupled receptors for crystallogenesis [15-18]. Although successful, the antibody approach is typically outside the scope of most laboratories because of the lengthy and challenging process of raising, selecting and producing the antibodies. A promising alternative to naturally produced antibodies is to use phage display to select binding partners [19,20]. A variation of the antibody technique involves protein engineering to introduce soluble protein domains that help extend the polar surface area for crystallization [21,22]. However, in the absence of extensive knowledge of the protein of interest, this method can potentially introduce several complications including protein instability and crystal disorder.
Another approach is to crystallize membrane proteins directly in a bilayer environment. Originally conceived by Landau and Rosenbusch, successful crystallization of bacteriorhodopsin from the lipidic cubic phase (LCP) demonstrated for the first time that membrane proteins could be crystallized from bilayers [23]. Structurally, the cubic phase consists of a single lipid bilayer bent into a highly organized three dimensional lattice pervaded by an interconnected aqueous channel system. Monoolein (9.9 MAG) is the lipid of choice for LCP, although success has also been obtained with related lipids such as monovaccenin (11.7 MAG) for sensory rhodopsing II-transducer complex [21] and 7.7 MAG for the recent crystal structure of β2 adrenergic receptor-Gs protein complex [17]. When embedded in an LCP, a protein can diffuse in three dimensions and feed any crystal nuclei that form. Crystals tend to be of the Type I variety, with extensive crystal contacts in both the soluble and membrane regions, thereby providing more potential for lattice formation than the Type II crystals usually obtained with detergents. Since its introduction in 1996, the LCP method has undergone numerous advancements [24] and has seen numerous successes including structures of the highly sought after G-protein-coupled receptors [25-28]. However, the method is still not routinely used in most laboratories due to several practical disadvantages associated with handling the highly viscous, semi-solid LCP, which requires specialized tools for all steps of crystallization from set-up to harvest.
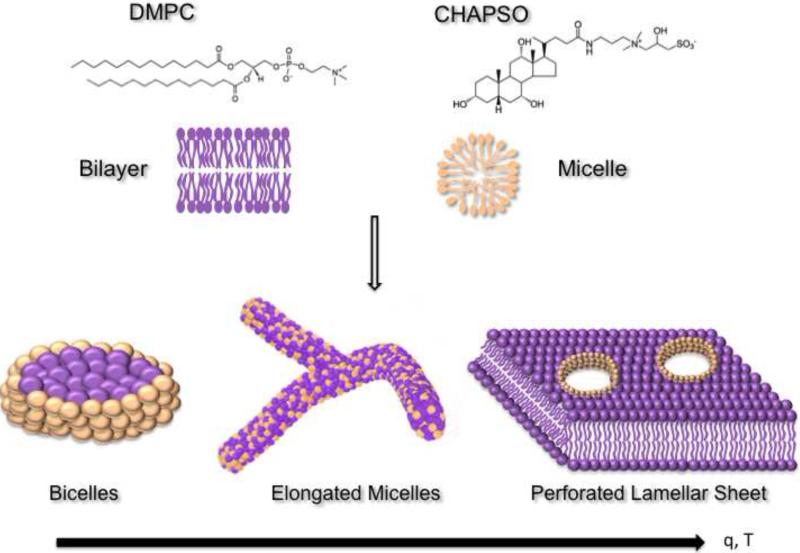
Bicelles also represent an attractive lipidic medium for crystallizing membrane proteins in a native-like bilayer environment. Bicelles were introduced in the mid 1990's as magnetically alignable model membranes and have since been extensively used for both solid and solution state NMR [29-31]. In 2002, owing to their unique properties as an excellent membrane mimetic system, bicelles were utilized to crystallize bacteriorhodopsin, showing their potential as a convenient lipidic medium for membrane protein crystallization [32]. Structurally, bicelles can be thought of as solubilized lipid bilayer disks that are formed by the addition of an amphiphile (detergent or a short-chain lipid) to a long-chain lipid (Figure 1) in an aqueous medium. In most cases, the long-chain lipid is a phosphatidylcholine (PC) lipid such as dimyristoyl phosphatidylcholine (DMPC) or ditridecanoyl phosphatidylcholine (DTPC). Examples of the amphiphile are short chain lipids like dihexanoyl-phosphatidylcholine (DHPC) or detergents like CHAPSO. These mixtures spontaneously form disc-shaped aggregates, with the long-chain phospholipid forming a central planar bilayer that is surrounded by a rim of amphiphile protecting the hydrophobic edge of the bilayer. The ratio of long-chain to short-chain lipids, also known as q, controls the physical diameter of the bicelles [33]. Importantly, it has been shown that membrane proteins can maintain their functionality upon reconstitution into bicelles [34].
