Abstract
The homeodomain-containing transcription factor Pitx2 (pituitary homeobox protein 2) is present in many developing embryonic tissues, including the heart. Its homeodomain is responsible for the recognition and binding to target DNA sequences and thus constitutes a major functional unit in the Pitx2 protein. NMR techniques were employed to determine the solution structure of the native Pitx2 homeodomain and a R24H mutant that causes the autosomal dominantly inherited ring dermoid of the cornea syndrome. The structures reveal that both isoforms possess the canonical homeodomain fold. However, the R24H mutation results in a 2-fold increase in DNA-binding affinity and a 5°C decrease in the thermal stability, while changing the dynamic environment of the homeodomain only locally. When introduced into full-length Pitx2c, the mutation results in only a 25% loss of transactivation activity. Our data correlate well with clinical observations suggesting a milder deficiency for the R24H mutation compared to other Pitx2 homeodomain mutations.
Keywords: Pitx2, homeodomain, NMR, protein dynamics, Axenfeld-Rieger
The homeodomain-containing pituitary homeobox 2 (Pitx2) protein is a pivotal transcription factor required for the development of embryonic left-right asymmetry, as well as maintenance of several adult tissues including the pituitary, eye, and heart (1–10). In humans, haploinsufficiency of the Pitx2 allele (through absence or mutation) causes a range of developmental disorders and disease states, ranging from developmental syndromes like Axenfeld-Rieger to glaucoma (8, 11–13). Loss of PITX2 function in mice is lethal, and causes severe cardiovascular defects such as atrial isomerism, double inlet left ventricle, transposition of the great arteries, persistent truncus arteriosus, and abnormal aortic arch remodeling (14). Consistent with the severity of Pitx2 phenotypes, it has been shown to be a central downstream transcriptional activator in the canonical WNT/β-catenin signaling pathway (15, 16) and plays an important role in TGF-β signaling (17–20).
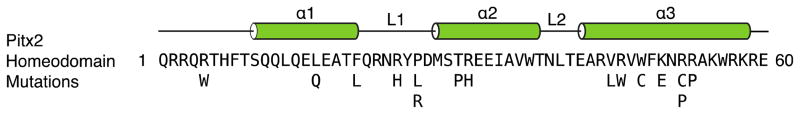
The homeodomain is an evolutionarily conserved protein fold present in transcription factors. Given the high prevalence of the homeodomain, it has served as a model system for probing the functional and structural aspects of protein-DNA interactions over the past two decades (21–39). The 60 amino acid homeodomain, canonically numbered 1–60, contains a helix-turn-helix motif that binds both DNA and RNA in prokaryotic and eukaryotic organisms (Figure 1) (40). The tertiary structure consists of three helical regions folded into a compact, globular structure with an N-terminal extension. Helix 1 (α1) is preceded by the N-terminal arm and is separated by a loose loop from the helix-turn-helix motif formed by helices 2 (α2) and 3 (α3) (Figure 2A). Helix 3, also called the recognition helix, lies in the major groove of the DNA and establishes specific contacts to the DNA bases. The N-terminal arm makes additional contacts with the bases in the DNA minor groove. In addition to the recognition helix and the N-terminal arm, the loop between α1 and α2 also interacts with the DNA backbone (21, 24). Typically, eight highly conserved hydrophobic amino acids (in the Pitx2 homeodomain: F8, L13, L16, F20, L40, V45, W48 and F49) spread throughout the 3 helices constitute the hydrophobic protein core (24).
Figure 1.
Human Pitx2 homeodomain amino acid sequence, secondary structure elements, and reported missense mutations. Numbering of the consensus sequence runs from 1–60. Mutations were compiled from: (8, 11–13, 41–49).
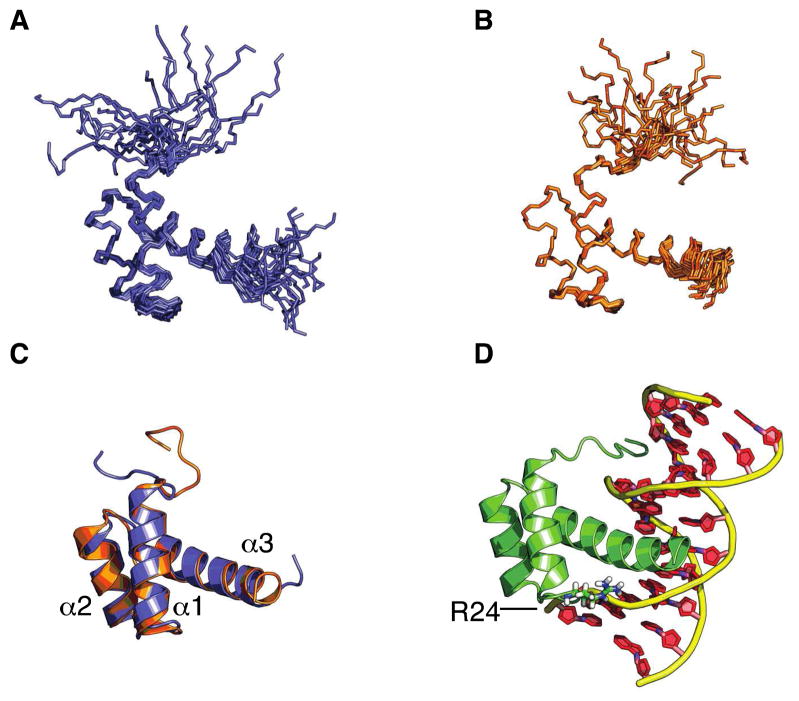
Figure 2.
Ribbon and cartoon representations of the Pitx2 homeodomain solution structures. Ensemble of the 20 lowest energy structures for A) the wild-type Pitx2 homeodomain (PDB: 2L7F) and B) the R24H mutant Pitx2 homeodomain (2L7M). C) Maximum likelihood superposition of closest to the average wild-type (blue) and R24H (orange) structures. D) Structure of the Pitx2 homeodomain – TAATCC consensus DNA complex(24) (PDB: 2lkx). Residue 24 is located in the loop connecting helices 1 and 2.
The homeodomain that is present in three out of four characterized Pitx2 isoforms has been identified as a hotspot for mutations. Of the known disease-causing mis-sense mutations, the majority can be found in the homeodomain (Figure 1) (8, 11–13, 41–49). The majority of these homeodomain mutations fall into three broad categories 1) modifying DNA-binding ability; 2) enhancing or diminishing transactivation capability; and 3) impacting protein stability. One such mutation that displays mild characteristics of all three categories, as we determined here, involves an amino acid substitution in position 24 of the homeodomain. This arginine to histidine substitution (R24H) corresponds to position 62 in the Pitx2a, position 108 in the Pitx2b, and position 115 in the Pitx2c isoforms and results in the autosomal dominantly inherited ring dermoid of the cornea syndrome (41). Unlike many other Pitx2 homeodomain mutations, the only clinical manifestations in R24H-affected patients are in the eyes (41), suggesting that from a clinical perspective this may be considered a mild mutation.
To characterize the effects of the R24H mutation in the Pitx2 homeodomain and to provide some insight into the origin of the clinical manifestations of this mutation, we have performed an in depth characterization of the structure, dynamics and functional properties of both the wild-type and R24H mutant homeodomains. The combined results of the NMR structure determinations, dynamics measurements, and other biophysical and functional studies indicate that for the most part the perturbations introduced by the R24H mutation are generally modest, and impart mild changes in DNA-binding affinity, transactivation activity, and conformational stability compared to wild-type Pitx2.
