Abstract
Biliary atresia is a pediatric liver disease of unknown underlying etiology, in which fibro-inflammatory destruction of the extrahepatic biliary system leads to obstructive cholestasis. MicroRNAs are a class of short (18–23 nucleotide), non-coding RNA molecules which act as negative regulators of target mRNA stability and translation. The importance of these molecules in normal and diseased liver has been demonstrated, but their potential role in the pathogenesis of biliary atresia has not been addressed. We have profiled changes in liver microRNA levels in an established mouse model of the disease, identified significantly altered transcripts, and defined the spatial expression patterns of selected microRNAs. Two of these, miR-29a/29b1 are up-regulated in experimental biliary atresia. Using antisense oligonucleotide-mediated inhibition in mice, we have delineated the full set of hepatic genes regulated by miR-29 and identified two mRNA targets of potential pathological relevance in experimental biliary atresia, Igf1 and Il1RAP. We have used reporter assays to confirm that Igf1 and Il1RAP are direct targets of miR-29.
Introduction
Biliary atresia (BA) is a fibro-inflammatory liver disease of infants in which a primary insult of unknown etiology leads to progressive T-cell mediated destruction of the extrahepatic biliary system (1–4). Loss of the large bile ducts that drain the liver results in severe, life-threatening cholestasis, and patients present with hyperbilirubinemia and acholic stools, typically by eight weeks of age. The only intervention currently available to restore bile flow from the native liver is the Kasai portoenterostomy, which is successful in less than 80% of cases. Of the patients with a successful short-term outcome from the Kasai procedure, 50% will ultimately require liver transplant for chronic complications. BA is the leading indicator for pediatric liver transplantation worldwide, accounting for 40–50% of pediatric liver transplants (5).
The Rhesus rotavirus (RRV)-BALB/c model of biliary atresia is an excellent model of the clinical disease (6). As in the human disease, there is a defined window of incidence restricted to the early neonatal period; mice infected with RRV within the first day of life progress rapidly from occlusion and destruction of the extrahepatic ducts, to cholestasis and death in approximately two weeks. As in the clinical disease, pathogenesis is T-cell mediated: interferon gamma (Ifng) mutant mice, which do not mount a Th1 response, are refractory to the RRV model, and conversely, inflammation of the extrahepatic ducts can be induced in naïve mice by transplantation of T cells isolated from RRV-infected mice (7, 8). Previous studies of gene expression in both clinical and experimental BA have demonstrated that the parallels between the mouse model and the human disease extend to the molecular level, and have been informative in guiding experimental design aimed at uncovering potential therapeutic interventions (9, 10).
Here we explore the potential pathogenic role of the small, non-coding, regulatory RNA molecules known as microRNAs (miRNAs), using the RRV-BALB/c model. We identify specific, significant alterations in the liver miRNA transcriptome during the progression of the disease model. Our results reveal dynamic changes in both spatial and temporal miRNA abundance, some of which correlate with alterations in cell population due to infiltration, while others exhibit widespread expression changes throughout the lobule, reflecting altered cellular states. Among the latter group we have focused on miR-29a (co-transcribed with the related miRNA, miR-29b1), which is significantly up-regulated in both infiltrating and parenchymal cells following RRV infection. We present the first in vivo identification of mRNA targets of miR-29 regulation, and directly link the overexpression of miR-29 to the down-regulation of two mRNA targets of potential relevance to BA pathogenesis.
Materials and Methods
RRV Model of biliary atresia
The animals used in this study were humanely housed, in accordance with state and federal guidelines under the supervision of the Institutional Animal Care and Use Committee at the Children’s Hospital of Philadelphia. Mice were fed a standard rodent chow diet and water ad libitum. Neonatal BALB/c pups were injected intraperitoneally either with 1 × 106 fluorescence-forming units of RRV, or with saline, within the first day of life as described previously (9).
miRNA expression profiling
For the miRNA microarray experiment, RRV and saline injected animals were euthanized at 3, 8, and 14 days post infection (n=5 per treatment, per time point), and total RNA was purified from liver using the mirVana™ miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. A pooled common control was assembled from an equal RNA mass from each of the samples, and samples and control were separately dye-labeled, and hybridized to a miRcury v9.0 miRNA microarray (Exiqon) at the University of Pennsylvania School of Medicine Microarray core facility. Sample hybridization intensities were scored relative to the common control, and raw intensity data were normalized and analyzed using the SAM add-in (11) for Microsoft Excel. MiRNAs exhibiting a fold-change of greater than 10% up or down, at a false discovery rate of 5% were chosen for further study. The microarray data have been deposited at the NCBI GEO repository under the accession number GSE33418.