Figure 1. Phase behavior of bicellar mixtures.
Bicelles are formed by mixing a phosphatidylcholine lipid such as DMPC with a detergent such as CHAPSO. The detergent shields the hydrophobic edges of the bilayer. With increasing temperatures and q, disc-shaped bicelles undergo phase transformation into worm-like cylindrical micelles and perforated lamellar sheet, which may also be beneficial for crystallization.
Bicellar mixtures show a variety of different morphologies depending on the ratio of its components “q” and the temperature (Figure 1). At low temperatures and q, the system exists as a fluid and forms characteristic bicelles with the long-chain lipid residing in the planar bilayer regions and the short chain lipid or detergent in the highly curved regions at the rim of the disc [35]. Higher temperatures, in the range of the fluid-to-gel transition temperature, can transform the system into flexible worm-like quasi-cylindrical elongated micelles known as the chiral nematic phase or the cholesteric phase [35-37]. These ribbon-like structures have high concentrations of the short-chain lipid along the edges of the micelles [38,39]. As the temperature is further increased, the system transitions into the perforated lamellar phase, which consists of multilamellar vesicles with pore-like defects in the lamellar sheets. The edges of the pores are lined with the short-chain lipid [38]. The lamellar phase is a gel and is also referred to as the swiss cheese form [40].
The ease of use of bicelles as a lipidic crystallization medium can be attributed to their unique phase behavior [41]. When maintained below the transition temperature, bicelles are in a convenient, pipettable liquid state. In practical terms, bicelle crystallization is similar to detergent-based protocols. There is simply an extra step of mixing the purified detergent-solubilized membrane protein with the bicelle stock solution prior to setting up crystallization trials. Standard crystallization techniques including robotics and all commercially available screens are compatible with the bicelle method. Thus, the bicelle method can be described as an intermediate between the traditional detergent only crystallization method and the rigid LCP method, while neatly combining the practical convenience of the detergent method with the advantages of a native-like lipidic environment.
Like the LCP technique, bacteriorhodopsin provided the proof-of-concept for the bicelle crystallization method [32]. After the initial success, bacteriorhodopsin was also crystallized in other crystal forms using different bicelle formulations and conditions [42]. However, the real strength of the bicelle method was demonstrated by the crystal structure of the human β2-adrenergic G-protein-coupled receptor in complex with a Fab fragment at 3.4 Å resolution [16]. Other major breakthroughs came with the report of the structure determination of voltage-dependent anion channel 1 (VDAC1), a eukaryotic β-barrel channel solved to 2.3 Å resolution [43], and xanthorhodopsin, a member of the bacterial rhodopsin family solved to 1.9 Å resolution using bicelle crystallization [44]. Recently, the structure of rhomboid protease was also determined to high resolution from crystals grown with bicelles [45]. Table 1 summarizes all the membrane proteins solved using bicelles to date and the crystallization conditions. The growing collection of proteins demonstrates the versatility of the bicelle method for crystallizing a variety of membrane proteins that already includes colored, colorless, alpha-helical and beta-sheet proteins from both prokaryotic and eukaryotic sources including humans.
Table 1.
Membrane protein structures solved using the bicelle method.