MATERIALS AND METHODS
Protein Expression and Purification
Proteins were expressed from a pet28 expression vector as a His6-TEV-GS-Pitx2homeodomain-EFIVTD fusion protein in Escherichia coli BL21DE3star cells (Invitrogen). Expression conditions were as described previously (24). Cells were harvested by centrifugation at 2500g for 15 mins. Harvested cells were resuspended in 137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic at pH 7.4 (PBS) with the addition of 10mM imidazole. Resuspended cells were lysed by passing through a chilled French Press twice at 12,000 PSI. The lysate was cleared by centrifugation at 25,000g for 30 minutes, at 4°C. Cleared lysate was applied to a pre-equilibrated 5ml HisTrap HP column (Gelifesciences). The column was washed with 10 column volumes (c.v.) of PBS+10mM imidazole. Next, an additional wash step of 10 c.v. with PBS+100mM imidazole was performed. Target fusion protein was eluted with 10 c.v. of PBS+500mM imidazole. Protein concentrations were estimated via A278 (ε 278 = 18350 cm−1 M−1) and the eluate was adjusted to contain 10% (v/v) glycerol, 5mM β-mercaptoethanol and 5mM EDTA. In-house produced TEV-protease was added for fusion tag removal at a 1:25 ratio and cleavage was performed at 4°C over 4 hours. Cleaved protein was then loaded on a 1ml HiTrap SP FF (Gelifesciences) cation exchange column, washed with washing buffer (10 mM NaH2PO4, 400 mM NaCl, pH 7.0), and eluted with buffer containing a higher salt concentration (10 mM NaH2PO4, 1 M NaCl, pH 7.0). Purity was determined to be >98% by SDS-PAGE. The eluted homeodomain was then dialyzed overnight at 4 °C into 10 mM NaH2PO4, 150mM Na2SO4, 1mM EDTA pH 7.0 for NMR or as indicated for the biophysical experiments. Proteins were then concentrated utilizing Amicon Ultra15 spin filters (Millipore) with a molecular cut-off of 3kDa.
Consensus response element DNA
All DNA was purchased from IDTDNA. The utilized constructs were sense 5′-GCTCTAATCCCCG-3′ and antisense 3′-CGAGATTAGGGGC-5′ with the complex referred to as TAATCC or consensus DNA. To obtain double-stranded DNA, the complementary DNA strands were annealed by heating for 15 minutes at 95°C followed by rapid cooling on ice.
Nuclear Magnetic Resonance Spectroscopy and Structural Calculations
For the structural studies, NMR sample concentrations ranged approximately from 0.25–1.5 mM, in 90% 10mM NaH2PO4, 150mM Na2SO4 pH 7.0 and 10% D2O. All NMR experiments were carried out on Varian Inova 400, 500, 600 and 800 MHz spectrometers. The sample temperature was set to 295K and spectra were referenced to an external DSS standard. Standard triple-resonance methodology using protein that was 13C/15N uniformly labeled was employed as described previously (21, 24). Raw data was processed utilizing NMRPipe (50) and spectra were analyzed via NMRViewJ (51). Dihedral angles were predicted utilizing Talos+ (52) and only good predictions were utilized during structure calculations. CYANA structures were calculated as previously described (21, 24). One hundred final cycle CYANA structures were subsequently refined using the generalized Born potential (53) implemented in AMBER 10 (54) to account for solvent effects as previously detailed (55). Twenty structures were determined by the FINDFAM software (56) to be sufficient to represent the conformational space consistent with the experimental data and were used to represent the NMR structure ensemble. Quality of the final NMR structures was assessed utilizing MolProbity (57).
For the histidine side-chain pH titrations, a 15N labeled sample was prepared at pH 4.0 as described above. Utilizing 1H-15N multiple bond correlation experiments (58), chemical shift values were measured in 0.5 pH unit increments. To calculate pKa values, the pH dependencies of the chemical shifts were fitted by non-linear regression to the Henderson-Hasselbalch equation (1) utilizing GraphPad Prism.
| (1) |
where δ is the measured chemical shift, δ 0 is the chemical shift of the deprotonated form and Δδ is the difference between the protonated and deprotonated forms.
15N Backbone Relaxation Data Collection and Analysis
Relaxation data for the wild-type Pitx2 homeodomain was recorded using a sample of 0.5 mM concentration. For the R24H mutant homeodomain, data was collected on four samples generated by serial dilutions, with concentrations of 1, 0.5, 0.25 and 0.125 mM. Backbone 15N relaxation rate constants R1 and R2 were determined from data recorded at 400 and 600 MHz, and {1H}-15N heteronuclear NOE values were determined from data recorded at 600 MHz via sensitivity-enhanced pulse sequences (59, 60). The R1 and R2 experiments did not employ a water flip-back scheme, whereas the NOE experiment did. The R2 measurements were made using a CPMG pulse train with a pulse spacing of 1 ms (center-to-center) and rf field strengths of 5 kHz (600 MHz data) and 4.2 kHz (400 MHz data); to avoid possible sample heating effects from the CPMG pulse train (61, 62), the delay between transients was set to 3 s and a constant duty cycle for the 15N irradiation was achieved by including a second CPMG block at the beginning of the pulse sequence that was adjusted such that the sum of the two CPMG blocks was a constant (63). For the R1 and R2 experiments the data for the various time points was collected in an interleaved fashion, the peak intensities were quantitated using the autoFit software in the NMRPipe package, and errors in intensities were determined from a combination of repeat measurements for several time points and evaluation of the baseplane noise. The rate constants and associated errors were determined using the Curvefit software (64). The NOE values were determined from pairs of 2D spectra – in one case (NOE spectrum) a train of 180° 1H pulses separated by 50 ms was applied (65) for a duration of 4 seconds at the beginning of the pulse sequence, and in the other case (reference spectrum) the same 1H pulse train was applied but with the power level at the minimum hardware setting and with the frequency shifted far off-resonance. For the reference spectrum an additional 2 seconds was included in the recovery period between transients, for a total of 6 seconds recycle delay. The NOE and reference data sets were recorded in an interleaved fashion to minimize systematic errors. Three repeats of the NOE measurement were performed and the results were averaged together.
Generalized order parameters, S2, were determined from the 15N R1 and R2 relaxation rate constants at 400 and 600 MHz and the 600 MHz {1H}-15N heteronuclear NOE using the Modelfree program (version 4.20) (64) and a protocol similar to that reported previously (66). Three dynamical models, described previously, were utilized in the data analysis. Using the common nomenclature, these were models 2, 4 and 5. In model 2, S2 is optimized along with an effective internal correlation time τe. For model 4 a chemical exchange term, Rex, is added to the model 2 parameters. Model 5 attempts to fit the data with two order parameters Sf2 and Ss2 (S2 = Sf2Ss2) and one internal correlation time τ s corresponding to the slower time scale reported on by Ss2. Initial estimates of the overall rotational diffusion tensor were determined from extensive grid searches for both an isotropic diffusion model and an axially symmetric model. Subsequently, the diffusion tensor parameters were optimized as part of the Modelfree analysis protocol. In the case of the axial diffusion model, one structure from the final ensemble of 20 lowest energy structures was selected using the program THESEUS (67), which uses a maximum likelihood method to superposition the ensemble and to select the structure closest to the ‘average’ structure for a family.
Circular Dichroism (CD) Spectroscopy
CD experiments were performed on an AVIV 215 CD spectrophotometer. Thermal melts were recorded in 1°C increments with 3 minute equilibration times at 5μM concentrations in a 1cm pathlength cell under 30 rpm stirring in PBS, pH 7.4 buffer with 5 second signal averaging. Spectra were collected offset (226nm) from the α-helical minimum (222nm) as the DNA signal at this wavelength was stable over the temperature range studied. Data analysis was performed as described elsewhere.(68, 69)
Isothermal Titration Calorimetry (ITC)
Measurements were performed in triplicate in 50mM NaH2PO4, 150 mM NaCl, pH 7.0 at 25°C using a MicroCal VP-ITC instrument. Protein concentrations ranged from 10–30μM with fixed dsDNA concentrations of 100 or 300μM. All DNA and protein samples were buffer matched and heat of dilution was measured in a separate experiment and corrected for. Raw data was analyzed assuming a single binding site model utilizing MicroCal ORIGIN 7 software.
Luciferase Reporter Transactivation Assay
The Cyclin D2 luciferase reporter construct contained 798 bps (−1 to −798) of the cyclin D2 promoter in a pGL4.24 luciferase vector (Promega, MI) (70). The PITX2c expression construct was situated within a pEGFP-N1 vector (Clontech, CA) expressing N-terminal flag-tagged Pitx2c protein (70, 71). Both vectors were generous gifts of Dr. Guo-Zhang Zhu (Marshall University). The R24H mutant was generated using a standard site-directed mutagenesis protocol. 24 h prior to transfection, HEK293 cells were plated in 96-well plates at a density of 2 × 104 cells/well. A total of 100 ng of reporter and Pitx2 plasmid mixes were transfected into HEK293 cells with Lipofectamine transfection reagent (Invitrogen, CA). 24 h or 48 h post-transfection, the luciferase activity was measured using the Dual-Glo™ luciferase assay system (Promega, WI). Results were analyzed using GraphPad Prism software. An empty vector control was performed and all values were normalized to the luciferase expression from the wild-type reporter to produce fold-induction values.