To validate the results of the microarray we performed real time quantitative PCR Taqman assays (Applied Biosystems) for candidate miRNAs.
Characterization of miRNA spatial expression
The spatial expression of miRNAs was characterized by in situ hybridization on frozen liver sections using 3′-, or 5′- and 3′-double digoxygenin-labeled antisense LNA probes (Exiqon) (12). To ensure detection of miRNA with low expression a tyramide signal amplification step was incorporated.
Bioinformatic analysis
Using the Partek software suite, we intersected previously published RRV model gene expression data (9, 10), with our RRV miRNA microarray data, and used the predictions from the Targetscan 5.1 algorithm to generate candidate miRNA-target pairs.
Gene expression microarray analysis of miR-29 mRNA targets
Adult BALB/c female mice were injected intraperitoneally with a single dose at 20 mg per kg of antisense oligonucleotide either against miR-29a (5′-TAACCGATTTCAGATGGTGCTA-3′) or against a scrambled sequence (5′-TCATTGGCATGTACCATGCAGCT-3′ Antisense oligonucleotides contained 2′-O-methoxyethyl (2′-MOE), 2′-flouro (2′-F) 2′-alpha-flouro units with a phosphorothioate backbone (Regulus Therapeutics). Six days following the injection, liver was isolated, total RNA was prepared as described above, and the RNA was amplified and biotinylated using the MessageAmp Premier kit (Ambion). Samples (n=4 each experimental and control) were hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays in the Children’s Hospital of Philadelphia Nucleic Acids Core Facility and analyzed with the assistance of the Penn Bioinformatics Core. Probe intensities were normalized using the GCRMA method (13) and the significance of the log2-transformed, GCRMA-normalized signal intensities was determined using SAM (11). The microarray data have been deposited at the NCBI GEO repository under accession number GPL14829. Gene Set Enrichment Analysis was performed using the DAVID package (14–16).
Identification of direct targets of miR-29a
DNA from the 3′ UTR of mouse Igf1 and Il1RAP was amplified by nested PCR from C57B/6 genomic DNA, and cloned into pMiRCheck2, a modified derivative of pSiCheck2 (Promega) in which the SV40 promoter/enhancer driving Renilla luciferase has been replaced by the weaker PGK promoter from pL451(17). Sequences of the oligonucleotide primers used, and pMiRCheck2 are listed in online-only Supplemental Table 4 (available at http://links.lww.com/MPG/A84). In the case of miR-29a overexpression 3×104 NIH3T3 cells were seeded into 24-well plates with 900ng of expression plasmid and 100ng of dual luciferase reporter plasmid per well, using 3ul of FugeneHD (Roche). For the antisense oligonucleotide assays, 2ul of Lipofectamine 2000 (Invitrogen) was used to co-transfect 200ng of reporter plasmid with antisense oligonucleotide (Regulus Therapeutics, as described above) at 20nM in 24-well format using the same seeding density. After 16 hrs the cells were rinsed once with 1x PBS, and media was replaced with fresh media. After 24hrs of additional outgrowth the cells were rinsed once with 1x PBS, lysed in 150ul of 1x Passive Lysis Buffer (Promega), and firefly and Renilla luciferase activities were measured from a 10ul aliquot, on a GloMax Multi luminometer (Promega) using Stop and Glo reagents (Promega), according the manufacturer’s instructions.
Relative light units were calculated as the ratio of Renilla to firefly luciferase activity, and the reporters were normalized between the control expression plasmids or ASOs to correct for non-specific effects of the differences between the experimental UTRs, and the empty pMiRCheck2. Values from the empty pMirCheck2 samples for a given control treatment were used to correct for non-specific effects of the treatment on the normalizer.