| No. | Protein | Source | Resolution (Å) | Protein Concentration (mg/ml) | Bicelle Formulation | Crystallization Condition | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Bacteriorhodopsin | Halobacterium salinarum | 2.0 | 8 | 8% DMPC:CHAPSO (2.8:1) | 2.8M Sodium Phosphate pH 3.7, 180mM Hexanediol | Faham and Bowie, 2002 [32] |
| 1.8 | 8 | 8% DTPC:CHAPSO (3:1) | 100mM Sodium Formate pH 4.3, 28.5% PEG2000, 280mM Ammonium Sulfate, 180mM Hexanediol | Faham et al., 2005 [42] | |||
| 2 | β2-Adrenergic receptor | Homo sapiens | 3.4/3.7 | 10 | 8.3% DMPC:CHAPSO (3:1) | 1.85-2M Ammonium Sulfate, 180mM Sodium Acetate, 5mM EDTA, 100mM MES or HEPES pH 6.5-7.5 | Rasmussen et al., 2007 [16] |
| 3 | Voltage-dependent anion channel 1 | Mus musculus | 2.3 | 12 | 7% DMPC:CHAPSO (2.8:1) | 18% MPD, 0.1 M Tris pH 8.5, 10% PEG400 | Ujwal et al., 2008 [43] |
| 4 | Xanthorhodopsin | Salinibacter ruber | 1.9 | 4 | 4.2% DMPC, 5% NM | 2.5-3.5M Sodium Phosphate pH 5.6, 2.5mM Sodium Azide | Luecke et al., 2009 [44] |
| 5 | Rhomboid protease | Escherichia coli | 1.7 | 9 | 2% DMPC:CHAPSO (2.6:1) | 1.5 M NaCl, 0.1 M Bis-Tris (pH 7) | Vinothkumar, 2011 [45] |
The next section focuses on the experimental details of the bicelle methodology. A step-by-step protocol is provided for setting up high-throughput crystallization trials of purified membrane proteins using standard liquid handling robots.
2. Experimental Details
The bicelle methodology comprises four main steps: i) preparation of the lipidic bicellar mixture; ii) reconstitution of purified detergent-solubilized membrane protein into bicelles; iii) manual or robotic setup of crystallization trials and crystal optimization; and iv) visualization and extraction. Each step is discussed in detail below.
2.1 Bicelle Preparation
Bicelles can be prepared from a number of lipid:amphiphile combinations and at different concentrations [42,31]. Since the majority of the proteins crystallized using bicelles have been obtained from the DMPC:CHAPSO formulation (Table 1), this is a good mixture to begin with. DMPC:CHAPSO bicelles can either be purchased commercially as a premixed ready-to-use formulation [46] or prepared in the lab as described below:
2.1.1 Weigh out DMPC and CHAPSO
Add deionized water to the final volume and vortex. The molar ratio of DMPC:CHAPSO can be chosen to be between 2.6 and 3.0 (Table 1). The final bicelle concentration can be chosen to range between 10%-40%. Keep in mind that the higher the bicelle concentration, the greater the difficulty in dissolving the lipid and the higher the viscosity of the solution. A concentrated bicelle formulation can be advantageous when the protein concentration is low, however.
2.1.2 Dissolving the lipid to obtain a homogenous mixture requires some effort
Cycles of brief heating (~40-50°C), cooling on ice, vortexing and freezing should be performed to help the lipid go into solution. This can take several hours, but it only needs to be done once and then the stock can be used for many protein samples.
2.1.3
Upon completion, the mixture should be a viscous liquid on ice, but gel upon raising the temperature to ~30 °C, which reflects the change in phase from bicelles to more organized phases such as the perforated lamellar phase (Figure 1). It should be stored frozen at -20°C to prevent hydrolysis of the phospholipid head groups.
2.2 Protein incorporation into bicelles
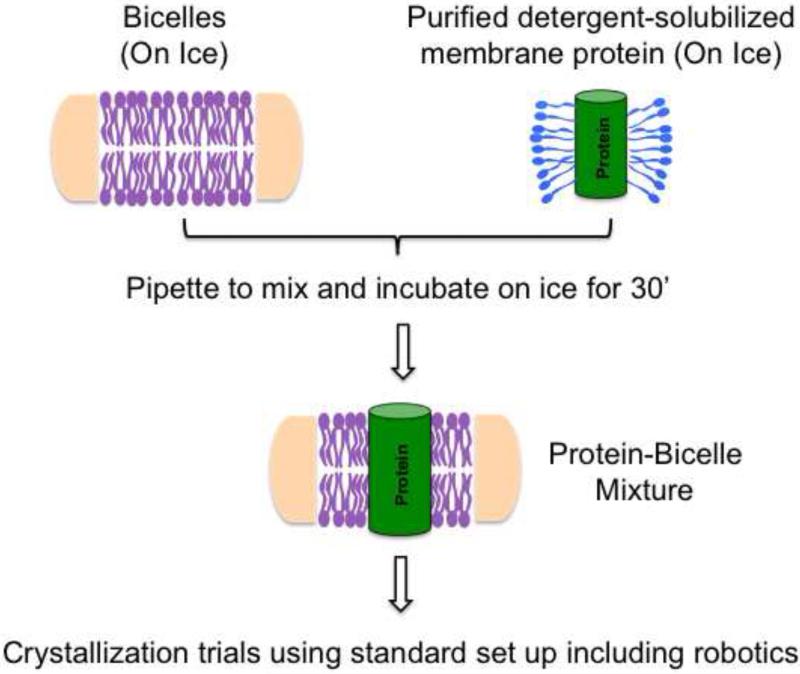
Since most bicelle structures have been obtained from ~8% DMPC:CHAPSO bicelle concentration with 8-12 mg/ml final protein concentration (Table 1), a good starting point would be to set up initial screens under these conditions and then screen other concentrations at the optimization stage or if initial screens do not yield any hits. As detailed below, mixing the protein sample is much easier with bicelles compared with other lipidic crystallization methods (Figure 2). The protein-bicelle mixture should be made fresh for same day use in crystal trials.