Western analysis
24 h prior to transfection, HEK293 cells were plated in 6-well plates at a density of 5 × 104 HEK293 cells/well. Transfections were performed using Lipofectamine™ 2000 (Invitrogen). 24 h post-transfection, HEK293 cells were washed once with phosphate-buffered saline and then incubated for 10 min at 4 °C in 100 μl of TNT lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 1% Triton X-100) and a complete miniprotease inhibitor mixture (Roche Applied Science, IN). Samples were then harvested into 1.5-ml microcentrifuge tubes, vortexed for 30 s, and then centrifuged. Protein levels in the supernatants were determined using a Coomassie protein assay kit (BioRad, CA), and 10 μg of protein from each sample was separated by 10% SDSPAGE (BioRad, CA) and then transferred to a polyvinylidene difluoride membrane (Millipore, MA) and immunoblotted with primary antibodies: α-Flag (Sigma, MO) or α-tubulin (Sigma, MO) and horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch, PA). Detection of the bound antibody by enhanced chemiluminescence was performed according to the manufacturer’s instructions (Santa Cruz, CA).
RESULTS
NMR Solution Structures of wild-type and R24H mutant Pitx2 homeodomains
Analysis of 15N and 13C heteronuclear-edited NOESY spectra yielded a total of 871 protein distance restraints for the wild-type and 1467 restraints for the R24H mutant protein (Table 1). On average, there were 13 and 22 nuclear Overhauser effect (NOE) restraints per residue for wild-type and R24H mutant proteins, respectively. The large difference in the number of NOE constraints between the two forms of the homeodomain is due to a factor of four difference in sample concentration employed for the NOE-based experiments and, secondarily, the fact that one of the NOE experiments for the wild-type protein was collected at 600 MHz on a conventional probe, whereas all of the NOE data for the R24H mutant was collected at 800 MHz using a cryoprobe. NOE and TALOS-predicted dihedral angle restraints were utilized for subsequent CYANA structure calculations. One hundred CYANA structures were subsequently refined in AMBER, and the NMR structural ensemble consisting of the 20 lowest energy structures exhibited mean AMBER energies of −4316 kcal mol−1 for the wild-type and −4095 kcal mol−1 for the mutant protein. When superimposed, the ensemble of the 20 lowest energy NMR structures exhibited a backbone average root mean square deviation (rmsd) of 1.33 Å and 0.61 Å, respectively, for the well-ordered regions (8–58) of the wild-type (Figure 2A) and R24H mutant (Figure 2B) homeodomains.
Table 1.
Structural statistics for the Pitx2 homeodomain structure ensembles (n=20)
| Wild-type | R24H mutant | |
|---|---|---|
| NMR constraints | ||
| Distance total | 871 | 1467 |
| Intra-residue (i=j) | 253 | 402 |
| Sequential (|i–j|=1) | 229 | 376 |
| Medium range (1 < |i–j| ≤ 4) | 263 | 420 |
| Long range (|i–j| ≥ 5) | 126 | 269 |
| Dihedral angles (ϕ and ψ) | 80 | 98 |
| Amber constraint violations | ||
| Average number of distance constraint violations per structure | ||
| 0.1Å < d < 0.2Å | 0 | 0.25 |
| 0.2Å < d | 0 | 0 |
| Maximum average distance violation (Å) | 0.093 | 0.156 |
| Average number of dihedral angle violations per structure (θ > 0°) | 0 | 0 |
| Maximum average dihedral angle violation (degrees) | 0 | 0 |
| Ensemble superposition statistics (Å)a | ||
| All (residues 1–60) | ||
| Backbone atoms | 8.34 ± 0.23 (3.32) | 4.47 ± 0.05 (1.78) |
| Heavy atoms | 8.43 ± 0.29 (3.36) | 4.44 ± 0.06 (1.77) |
| Well-ordered regions (residues 8–58) | ||
| Backbone atoms | 1.33 ± 0.20 (0.53) | 0.61 ± 0.05 (0.24) |
| Heavy atoms | 1.89 ± 0.24 (0.75) | 1.01 ± 0.05 (0.40) |
| AMBER energies (kcal mol−1) | ||
| Constraint | 1.320 ± 0.132 | 5.036 ± 0.269 |
| van der Waals | −499.1 ± 9.2 | −502.4 ± 6.6 |
| Total | −4316.4 ± 16.5 | −4095.4 ± 12.2 |
| MolProbity statisticsb | ||
| MolProbity score | 1.20 ± 0.30 | 1.16 ± 0.15 |
| MolProbity percentile rank | 97.70 ± 3.24 | 99.10 ± 1.18 |
| Clash score | 0.21 ± 0.36 | 0.09 ± 0.26 |
| Clash score percentile | 99.75 ± 0.43 | 99.90 ± 0.30 |
| Ramachandran space (%) | ||
| Favored | 96.31 ± 2.68 | 98.44 ± 1.31 |
| Allowed | 2.69 ± 2.06 | 1.09 ± 1.22 |
| Outliers | 1.00 ± 1.22 | 0.47 ± 0.72 |
Ensemble superposition statistics were calculated using THESEUS, reported as classical least squares pairwise <RMSD> ± ML<σ> (LS<σ>)
MolProbity clash score is the number of serious steric overlaps (> 0.4 Å) per 1000 atoms. Clash score percentile: 100th is the best among structures of comparable resolution, whereas 0th is the worst.
The well ordered regions are composed primarily of 3 helices spanning residues 10–22, 28–38 and 42–58 for helices α1, α2, and α3, respectively. The helices are connected via two short loops, L1 and L2. The 8 conserved hydrophobic residues in combination with P26 and I34 form the protein core and helices α1 and α2 run roughly parallel to each other and perpendicular to α3. This tertiary structure is representative of the canonical homeodomain fold. An overlay of the average wild-type and R24H mutant Pitx2 homeodomain structures (Figure 2C) indicates that the overall fold is minimally perturbed by the R24H mutation. With respect to the DNA-bound Pitx2 homeodomain complex(24) (Figure 2D), a superposition with the well-structured portion of the free (wild-type) homeodomain reveals a close alignment, with a backbone RMSD (as determined by PyMol) of 0.713 Å.
In contrast to the rigid and well-structured protein core, it is clearly evident from the superposition of the 20 structure conformers, as well as the backbone relaxation measurements (vide infra), that the N-terminal arm (1–9) samples a broad conformational space in the absence of a binding partner. In the presence of the TAATCC consensus DNA, the N-terminal arm binds in the minor groove of DNA (Figure 2D) and becomes more rigid as specific interactions between the N-terminal arginines and the DNA bases are formed (24).
The arginine in position 24 is solvent-exposed and does not exhibit any long-range NOE constraints in the absence of DNA. However, in the presence of its consensus TAATCC DNA binding site, R24 is oriented in close proximity to the phosphate backbone and likely makes a non-specific electrostatic contact. A histidine in this position (R24H) could make a similar contact, depending on the pKa of its cyclic imidazole side chain.
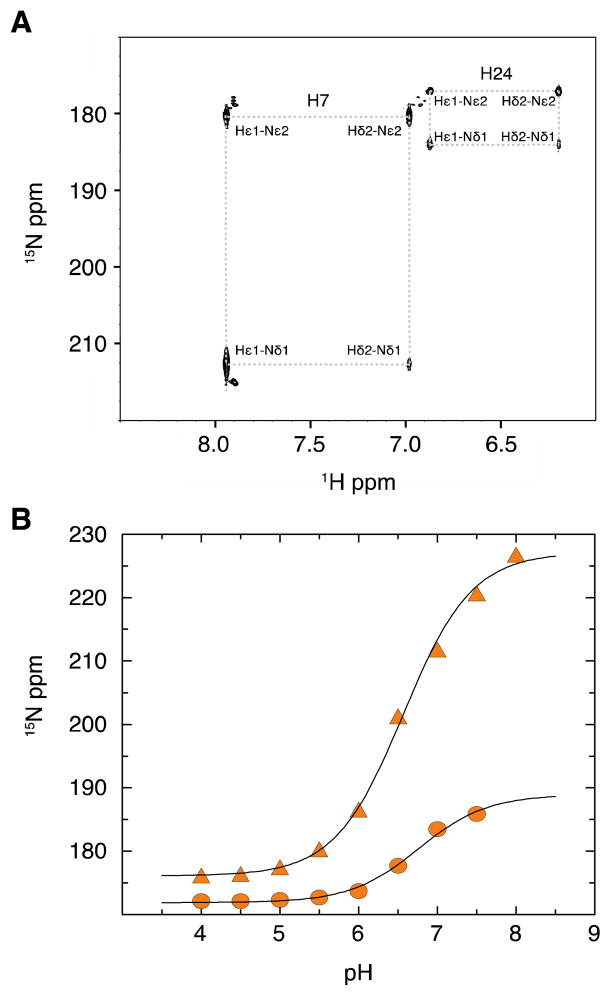
NMR Histidine pH Titration
The typical arginine side chain pKa is ~12.5, whereas the pKa of a histdine side chain is typically much lower (~ 6.0). To address a possible charge effect of the arginine to histidine mutation, 2D 1H-15N HSQC spectra of a uniformly 15N-labeled R24H mutant Pitx2 homeodomain were collected between pH 4.0 and 8.0 in 0.5 pH increments. In the spectra collected below pH 7.5, four cross-peaks corresponding to the Hε1-Nδ1, Hε1-Nε2, Hδ2-Nε2, and Hδ2-Nδ1 were observed for both H7 and H24 (Figure 3A). At and above pH 7.5, only three cross-peaks corresponding to the H7 Hε1-Nδ1, Hε1-Nε2, and Hδ2-Nε2 resonances were observed. Figure 3B shows the pH titration curves for the Nδ1 chemical shifts. pKa values of 6.57±0.04 (H7) and 6.76±0.08 (H24) were calculated by fitting the pH titration data to the Henderson-Hasselbalch equation. Thus, at an experimental pH of 7.0, the histidine side chains are approximately 63% and 73% deprotonated for H24 and H7, respectively.