Results
MiRNAs are differentially expressed with time and treatment in experimental BA
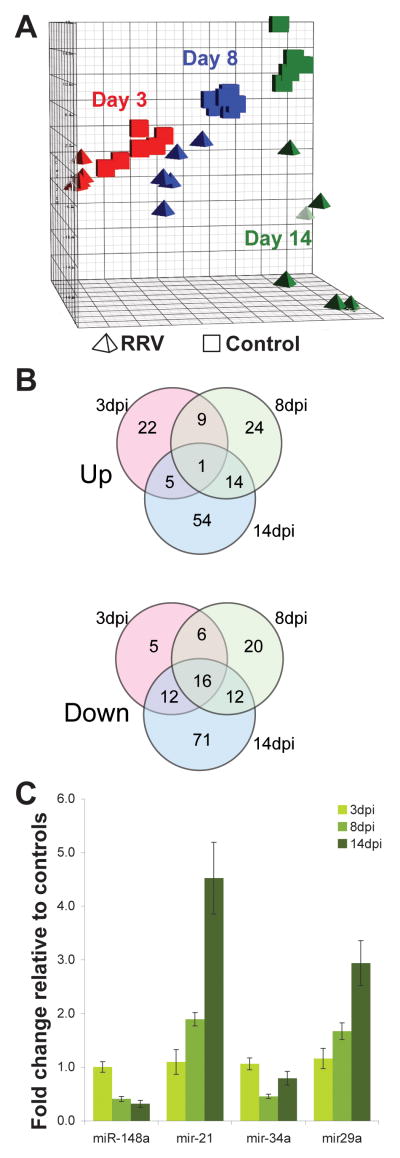
We isolated RNA from livers of RRV or saline injected BALB/c neonatal mouse pups at 3, 8, and 14 days post injection (dpi), and profiled the miRNA transcriptome by hybridization microarray. Principal component analysis illustrates that at all three time points the RRV-infected samples are clearly separated from their control counterparts, and become not only more distinct from the control group but also more divergent within the infected group with time, reflecting the variable nature of disease progression (Figure 1A). In contrast, the control samples cluster together at all three time points.
Figure 1.
MicroRNA microarray summary and validation. (A) Principal component analysis of miRNA microarray data. Samples from saline-injected mice, cubes; RRV-injected, tetrahedra. Red, blue, and green represent 3dpi, 8dpi and 14dpi time points respectively. (B) Number of significantly increased and decreased miRNAs at each time point. See Supplemental Table 1 for listing of miRNAs in Fig 1B. (C) Confirmatory qPCR of selected miRNAs at the indicated time points.
We used statistical analysis of microarrays to identify miRNAs whose levels changed significantly with treatment, time, or both (Figure 1B and online-only Supplemental Table 1, available at http://links.lww.com/MPG/A81). Because more abundant transcripts in parenchymal cells may mask changes in miRNA levels in subpopulations of cells within the liver, we choose inclusive criteria for further study: miRNA whose abundance changed by +/−10% relative to the corresponding controls, with a p-value of <0.05 after correction for multiple testing. We validated the results of the hybridization microarray by confirming changes in the abundance of selected miRNAs using individual Taqman assays (Figure 1C).
Spatial changes in miRNA expression in experimental BA
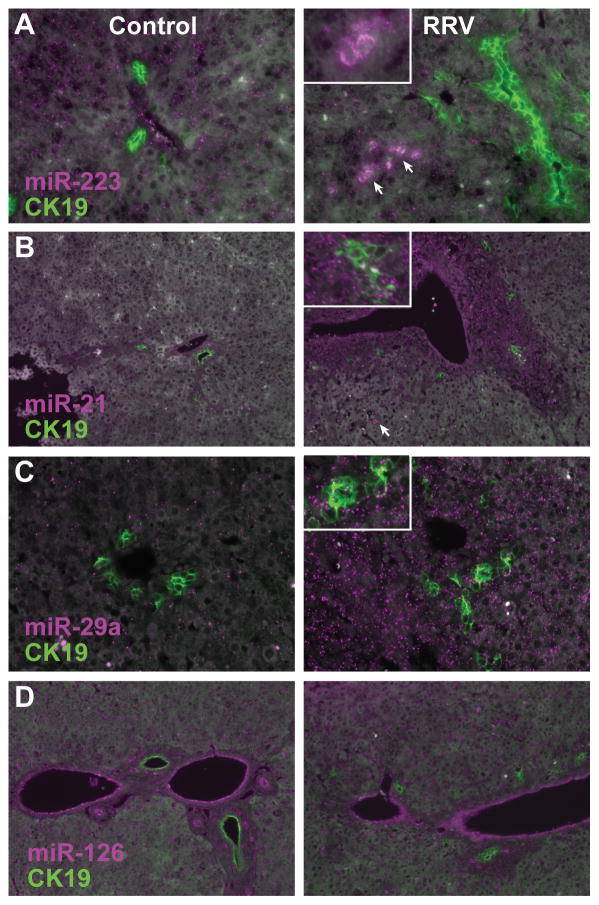
Increased levels of miRNAs in infected livers may reflect induction of higher transcription in response to infection or cholestasis, a shift in cell population due to increased numbers of infiltrating cells, or a combination of both factors. To explore the source of the miRNAs most significantly increased in RRV samples, we performed in situ hybridization to determine their expression patterns in infected and control liver sections throughout infection. Consistent with previously published work (18), we observed strong miR-223 expression in infiltrating mononuclear cells (Figure 2A, arrows and inset). These are likely to be granulocytes, in light of their morphology and the specific expression of miR-223 in this cell type (12, 19). In contrast, miR-21 and miR-29a levels are elevated both hepatocytes and cholangiocytes, with miR-29 also present in rare infiltrating mononuclear cells (Figure 2B–C). The pattern of increased expression is not uniform throughout sections, with a more apparent increase surrounding portal tracts in the case of both miR-21 and miR-29a, suggesting an underlying local effect that may originate with either the periportal mesenchyme or the inflammatory infiltrate.