Figure 2. Bicelle crystallization.
Detergent-solubilized purified membrane proteins can be mixed directly with bicelles on ice. Homogenous mixing can be achieved by simply pipetting the contents up and down. After incubating the protein/bicelle mixture on ice for ~30 minutes, crystallization trials can be carried out using any standard format including robotics.
2.2.1
Thaw the bicelles at room temperature until the phase changes to a clear gel. Multiple freeze-thaws will not affect bicelle behavior.
2.2.2 Place the bicelle mixture on ice to liquefy
Vortex briefly to reestablish a homogenous phase and place it back on ice. While chilled on ice, it will remain in the liquid phase making it amenable to pipetting.
2.2.3
Add the bicelle solution to the purified detergent-solubilized protein (ideally >10 mg/ml) in a 1:4 (V/V) ratio. Depending on the concentration of the bicelle stock solution (10-40%), this will give a final bicelle concentration of 2-8%.
2.2.4
Gently pipette the contents up and down until the solution becomes clear and homogenous. A quick spin with a tabletop centrifuge can help remove bubbles that may appear during mixing.
2.2.5
Incubate the mixture on ice for about 30 min to allow for complete reconstitution of protein into bicelles. The proteo-bicelle mixture is now ready for crystallization trials. Keep the protein-bicelle mixture on ice until ready to set up crystal trials.
2.3 Crystallization Trials
Trials can be carried out in either hanging drop or sitting drop format using any commercially available screen. Manual crystallization trials can be set up in the same manner as detergent based techniques. In most cases, high-throughput crystallization trials can also be performed routinely. Robots like the popular Mosquito (TTP Labtech) can easily pipette protein-bicelle mixtures without requiring any special handling for setting up crystal trials. Other robots that lack positive displacement system can also be used for bicelle crystallization with the following suggestions that will help keep the protein-bicelle mixture cool, thus lowering the viscosity and ensuring accurate pipetting.
2.3.1
Keep the plate that holds the protein-bicelle sample on ice during set-up and in between multiple runs. This plate should be the last to go on the robot before starting the run.
2.3.2
Do not program the robot to mix the reservoir and the protein-bicelle solution. This will result in heating and hence increased viscosity, which may affect dispensing of the drops.
Since the phase behavior of bicelles is temperature dependent, it may be useful to incubate the crystallization trials at different temperatures. A good starting point for the initial crystal trials is 20°C. Higher temperatures, such as 30°C and 37°C can also be tested, as they induce the lamellar phase (Figure 1) [40], which has the advantage of pre-organizing the protein in layers. Temperatures below 20°C can be tested but should not be less than 4°C since this will cause the lipids to precipitate over extended period of time.
As with traditional crystallization trials, the bicelle trials should be monitored on a regular basis for crystal appearance and growth. A general schedule can be checking the tray on the 1st and 3rd day followed by weekly inspection after setup.
Optimization of initial crystals can be carried out using methods routinely used for detergent based crystals such as grid screening, additive screening and different temperatures. In addition, the final bicelle percentage and the ratio of protein:bicelle can be varied. Importantly, bicelles can also be doped with specific lipids that may be necessary for protein stability and function [47-52].
2.4 Visualization and Crystal Extraction
Unlike other lipidic media that may hinder crystal visibility, even colorless protein crystals can be easily viewed using standard microscopes with the bicelle method. However, as with other lipidic media, bicelle based trials form crystalline shapes that may be confused with protein crystals. Consequently, a UV-microscope can very useful for identifying these non-proteinaceous false positives [53].