Figure 3.
NMR titration of the histidine side chain. A) pH 7.0 snapshot of a long-range 1H-15N HSQC experiment to detect the pH dependent protonation of the two histidine side chains in the R24H Pitx2 homeodomain mutant. B) Chemical shift titration curves of the 15N chemical shifts of the histidine Nδ1 side chain resonances with fits to equation 1. Filled triangles correspond to H7 and filled circles to H24.
15N Backbone Relaxation Analysis
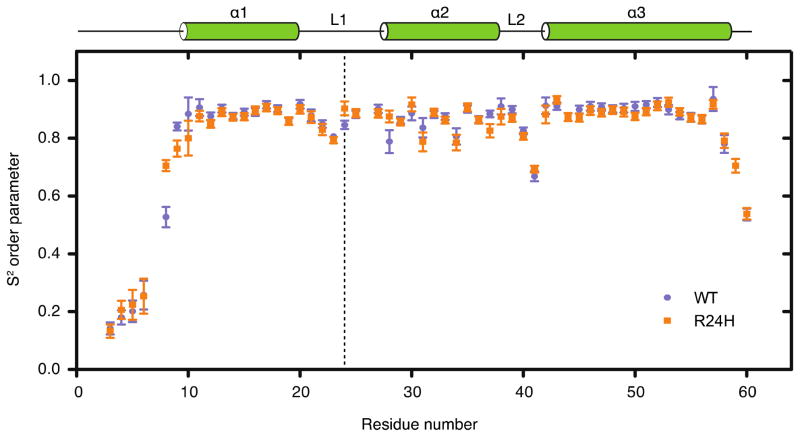
15N R1 and R2 relaxation parameters were determined at proton frequencies of 600 and 400 MHz and {1H}-15N heteronuclear NOE data at 600 MHz, at a temperature of 295 K, in order to compare the conformational flexibility of the wild-type and R24H mutant Pitx2 homeodomains. The relaxation data for the wild-type Pitx2 homeodomain was collected on a sample of 0.5 mM concentration. For the R24H mutant homeodomain, the R1 and R2 relaxation data were collected for four samples, with sample concentrations of 1, 0.5, 0.25 and 0.125 mM. Following extraction of the individual relaxation parameters (presented graphically in Supplementary Figure 1), the complete data sets at 0.5 mM concentration were subjected to ModelFree analysis (72) to determine S2 generalized order parameters (Figure 4). The backbone 15N order parameters – a measure that distinguishes ordered from flexible regions in a protein – can range from a value of 1, indicative of completely restricted internal motion (on the picosecond-nanosecond timescale), to a value of 0, indicative of completely unrestricted motion. The relaxation data was fit using an isotropic and an axially symmetric diffusion tensor for both the wild-type and R24H mutant Pitx2 homeodomains, and for both homeodomains a χ2 analysis lead to the selection of the symmetric diffusion model. This data analysis yielded values of 7.9 and 7.5 ns for the overall rotational correlation times and values for the rotational anisotropy Dr of 0.78 and 0.83 for the wild-type and R24H mutant homeodomains, respectively. The values determined for the overall rotational correlation times are larger than one would expect for homeodomain monomers tumbling freely in solution, thus indicating the presence of some degree of self-association, presumably a monomer-dimer equilibrium. This observation is corroborated by the concentration dependence of the 15N R2 rate constants (Supplementary Figure 1). The self-association appears to be relatively weak and non-specific, as no significant changes in amide 1H and 15N chemical shifts were observed in the concentration series of data. Although S2 values can be over-estimated for proteins that self-associate, previous experimental studies have found increases in S2 that were not dramatic (73, 74). In the analysis presented below we limit ourselves to qualitative rather than detailed quantitative comparisons regarding the S2 values of the Pitx2 homeodomains.
Figure 4.
S2 generalized order parameters for the wild-type (●) and R24H mutant (■) Pitx2 homeodomain. The N-terminal arm is highly flexible in the absence of a DNA binding partner. The three helices exhibit average S2 values of ~0.85 suggesting only a limited degree of internal motion. The connecting loop regions L1 and L2 generally have lower S2 values indicative of increased ps to ns internal motion in these regions. Similar to the N-terminus, the C-terminus is highly mobile (including the 6 amino acid artificial tale – data not shown). The vertical dashed line indicates the position of residue 24.
The generalized order parameter S2, heteronuclear NOE, and R2 data all suggest that the N-terminal arm is highly flexible in the absence of DNA (Figure 4 and Supplementary Figure 1). This finding is also reflected in our solution structures that lack any long-range structural restraints in the first nine residues and is consistent with previous reports on the backbone dynamics of the vnd/NK-2 and the TTF-1 homeodomains (75, 76). In the presence of its consensus DNA, the N-terminal arm binds in the minor groove of DNA and loses some of its mobility characteristics (21, 24). This change in the dynamic nature of the N-terminal arm is thought to contribute significantly to the process of specific DNA-binding by the homeodomain motif (77–80). In addition, the loop regions L1 and L2 connecting α1 with α2 and α2 with α3 display a higher degree of motion than the rest of the protein, as indicated by the S2, NOE, R1 and R2 data. This effect, especially in the L2 region, is more striking than in previous studies of other homedomains (75, 76) and may allow for the L1 and L2 regions to serve as hinges for the homeodomain to adjust the positions of its helices relative to each other. The core fold (α1, α2, α3) of the Pitx2 homeodomain, by contrast, is relatively restricted, independent of a DNA-binding event.
In comparison to the wild-type data, around the point of the R24H mutation the dynamic behavior is generally little changed as measured by the S2 values. Residue H24 is modestly more restricted in comparison to the wild-type R24 residue. Residue M28 is also modestly more restricted in the R24H mutant versus wild-type homeodomain. The immediately downstream residues Y25 and D27 (P26 can not be observed due to the unique nature of the proline cyclic side chain) and upstream residues N23 and R22 also exhibit a comparable degree of internal motion suggesting that the local dynamic environment is not significantly perturbed by this mutation. Residues F8, T9, and W37 display S2 values that differ between the wild-type and mutant proteins. In these cases different motional models were selected for the wild-type versus mutant homeodomains, and thus the possibility exists that the observed differences arise due to model selection bias in the calculation of the generalized order parameters for these residues. Compared to helices 1 and 3, helix 2 exhibits somewhat greater variability of the S2 values along the helix, for both wild-type and mutant proteins. Also in common between the two isoforms is an increase in flexibility towards the end of helix 3. This observation likely explains the observed increase in disorder in this region of the structures shown in Figure 2. Data for residue R59 in the wild-type homeodomain is absent from Figure 4 as none of the motional models that were considered provided an adequate fit of the relaxation data. The apparent, increased disorder at the C-terminal end of helix 3 for the wild-type homeodomain versus the R24H mutant is likely due to the increased number of structural constraints that were obtained for the mutant (Table 1), as the relaxation data does not indicate significant differences in dynamic disorder.