Figure 2.
In situ hybridization of (A) miR-223, (B) miR-21, (C) miR-29a, and (D) miR-126 (magenta), with simultaneous immunofluorescent detection of CK19 to detect cholangiocytes (green). In (A), the inset shows a higher magnification view of infiltrating monocytes (arrows). In (B) and (C), the insets include higher magnification views of bile ductules. Liver sections from the 8 days post saline (control) or RRV injection were hybridized with locked nucleic acid miRNA probes and a rabbit polyclonal CK19 antiserum.
Endothelial miRNA levels decrease in response to RRV
Hybridization array data indicated that miR-126 is present at significantly lower levels at all three time points in RRV infected animals (Supplemental Table 1). In situ hybridization indicates that miR-126 is strongly expressed in vascular and sinusoidal endothelial cells relative to the rest of the tissue in control animals, but that the levels in infected animals are decreased (Figure 2D).
Inhibition of miR-29a in vivo identifies liver-expressed mRNA targets
Based on our array data, two miRNAs from a single transcript, miR-29b1 and miR-29a, were predicted to have liver expression that was both highly abundant and significantly induced in RRV-infected animals at 8 and 14 dpi. As miR-29 has previously been implicated both in fibrosis and liver disease (20–23) we sought to delineate the full set of hepatic miR-29 target genes by treating adult BALB/c mice with antisense oligonucleotides against either miR-29a or a control scrambled sequence. We measured body and liver masses six days after injection, and performed serological analysis for a panel of liver markers (ALT, AST, GGTP, direct bilirubin, albumin, cholesterol) and measured fasting blood glucose. The ASO29a mice had significant decreases in both liver mass (control: 0.94±0.02g; anti-29a: 0.83±0.03g, p<0.02) and serum cholesterol (control: 53.8 ±1.5 mg/dl, anti-29a: 43.8 ±2.4 mg/dl, p<0.02) relative to animals injected with control ASO. There were no significant differences between the two groups in any of the other metrics.
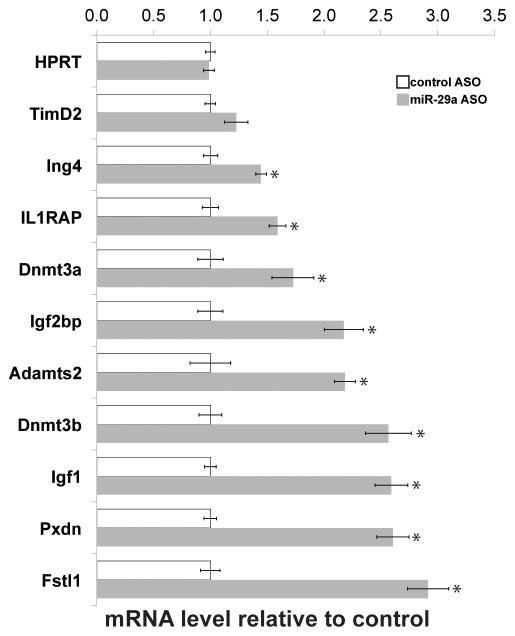
We isolated total liver RNA from antisense-injected animals and performed gene expression microarray analysis. Using Significance Analysis of Microarrays (24), we identified 104 transcripts up-regulated by 1.5-fold or greater, and 70 similarly down-regulated transcripts (at a false discovery of <10%) (online-only Supplemental Table 2, available at http://links.lww.com/MPG/A82). We validated the microarray data by quantitative PCR (Figure 3). Dnmt3a and Dnmt3b have previously been shown to be targets of miR-29a (25); the up-regulation of both these genes (Figure 3) confirms that the ASO-based approach was able to repress miR-29a targets. Consistent with recently published results implicating miR-29 in the regulation of fibrosis (20–22), there is an over-representation of collagen genes among the up-regulated transcripts. Pathways analysis using the DAVID functional classification tool (14–16) indicates significant enrichment of collagen genes (33.7-fold enrichment, p<9.6×10−12, and extracellular matrix (integrin) signaling pathway members (20.6-fold enrichment, p<4.4×10−17), including laminin, elastin, fibrillin, matrix metalloproteases, Sparc, and ADAM family genes. Pathways analysis on the down-regulated transcripts indicates over-representation of xenobiotic and P-450 pathway components, largely due to effects on multiple transcripts of glutathione-S-transferase family members.