Generally, it takes 2-3 days for crystals to form and about a week or more to grow to their maximum size, as was the case for bacteriorhodopsin [32] and mouse voltage-dependent anion channel 1 [unpublished observation]. However, for other membrane proteins it can take a longer time for crystal growth. For example, under some conditions, bacteriorhopsin crystals were only seen after ~1 month, so it is important to continue monitoring the crystal trials after the first few weeks [42].
Since the protein-bicelle mixture has a viscosity similar to detergent-based drops, crystal extraction can be carried out in a similar fashion to traditional setups. Thus, in contrast with the LCP method, no lipase or other such treatments are required for fishing crystals out of the lipidic media. In fact, the bicelle phase surrounding the protein crystal may be beneficial in providing some level of cryo-protection.
3. Conclusions
Bicelles are a unique lipidic media that provide membrane proteins with a native-like bilayer environment while offering the ease of use of detergents. Thus, in terms of ease of use, bicelles have a distinct edge over other lipidic crystallization protocols. Once bicelles are in hand, they can be mixed directly with purified detergent-solubilized membrane protein and from this step onwards crystallization setup proceeds almost exactly the same as traditional detergent-based methods.
Bicelles offer several other practical advantages compared with other lipid based crystallization methods like LCP. These include extended storage periods for bicelles, simple incorporation of proteins into bicelles, ability to use high-throughput robots, routine crystal analysis and harvesting. Furthermore, bicelles can be easily doped with lipids specific for the membrane protein of interest. In sum, bicelles are a versatile and user-friendly lipid media that should be part of all membrane protein crystallization projects.
Acknowledgments
We thank Dr. Salem Faham for providing technical expertise and guidance on the bicelle method. This work was supported by NIH grant R01GM063919 to JUB.
Abbreviations
- LCP
Lipidic Cubic Phase
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- CHAPSO
3-[(3-cholamidopropyl) dimethylammonio]-2-hydroxy-1-propanesulfonate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanabe M, Iverson TM. Current Topics in Membranes. Vol. 63. Academic Press; 2009. Chapter 10 A Practical Guide to X[hyphen (true graphic)]Ray Crystallography of [beta][hyphen (true graphic)]barrel Membrane Proteins: Expression, Purification, Detergent Selection, and Crystallization; pp. 229–267. [Google Scholar]
- 3.Michel H. Crystallization of membrane proteins. Trends in Biochemical Sciences. 1983;8:56–59. [Google Scholar]
- 4.Hunte C, Richers S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostermeier C, Iwata S, Ludwig B, Michel H. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat Struct Mol Biol. 1995;2:842–846. doi: 10.1038/nsb1095-842. [DOI] [PubMed] [Google Scholar]
- 7.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 10.Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 11.Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, et al. Structural Basis of Biological N2O Generation by Bacterial Nitric Oxide Reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 14.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen SGF, Choi H-J, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, et al. Crystal structure of the [bgr]2 adrenergic receptor-Gs protein complex. Nature. advance online publication. 2011 doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokoch MP, Zou Y, Rasmussen SGF, Liu CW, Nygaard R, Rosenbaum DM, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uysal S, Vásquez V, Tereshko V, Esaki K, Fellouse FA, Sidhu SS, et al. Crystal structure of full-length KcsA in its closed conformation. Proceedings of the National Academy of Sciences. 2009;106:6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, et al. High-throughput Generation of Synthetic Antibodies from Highly Functional Minimalist Phage-displayed Libraries. Journal of Molecular Biology. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 22.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 23.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 25.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, et al. High-Resolution Crystal Structure of an Engineered Human β2-Adrenergic G Protein–Coupled Receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, et al. Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, et al. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders CR, Prosser RS. Bicelles: a model membrane system for all seasons? Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 30.Sanders CR, 2nd, Hare Brian J., Howard Kathleen P., Prestegard James H. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog. NMR Spectrosc. 1994;26:421–444. [Google Scholar]