Regarding slower motional processes on the microsecond-millisecond timescale, only residue T38 at the beginning of loop 2 appears to be significantly affected as measured by the Rex term of the Modelfree data analysis, and the wild-type and R24H homeodomains exhibit similar values (3.5±0.6 Hz versus 4.6±0.5 Hz, respectively). There is strong evidence for even slower motions for most of the N-terminal arm residues, again for both homeodomains, as judged by the significantly weakened intensities for these residues in the NMR spectra (Supplementary Figure 2).
Stability of the free and DNA-bound homeodomain
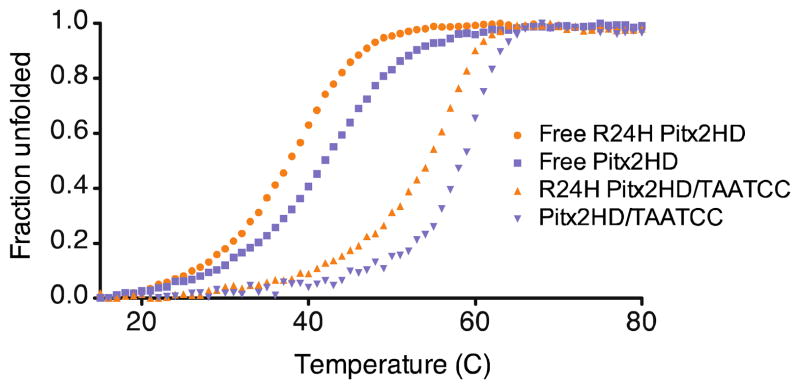
To address whether the R24H mutation and the resultant changes in helix orientation impact protein stability, we performed circular dichroism (CD) spectroscopy on the free and DNA-bound forms of the Pitx2 homeodomains. The solution structures confirmed that both isoforms possess the typical homeodomain fold, yet the thermal denaturation profile reveals that the R24H mutant is significantly less stable (Tm= 37.8°C) than the wild-type homeodomain (Tm= 42°C) (Figure 5). The wild-type Tm is comparable to that of the K-50 class Bicoid homeodomain (Tm= 44°C) (81) and many other characterized homeodomains (81–83). Both isoforms display a two-state folding transition that is stabilized by binding to the TAATCC consensus DNA site (Figure 5). Interestingly, the degree of stabilization arising from binding DNA appears to be independent of the mutation, +16.4°C vs. 16.5°C, respectively for the mutant and wild-type proteins. This same observation holds true for a non-consensus DNA sequence that stabilizes both homeodomain isoforms by about 10°C (data not shown). The functional consequences of the reduced thermal stability of the R24H mutant are currently unknown; it is possible that this reduction could lead to more rapid degradation of the mutant in vivo.
Figure 5.
Thermal unfolding profiles of the Pitx2 homeodomain. Unfolded fractions of Pitx2 homeodomain wild-type and R24H plotted as a function of temperature. DNA free: wild-type (■), R24H (●). TAATCC bound: wild-type (▼), R24H (▲). Apparent Tm values for the wild-type free 42.0°C, R24H free 37/8°C, wild-type/TAATCC 58.5°C, R24H/TAATCC 54.2°C.
Thermodynamics of Pitx2 Homeodomain – consensus DNA binding
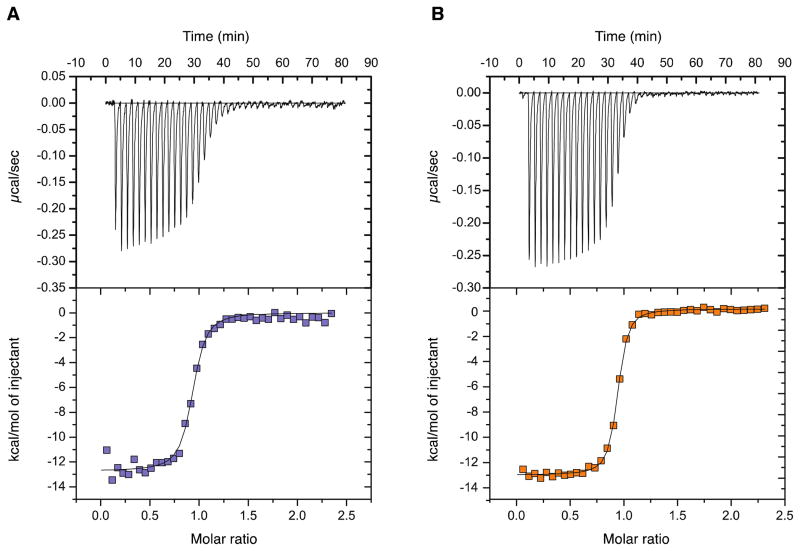
To investigate the thermodynamic parameters involved in the Pitx2 homeodomain/consensus DNA complex formation we performed isothermal titration calorimetry (ITC). All ITC experiments were performed with our purified recombinant protein in the reaction chamber and titrating in duplex TAATCC consensus DNA. As shown in Table 2, the interactions of the wild-type and R24H mutant Pitx2 homeodomain with the consensus DNA duplex are exothermic and exhibit 1:1 stoichiometry. The reactions are enthalpically driven with a significant entropic contribution. Interestingly, the mutant exhibits a greater entropic contribution that makes the interaction more favorable under the conditions studied. At 25°C in the presence of 50mM NaH2PO4, 150 mM NaCl, pH 7.0 the dissociation constants (Kd) for the complexes are ~26 nM for the R24H mutant and ~66 nM for the wild-type protein (Figure 6). These values are well within the typical range found for DNA-binding proteins and homeodomain complexes in particular (84–86). Given the strong salt dependence of binding for the K-50 class homeodomains (unpublished observations) as well as for homeodomains in general (84), the association and dissociation parameters vary greatly depending on buffer composition and it is not surprising that previous gel shift experiments with lower salt buffer yielded lower Kd values (24). In the absence of a high-resolution structure of a complex of the R24H mutant homeodomain with the consensus DNA sequence, we can only speculate on the physical orign of the higher affinity of the R2$H mutant for the DNA. One possibility is that the R24H mutation introduces small rearrangements of nearby side chains that lead to more optimal interactions with the DNA.
Table 2.
Results of ITC measurements on Pitx2 homeodomain/DNA complexes
| K(M−1) | Kd (nM) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS° (kcal/mol) | |
|---|---|---|---|---|---|
| Pitx2HD wt / TAATCC | 1.51(±0.29) × 107 | 66 | −9.84±0.27 | −7.91±0.17 | −1.93±0.10 |
| Pitx2HD R24H / TAATCC | 3.79(±0.37) ×107 | 26 | −10.32±0.12 | −7.53±0.30 | −2.79±0.36 |
Figure 6.
ITC binding data for the Pitx2 homedomain/consensus DNA interaction. Panel A. A representative isothermal titration of the Pitx2 homeodomain wild-type with TAATCC duplex DNA at 25°C. Panel B. A representative isothermal titration of the R24H Pitx2 homeodomain with TAATCC duplex DNA at 25°C displaying ~2 fold higher binding affinity.
Pitx2c Luciferase Reporter
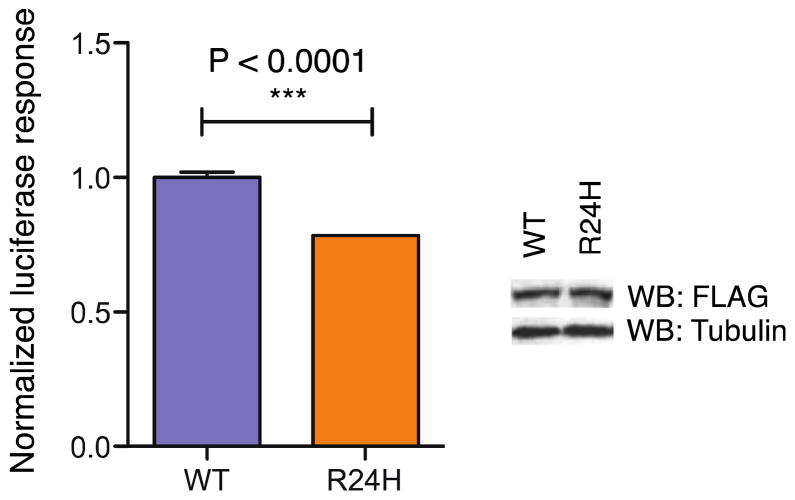
To understand better the impact of the R24H mutation in the context of the full-length protein, we transiently transfected HEK293 cells with an expression plasmid for human Pitx2c (wild-type or R24H mutant) and a luciferase reporter under the control of the Pitx2c target gene CCND2 (Figure 7). Both wild-type and R24H mutant Pitx2c proteins are expressed at comparable protein levels. Compared to wild-type protein, the transactivation response of the R24H mutant is approximately 25% lower, indicating that the mutation does not entirely abolish the transactivation response as in the case of other Pitx2 homeodomain mutations (Semina et al., 1996; Perveen et al., 2000; Amendt, 2005).