Figure 3.
In vivo inhibition of miR-29a: gene expression microarray validation. Relative transcript abundance, normalized to 28S rRNA transcript levels, of genes deregulated in response to miR-29a antisense injection, assayed by RT-qPCR of liver RNA (n=4 per group). *p<0.05.
Gene Set Enrichment Analysis (GSEA) indicates significant enrichment of the predicted miR-29a/b/c gene set (normalized enrichment score [NES] 1.54; p-value <0.001; FDR, 0.35), and focal adhesion signaling (NES, 1.45; p-value <0.001; FDR, 0.40) gene sets. In addition, gene sets representing Myc- and p53-targets were enriched (Myc: NES, 1.52; p-value <0.001; FDR, 0.34; p53: NES, 1.59; p-value <0.001; FDR, 0.41). Both Myc and p53 regulate miR-29 (26, 27); these results suggest that there is a feedback relationship in which miR-29 represses p53 and Myc-related pathways. The GSEA results are summarized in online-only Supplemental Table 3 (available at http://links.lww.com/MPG/A83).
We examined the list of deregulated genes to identify candidates of potential importance in BA pathogenesis. We focused on two genes: Igf1, previously shown to be important for cholangiocyte survival, and Il1RAP, a modulator of IL1 signaling in the liver (28, 29), both of which contained putative miR-29 binding sites in their 3′ UTRs. We first confirmed that both genes are up-regulated in experimental biliary atresia (Figure 3).
Igf1 and Il1RAP are direct targets of miR-29
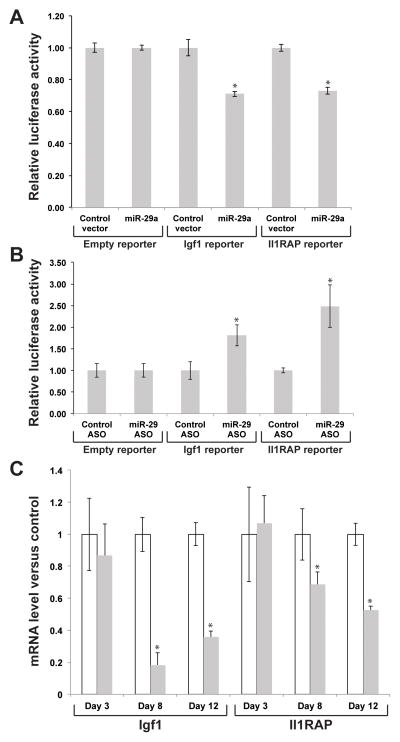
To test whether Igf1 and Il1RAP are direct targets of miR-29 regulation, we cloned the 3′ UTRs of each gene into a reporter plasmid (pMirCheck2) and performed dual luciferase assays in NIH3T3 cells, while over-expressing miR-29a (Figure 4A). When miR-29a was overexpressed, the reporters containing the Igf1 and Il1RAP 3′ UTRs were significantly down-regulated relative to the empty reporter (Igf1, 1.41-fold, p<0.002; Il1RAP, 1.36-fold, p<0.0002). Conversely, when the reporters were co-transfected with an ASO directed against miR-29a, the normalized Renilla luciferase activity was increased relative to control vector (Igf1, 1.81-fold, p<0.04; Il1RAP, 2.49-fold, p<0.02) (Figure 4B). Taken together, these data strongly suggest direct regulation of both genes by the miR-29 family.