- 31.Sanders CR, 2nd, Landis GC. Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry. 1995;34:4030–4040. doi: 10.1021/bi00012a022. [DOI] [PubMed] [Google Scholar]
- 32.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 33.Tiburu EK, Moton DM, Lorigan GA. Development of magnetically aligned phospholipid bilayers in mixtures of palmitoylstearoylphosphatidylcholine and dihexanoylphosphatidylcholine by solid-state NMR spectroscopy. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2001;1512:206–214. doi: 10.1016/s0005-2736(01)00320-0. [DOI] [PubMed] [Google Scholar]
- 34.Czerski L, Sanders CR. Functionality of a membrane protein in bicelles. Anal. Biochem. 2000;284:327–333. doi: 10.1006/abio.2000.4720. [DOI] [PubMed] [Google Scholar]
- 35.Harroun TA, Koslowsky M, Nieh M-P, de Lannoy C-F, Raghunathan VA, Katsaras J. Comprehensive Examination of Mesophases Formed by DMPC and DHPC Mixtures. Langmuir. 2005;21:5356–5361. doi: 10.1021/la050018t. [DOI] [PubMed] [Google Scholar]
- 36.Katsaras J, Harroun TA, Pencer J, Nieh M-P. “Bicellar” Lipid Mixtures as used in Biochemical and Biophysical Studies. Naturwissenschaften. 2005;92:355–366. doi: 10.1007/s00114-005-0641-1. [DOI] [PubMed] [Google Scholar]
- 37.van Dam L, Karlsson G, Edwards K. Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2004;1664:241–256. doi: 10.1016/j.bbamem.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Nieh M-P, Raghunathan VA, Glinka CJ, Harroun TA, Pabst G, Katsaras J. Magnetically Alignable Phase of Phospholipid “Bicelle” Mixtures Is a Chiral Nematic Made Up of Wormlike Micelles. Langmuir. 2004;20:7893–7897. doi: 10.1021/la048641l. [DOI] [PubMed] [Google Scholar]
- 39.van Dam L, Karlsson G, Edwards K. Morphology of Magnetically Aligning DMPC/DHPC AggregatesPerforated Sheets, Not Disks. Langmuir. 2006;22:3280–3285. doi: 10.1021/la052988m. [DOI] [PubMed] [Google Scholar]
- 40.Prosser RS, Hwang JS, Vold RR. Magnetically aligned phospholipid bilayers with positive ordering: a new model membrane system. Biophys. J. 1998;74:2405–2418. doi: 10.1016/S0006-3495(98)77949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sternin E, Nizza D, Gawrisch K. Temperature Dependence of DMPC/DHPC Mixing in a Bicellar Solution and Its Structural Implications. Langmuir. 2001;17:2610–2616. [Google Scholar]
- 42.Faham S, Boulting GL, Massey EA, Yohannan S, Yang D, Bowie JU. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ujwal R, Cascio D, Colletier J-P, Faham S, Zhang J, Toro L, et al. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luecke H, Schobert B, Stagno J, Imasheva ES, Wang JM, Balashov SP, et al. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinothkumar KR. Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 2011;407:232–247. doi: 10.1016/j.jmb.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.2011 MemX Biosciences http://www.memxbio.com/
- 47.Struppe J, Whiles JA, Vold RR. Acidic phospholipid bicelles: a versatile model membrane system. Biophys. J. 2000;78:281–289. doi: 10.1016/S0006-3495(00)76591-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soong R, Macdonald PM. PEG molecular weight and lateral diffusion of PEG-ylated lipids in magnetically aligned bicelles. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2007;1768:1805–1814. doi: 10.1016/j.bbamem.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 49.Diller A, Loudet C, Aussenac F, Raffard G, Fournier S, Laguerre M, et al. Bicelles: A natural [‘]molecular goniometer’ for structural, dynamical and topological studies of molecules in membranes. Biochimie. 2009;91:744–751. doi: 10.1016/j.biochi.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dufourc EJ. Sterols and membrane dynamics. J Chem Biol. 2008;1:63–77. doi: 10.1007/s12154-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loudet C, Diller A, Grélard A, Oda R, Dufourc EJ. Biphenyl phosphatidylcholine: a promoter of liposome deformation and bicelle collective orientation by magnetic fields. Prog. Lipid Res. 2010;49:289–297. doi: 10.1016/j.plipres.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Kaya AI, Thaker TM, Preininger AM, Iverson TM, Hamm HE. Coupling Efficiency of Rhodopsin and Transducin in Bicelles. Biochemistry. 2011;50:3193–3203. doi: 10.1021/bi200037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Judge RA, Swift K, Gonzalez C. An ultraviolet fluorescence-based method for identifying and distinguishing protein crystals. 2005. [DOI] [PubMed]