Figure 7.
Luciferase reporter response to Pitx2 expression in HEK293 cells. A Cyclin D2 luciferase reporter was transiently transfected with either full-length wild-type Pitx2c or full-length protein harboring the R24H mutation in the homeodomain. Data represents as mean ± SEM with ***P<0.0001. Western Blot analysis confirms that mutant protein is expressed at levels comparable to the wild-type protein.
DISCUSSION
Solution structures
Overall, the Pitx2 homeodomain fold represents a typical homeodomain motif, with three helical regions in a compactly folded structure, and a flexible N-terminal tail. No major structural differences were observed between the wild-type and R24H mutants. This observation is consistent with the fact that the R24H mutation does not lead to severe disfunction of Pitx2. In contrast, a 1H-15N HSQC 2D NMR spectrum of the V45L mutant of Pitx2 (Supplementary Figure 3), which is also a clinically observed mutation, indicates a significant perturbation of the stable fold that is consistent with the reduced DNA binding that has been reported for this mutant (44). Unlike V45, R24 is not part of the hydrophobic core of the homeodomain, and thus its mutation leads to more subtle effects on the stucture.
Functional effect of the R24H mutation
The experimentally determined pKa value of 6.76 for the R24H mutant differs significantly from the typical arginine pKa of 12.5. In a biological environment the arginine is likely charged at all times whereas the histidine residue will exist in a protonated/deprotonated equilibrium that may range from ~73% deprotonated at a cytosolic pH of 7.2 to ~90% deprotonated at a nuclear pH of 7.7 (87). The absence of a base-specific contact of R24 in the Pitx2 homeodomain/DNA complex structure suggests that the charge on the arginine is not needed to confer DNA-binding specificity. Indeed our ITC experiments at pH 7.0 indicate that the mutant has a 2-fold higher binding affinity than the wild-type homeodomain. The observation in our circular dichroism experiments that addition of different target DNA sequences stabilized wild-type and mutant homeodomain to identical degrees further implies that a positive side chain charge in position 24 is not imperative to confer DNA-binding specificity. In the absence of a Pitx2 homeodomain R24H/DNA complex structure, our results suggest that the lost charge does not impair DNA-binding activity. In fact, the Pitx2 R24H solution structure and the experimentally determined histidine pKa (which closely mimics the reported pKa value of 6.9 for a solvent exposed imidazole ring in a Gly-His-Gly model peptide (88)) suggest that residue 24 is generally solvent exposed.
L1 and α2 region as transactivation regulators
With very few exceptions, most of the Pitx2 transactivation activity has previously been attributed to its N- and C-termini (89). Interestingly, two reported exceptions include the T30P and R31H homeodomain mutations at the beginning of α2. These mutations both result in only slightly reduced levels of DNA-binding, but defective transactivation activity in the case of the T30P mutation (9, 90) and a reduction to 12.5% of the wild-type levels for the R31H mutation (91). Similar to the R24 and mutant R24H side chains, the T30 side chain lacks long-range contacts with other amino acids within the homeodomain in all of our NMR structures. In contrast, the R31 side chain is observed in long-range contacts with residues F49 and R46 in helix 3. Unlike the T30P and R31H mutations, the DNA-binding affinity for the R24H mutation is slightly higher and transactivation activity is not completely ablated. However, the increase in DNA-binding affinity and the simultaneous decrease in transactivation activity suggest that R24 is important for transactivation activity as well.
A prevailing model in the field describes the C-terminal tail of full-length Pitx2 looping back onto the homeodomain and N-terminus to inactivate the protein (89). As all our homeodomain structures indicate that residues 24, 30 and 31 are solvent exposed, these residues could potentially be available to make contacts with the Pitx2 N- and C-termini or other protein binding partners important for transactivation. The described mutations in L1 or α2 may cause Pitx2 to effectively displace its C-terminal tail, allowing for DNA-binding, but not effective transactivation. The fact that most other residues in close vicinity, with the exception of P26, extend away from the homeodomain core, and the altered dynamic behavior of the L1 region in the R24H mutant may further imply that this region is involved in intra- or intermolecular protein-protein interactions. Pitx2 is known to interact with other proteins, such as PAWR(92) and the Wnt/DvI/β-catenin pathway,(15) but to our knowledge the role of R24 in such interactions has not been determined. However, it is well established that residues in the L1 loop have important functional consequences in homeodomains.(93)
Dynamic nature of the Pitx2 homeodomain
Measurement of 15N backbone relaxation rates provide detailed information about molecular motion on the picosecond-nanosecond and microsecond-millisecond time-scales. A disordered state, such as that described above for the N-terminal arm of the homeodomain, or motions on the slower μs-ms time-scale often correlate to regions involved in protein function, including protein-protein and protein-DNA interactions (94, 95). In addition to the characterized ps-ns motion we have evidence that the N-terminal arm undergoes more complex motion on the μs-ms time scale. A comparison of the intensities of the amide backbone resonances (96) of the N-terminal arm with the resonances of the artificial C-terminal tail (Supplementary Figure 2) indicates strong peaks with narrow linewidths for the C-terminal tail residues, but a very heterogeneous distribution for the N-terminal arm. Such resonance broadening, especially for residues 1 and 7 is the result of slower motions and warrants further examination to better understand the dynamic nature of the N-terminal arm and its role in DNA and protein recognition and binding.
Supplementary Material
Acknowledgments
Funding: Funding for this project was provided through NIH grants GM063855 to M. R., RR19077 and RR027755 to the UC College of Medicine NMR facility, ES007250 and AI055406 to D.K., HL007382 to J.B.T. and an American Heart Association Fellowship and Fight for Sight Summer Student Fellowship to T.D.
We thank Jun Ma (Cincinnati Children’s Hospital Research Foundation) for providing the original Pitx2 homeodomain plasmid, Al Combs for subsequent construct optimization, and Guo-Zhang Zhu (Marshall University) for generously supplying the PITX2c vectors. We thank Walter Chazin and Jarrod Smith (Vanderbilt University) for kindly providing the FINDFAM software. We thank Andrew Herr, Rhett Kovall and Paul Rosevear for helpful comments.
ABBREVIATIONS
- TEV
tobacco etch virus
- PBS
phosphate-buffered saline
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- CPMG
Carr-Purcell-Meiboom-Gill
- CD
circular dichroism
- ITC
isothermal titration calorimetry
- NOESY
nuclear Overhauser effect spectroscopy
- HSQC
heteronuclear single quantum correlation
Footnotes
Supporting Information Available: Figures showing R1, R2 and NOE 15N relaxation parameters (Figure S1) and line broadening for residues in the N-terminal arm (Figure S2) for the wild-type Pitx2 homeodomain and the R24H mutant, and two-dimensional 1H-15N HSQC spectra (Figure S3) for the wild-type, R24H mutant, and V45L mutant Pitx2 homeodomains. Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 2.Ryan AK, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, Norris DP, Robertson EJ, Evans RM, Rosenfeld MG, Izpisua Belmonte JC. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 3.Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- 4.Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- 5.Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, Hamada H, Noji S. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 6.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjodal M, Gunhaga L. Expression patterns of Shh, Ptc2, Raldh3, Pitx2, Isl1, Lim3 and Pax6 in the developing chick hypophyseal placode and Rathke’s pouch. Gene Expr Patterns. 2008;8:481–485. doi: 10.1016/j.gep.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 9.Saadi I, Semina EV, Amendt BA, Harris DJ, Murphy KP, Murray JC, Russo AF. Identification of a dominant negative homeodomain mutation in Rieger syndrome. J Biol Chem. 2001;276:23034–23041. doi: 10.1074/jbc.M008592200. [DOI] [PubMed] [Google Scholar]
- 10.Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 11.Alward WL, Semina EV, Kalenak JW, Heon E, Sheth BP, Stone EM, Murray JC. Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. Am J Ophthalmol. 1998;125:98–100. doi: 10.1016/s0002-9394(99)80242-6. [DOI] [PubMed] [Google Scholar]
- 12.Kulak SC, Kozlowski K, Semina EV, Pearce WG, Walter MA. Mutation in the RIEG1 gene in patients with iridogoniodysgenesis syndrome. Hum Mol Genet. 1998;7:1113–1117. doi: 10.1093/hmg/7.7.1113. [DOI] [PubMed] [Google Scholar]
- 13.Tumer Z, Bach-Holm D. Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet. 2009;17:1527–1539. doi: 10.1038/ejhg.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 15.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 16.Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2005;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 17.Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 18.Xiao H, Zhang YY. Understanding the role of transforming growth factor-beta signalling in the heart: overview of studies using genetic mouse models. Clin Exp Pharmacol Physiol. 2008;35:335–341. doi: 10.1111/j.1440-1681.2007.04876.x. [DOI] [PubMed] [Google Scholar]
- 19.Glasgow E, Mishra L. Transforming growth factor-beta signaling and ubiquitinators in cancer. Endocr Relat Cancer. 2008;15:59–72. doi: 10.1677/ERC-07-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suszko MI, Antenos M, Balkin DM, Woodruff TK. Smad3 and Pitx2 cooperate in stimulation of FSHbeta gene transcription. Mol Cell Endocrinol. 2008;281:27–36. doi: 10.1016/j.mce.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Baird-Titus JM, Clark-Baldwin K, Dave V, Caperelli CA, Ma J, Rance M. The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site. Journal of molecular biology. 2006;356:1137–1151. doi: 10.1016/j.jmb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Billeter M, Guntert P, Luginbuhl P, Wuthrich K. Hydration and DNA recognition by homeodomains. Cell. 1996;85:1057–1065. doi: 10.1016/s0092-8674(00)81306-9. [DOI] [PubMed] [Google Scholar]
- 23.Scott MP, Tamkun JW, Hartzell GW., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 24.Chaney BA, Clark-Baldwin K, Dave V, Ma J, Rance M. Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome. Biochemistry. 2005;44:7497–7511. doi: 10.1021/bi0473253. [DOI] [PubMed] [Google Scholar]
- 25.Ades SE, Sauer RT. Differential DNA-binding specificity of the engrailed homeodomain: the role of residue 50. Biochemistry. 1994;33:9187–9194. doi: 10.1021/bi00197a022. [DOI] [PubMed] [Google Scholar]
- 26.Billeter M, Qian YQ, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. Journal of molecular biology. 1993;234:1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- 27.Fraenkel E, Rould MA, Chambers KA, Pabo CO. Engrailed homeodomain-DNA complex at 2.2 A resolution: a detailed view of the interface and comparison with other engrailed structures. Journal of molecular biology. 1998;284:351–361. doi: 10.1006/jmbi.1998.2147. [DOI] [PubMed] [Google Scholar]
- 28.Gruschus JM, Tsao DH, Wang LH, Nirenberg M, Ferretti JA. The three-dimensional structure of the vnd/NK-2 homeodomain-DNA complex by NMR spectroscopy. Journal of molecular biology. 1999;289:529–545. doi: 10.1006/jmbi.1999.2774. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch JA, Aggarwal AK. Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity. Embo J. 1995;14:6280–6291. doi: 10.1002/j.1460-2075.1995.tb00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hovde S, Abate-Shen C, Geiger JH. Crystal structure of the Msx-1 homeodomain/DNA complex. Biochemistry. 2001;40:12013–12021. doi: 10.1021/bi0108148. [DOI] [PubMed] [Google Scholar]
- 31.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Stark MR, Johnson AD, Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 33.Longo A, Guanga GP, Rose RB. Structural basis for induced fit mechanisms in DNA recognition by the Pdx1 homeodomain. Biochemistry. 2007;46:2948–2957. doi: 10.1021/bi060969l. [DOI] [PubMed] [Google Scholar]
- 34.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 35.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 36.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 37.Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 38.Wilson DS, Sheng G, Jun S, Desplan C. Conservation and diversification in homeodomain-DNA interactions: a comparative genetic analysis. Proc Natl Acad Sci U S A. 1996;93:6886–6891. doi: 10.1073/pnas.93.14.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 40.Dave V, Zhao C, Yang F, Tung CS, Ma J. Reprogrammable recognition codes in bicoid homeodomain-DNA interaction. Mol Cell Biol. 2000;20:7673–7684. doi: 10.1128/mcb.20.20.7673-7684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia K, Wu L, Liu X, Xi X, Liang D, Zheng D, Cai F, Pan Q, Long Z, Dai H, Hu Z, Tang B, Zhang Z, Xia J. Mutation in PITX2 is associated with ring dermoid of the cornea. J Med Genet. 2004;41:e129. doi: 10.1136/jmg.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips JC. Four novel mutations in the PITX2 gene in patients with Axenfeld-Rieger syndrome. Ophthalmic Res. 2002;34:324–326. doi: 10.1159/000065602. [DOI] [PubMed] [Google Scholar]
- 43.Weisschuh N, Dressler P, Schuettauf F, Wolf C, Wissinger B, Gramer E. Novel mutations of FOXC1 and PITX2 in patients with Axenfeld-Rieger malformations. Invest Ophthalmol Vis Sci. 2006;47:3846–3852. doi: 10.1167/iovs.06-0343. [DOI] [PubMed] [Google Scholar]
- 44.Priston M, Kozlowski K, Gill D, Letwin K, Buys Y, Levin AV, Walter MA, Heon E. Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld-Rieger syndrome. Hum Mol Genet. 2001;10:1631–1638. doi: 10.1093/hmg/10.16.1631. [DOI] [PubMed] [Google Scholar]
- 45.Li D, Zhu Q, Lin H, Zhou N, Qi Y. A novel PITX2 mutation in a Chinese family with Axenfeld-Rieger syndrome. Mol Vis. 2008;14:2205–2210. [PMC free article] [PubMed] [Google Scholar]
- 46.Perveen R, Lloyd IC, Clayton-Smith J, Churchill A, van Heyningen V, Hanson I, Taylor D, McKeown C, Super M, Kerr B, Winter R, Black GC. Phenotypic variability and asymmetry of Rieger syndrome associated with PITX2 mutations. Invest Ophthalmol Vis Sci. 2000;41:2456–2460. [PubMed] [Google Scholar]
- 47.Idrees F, Bloch-Zupan A, Free SL, Vaideanu D, Thompson PJ, Ashley P, Brice G, Rutland P, Bitner-Glindzicz M, Khaw PT, Fraser S, Sisodiya SM, Sowden JC. A novel homeobox mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome associated with brain, ocular, and dental phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:184–191. doi: 10.1002/ajmg.b.30237. [DOI] [PubMed] [Google Scholar]
- 48.Footz T, Idrees F, Acharya M, Kozlowski K, Walter MA. Analysis of mutations of the PITX2 transcription factor found in patients with Axenfeld-Rieger syndrome. Invest Ophthalmol Vis Sci. 2009;50:2599–2606. doi: 10.1167/iovs.08-3251. [DOI] [PubMed] [Google Scholar]
- 49.Vieira V, David G, Roche O, de la Houssaye G, Boutboul S, Arbogast L, Kobetz A, Orssaud C, Camand O, Schorderet DF, Munier F, Rossi A, Delezoide AL, Marsac C, Ricquier D, Dufier JL, Menasche M, Abitbol M. Identification of four new PITX2 gene mutations in patients with Axenfeld-Rieger syndrome. Mol Vis. 2006;12:1448–1460. [PubMed] [Google Scholar]
- 50.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 52.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 54.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia B, Tsui V, Case DA, Dyson HJ, Wright PE. Comparison of protein solution structures refined by molecular dynamics simulation in vacuum, with a generalized Born model, and with explicit water. J Biomol NMR. 2002;22:317–331. doi: 10.1023/a:1014929925008. [DOI] [PubMed] [Google Scholar]
- 56.Smith JA, Gomez-Paloma L, Case DA, Chazin WJ. Molecular dynamics docking driven by NMR-derived restraints to determine the structure of the calicheamicin ®1I oligosaccharide domain complexed to duplex DNA. Magn Reson Chem. 1996;34:S147–S155. [Google Scholar]
- 57.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hass MA, Hansen DF, Christensen HE, Led JJ, Kay LE. Characterization of conformational exchange of a histidine side chain: protonation, rotamerization, and tautomerization of His61 in plastocyanin from Anabaena variabilis. J Am Chem Soc. 2008;130:8460–8470. doi: 10.1021/ja801330h. [DOI] [PubMed] [Google Scholar]
- 59.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 60.Skelton NJ, Palmer AG, Akke M, Kordel J, Rance M, Chazin WJ. Practical Aspects of 2-Dimensional Proton-Detected N-15 Spin Relaxation Measurements. J Magn Reson Ser B. 1993;102:253–264. [Google Scholar]
- 61.Wang AC, Bax A. Minimizing the Effects of Radiofrequency Heating in Multidimensional Nmr Experiments. J Biomol NMR. 1993;3:715–720. doi: 10.1007/BF00198374. [DOI] [PubMed] [Google Scholar]
- 62.Yip GNB, Zuiderweg ERP. Improvement of duty-cycle heating compensation in NMR spin relaxation experiments. J Magn Reson. 2005;176:171–178. doi: 10.1016/j.jmr.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 63.Mulder FA, van Tilborg PJ, Kaptein R, Boelens R. Microsecond time scale dynamics in the RXR DNA-binding domain from a combination of spin-echo and off-resonance rotating frame relaxation measurements. J Biomol NMR. 1999;13:275–288. doi: 10.1023/a:1008354232281. [DOI] [PubMed] [Google Scholar]
- 64.Palmer AG, Rance M, Wright PE. Intramolecular Motions of a Zinc Finger DNA-Binding Domain from Xfin Characterized by Proton-Detected Natural Abundance C-12 Heteronuclear Nmr-Spectroscopy. J Am Chem Soc. 1991;113:4371–4380. [Google Scholar]
- 65.Ferrage F, Cowburn D, Ghose R. Accurate sampling of high-frequency motions in proteins by steady-state (15)N-{(1)H} nuclear Overhauser effect measurements in the presence of cross-correlated relaxation. J Am Chem Soc. 2009;131:6048–6049. doi: 10.1021/ja809526q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vugmeyster L, Trott O, McKnight CJ, Raleigh DP, Palmer AG., 3rd Temperature-dependent dynamics of the villin headpiece helical subdomain, an unusually small thermostable protein. Journal of molecular biology. 2002;320:841–854. doi: 10.1016/S0022-2836(02)00537-5. [DOI] [PubMed] [Google Scholar]
- 67.Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campanini B, Bologna S, Cannone F, Chirico G, Mozzarelli A, Bettati S. Unfolding of Green Fluorescent Protein mut2 in wet nanoporous silica gels. Protein Sci. 2005;14:1125–1133. doi: 10.1110/ps.041190805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang Y, Guigon CJ, Fan J, Cheng SY, Zhu GZ. Pituitary homeobox 2 (PITX2) promotes thyroid carcinogenesis by activation of cyclin D2. Cell Cycle. 2010;9:1333–1341. doi: 10.4161/cc.9.7.11126. [DOI] [PubMed] [Google Scholar]
- 71.Huang Y, Huang K, Boskovic G, Dementieva Y, Denvir J, Primerano DA, Zhu GZ. Proteomic and genomic analysis of PITX2 interacting and regulating networks. FEBS Lett. 2009;583:638–642. doi: 10.1016/j.febslet.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules. 1. Theory and Range of Validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 73.Baryshnikova OK, Sykes BD. Backbone dynamics of SDF-1alpha determined by NMR: interpretation in the presence of monomer-dimer equilibrium. Protein Sci. 2006;15:2568–2578. doi: 10.1110/ps.062255806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pfuhl M, Chen HA, Kristensen SM, Driscoll PC. NMR exchange broadening arising from specific low affinity protein self-association: analysis of nitrogen-15 nuclear relaxation for rat CD2 domain 1. J Biomol NMR. 1999;14:307–320. doi: 10.1023/a:1008319917267. [DOI] [PubMed] [Google Scholar]
- 75.Gumral D, Nadalin L, Corazza A, Fogolari F, Damante G, Viglino P, Esposito G. Helix mobility and recognition function of the rat thyroid transcription factor 1 homeodomain - hints from 15N-NMR relaxation studies. Febs J. 2008;275:435–448. doi: 10.1111/j.1742-4658.2007.06208.x. [DOI] [PubMed] [Google Scholar]
- 76.Fausti S, Weiler S, Cuniberti C, Hwang KJ, No KT, Gruschus JM, Perico A, Nirenberg M, Ferretti JA. Backbone dynamics for the wild type and a double H52R/T56W mutant of the vnd/NK-2 homeodomain from Drosophila melanogaster. Biochemistry. 2001;40:12004–12012. doi: 10.1021/bi010398r. [DOI] [PubMed] [Google Scholar]
- 77.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 78.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J Am Chem Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 79.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci U S A. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vuzman D, Azia A, Levy Y. Searching DNA via a “Monkey Bar” mechanism: the significance of disordered tails. Journal of molecular biology. 2010;396:674–684. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 81.Subramaniam V, Jovin TM, Rivera-Pomar RV. Aromatic amino acids are critical for stability of the bicoid homeodomain. J Biol Chem. 2001;276:21506–21511. doi: 10.1074/jbc.M102292200. [DOI] [PubMed] [Google Scholar]
- 82.Damante G, Tell G, Leonardi A, Fogolari F, Bortolotti N, Di Lauro R, Formisano S. Analysis of the conformation and stability of rat TTF-1 homeodomain by circular dichroism. FEBS Lett. 1994;354:293–296. doi: 10.1016/0014-5793(94)01145-1. [DOI] [PubMed] [Google Scholar]
- 83.Otting G, Qian YQ, Muller M, Affolter M, Gehring W, Wuthrich K. Secondary structure determination for the Antennapedia homeodomain by nuclear magnetic resonance and evidence for a helix-turn-helix motif. Embo J. 1988;7:4305–4309. doi: 10.1002/j.1460-2075.1988.tb03329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dragan AI, Li Z, Makeyeva EN, Milgotina EI, Liu Y, Crane-Robinson C, Privalov PL. Forces driving the binding of homeodomains to DNA. Biochemistry. 2006;45:141–151. doi: 10.1021/bi051705m. [DOI] [PubMed] [Google Scholar]
- 85.Fodor E, Mack JW, Maeng JS, Ju JH, Lee HS, Gruschus JM, Ferretti JA, Ginsburg A. Cardiac-specific Nkx2.5 homeodomain: conformational stability and specific DNA binding of Nkx2.5(C56S) Biochemistry. 2005;44:12480–12490. doi: 10.1021/bi050835s. [DOI] [PubMed] [Google Scholar]
- 86.Gonzalez M, Weiler S, Ferretti JA, Ginsburg A. The vnd/NK-2 homeodomain: thermodynamics of reversible unfolding and DNA binding for wild-type and with residue replacements H52R and H52R/T56W in helix III. Biochemistry. 2001;40:4923–4931. doi: 10.1021/bi0022341. [DOI] [PubMed] [Google Scholar]
- 87.Seksek O, Bolard J. Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry. J Cell Sci. 1996;109(Pt 1):257–262. doi: 10.1242/jcs.109.1.257. [DOI] [PubMed] [Google Scholar]
- 88.Markley JL. Observation of Histidine Residues in Proteins by Means of Nuclear Magnetic-Resonance Spectroscopy. Accounts Chem Res. 1975;8:70–80. [Google Scholar]
- 89.Amendt BA. The molecular and biochemical basis of Axenfeld-Rieger syndrome. In: Amendt BA, editor. The molecular mechanisms of Axenfeld-Rieger Syndrome. Landes Bioscience; New York: 2005. pp. 32–53. [Google Scholar]
- 90.Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J Biol Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 91.Kozlowski K, Walter MA. Variation in residual PITX2 activity underlies the phenotypic spectrum of anterior segment developmental disorders. Hum Mol Genet. 2000;9:2131–2139. doi: 10.1093/hmg/9.14.2131. [DOI] [PubMed] [Google Scholar]
- 92.Acharya M, Lingenfelter DJ, Huang L, Gage PJ, Walter MA. Human PRKC apoptosis WT1 regulator is a novel PITX2-interacting protein that regulates PITX2 transcriptional activity in ocular cells. J Biol Chem. 2009;284:34829–34838. doi: 10.1074/jbc.M109.006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan SK, Mann RS. The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 94.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grunberg R, Nilges M, Leckner J. Flexibility and conformational entropy in protein-protein binding. Structure. 2006;14:683–693. doi: 10.1016/j.str.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 96.Doerdelmann T, Kojetin DJ, Baird-Titus JM, Rance M. 1H, 13C and 15N chemical shift assignments for the human Pitx2 homeodomain and a R24H homeodomain mutant. Biomol NMR Assign. 2011;5:105–107. doi: 10.1007/s12104-010-9278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.