Figure 4.
Dual luciferase reporter assays: Igf1 and Il1RAP are direct targets of miR-29a. Igf1 and Il1RAP expression in RRV versus saline. (A) Luciferase reporter assays to detect repression of Igf1 and Il1RAP 3′ UTR sequences containing candidate miR-29a target sites by exogenous miR-29a. Results are expressed relative to a control reporter without additional UTR sequences. (B) Luciferase reporter assays to detect de-repression of Igf1 and Il1RAP 3′ UTR sequences containing candidate miR-29a target sites by a locked nucleic acid antisense oligonucleotide-mediated miR-29a inhibition. Results are expressed relative to a control oligonucleotide. (C) Relative abundance of Igf1 and Il1RAP in RRV infected mice, relative to saline controls, assayed by RT-qPCR on liver RNA at the indicated time points (n=4 per group). *p<0.05.
We have measured the expression of Igf1 and Il1RAP in experimental BA, and found that Igf1 expression is significantly decreased at 8 and 12 dpi (5.4 and 2.8-fold respectively), and Il1RAP is significantly decreased 1.9-fold at 12 dpi (Figure 4C). These results indicate that the changes in miR-29 expression observed in the BA model are likely to be reflected in multiple downstream pathways.
Discussion
We have described for the first time the changes in the hepatic miRNA transcriptome in the experimental model of biliary atresia. Changes in the abundance of miRNAs early in disease progression reflect expected changes in the liver following viral infection: miRNAs in clusters known to be associated with inflammation, cancer, cell proliferation, and apoptosis (28–32) (miR-15a, miR-106a, miR-17, miR-93), and monocyte miRNAs (33) (miR-223, miR-142-3p, which are abundant in granulocytes and T cells respectively) are increased in abundance. In contrast, known epithelial cell miRNAs (34, 35) (miR-192, -194 and -215), and previously described liver-expressed miRNAs (12, 36–38) (miR-30a, miR-30b, miR-29a) are decreased in relative abundance. A similar pattern persists in later time-points with increased abundance of miRNAs associated with cell proliferation (39, 40) (miR-21), and immunity (31) (miR-16, miR-21, miR-142-5p, miR-15b) relative to healthy controls, and decreased levels of epithelial cell miRNAs. The expression of miR-21, which is strongly induced, likely reflects its importance in cellular growth, and its known expression in both immune cells and cholangiocytes (39, 41). We have found decreased levels in miR-126, a known endothelial miRNA (42, 43). Although endothelial cells are not typically viewed as central to BA pathology, previous studies have identified gene expression changes in endothelial cells in BA clinical samples. Decreased miRNA levels might result from loss of miRNA due to cell death, to skewing of the normalized miRNA levels due to the shift in the cellular population, to the recently described phenomenon of stress-related export of miRNAs (44), or to a combination of these processes.
In addition to these changes, we observed increases of members of the miR-29 family, miR-29a and miR-29b1. Of the two, miR-29a is both more abundant and more strongly induced, and since the miRNAs are co-transcribed and are likely to have almost complete overlap of target genes, we have used miR-29a as a surrogate for studying both.
Using in situ hybridization, we have localized the expression of miR-29a in control and infected liver sections and shown that, while the miRNA is widely expressed throughout the lobule, in infected livers there is both an overall increase in the level of the miRNA and a greater increase in periportal region. Coincident with the increase in miR-29 levels is a drop in the levels of a known miR-29 direct target, Dnmt3a. The reciprocal relationship between miR-29 and methyl transferase gene expression in experimental BA is remarkable given the recent observation that DNA hypomethylation leads to bile defects in a zebrafish model, and correlates with clinical BA (45). To explore the potential role of miR-29 in BA pathogenesis, we first delineated the hepatic targets of miR-29 in vivo by antisense oligonucleotide mediated inhibition of miR-29a in healthy adult mice. Using the list thus obtained, we have identified two direct targets of miR-29, Igf1 and Il1RAP, with roles in cholangiocyte survival and the modulation of inflammation, respectively. This represents the first genome-wide detection of miR-29a targets in vivo.
Consistent with previous in silico predictions and reporter assays, the genes whose expression is increased when the repressive effect of miR-29 is blocked included multiple collagen and extracellular matrix genes, as well as the DNA methyl transferase genes Dnmt3a and Dnmt3b (22, 25, 46–48). Although miR-29a has been studied in stellate cells, our results indicate that miR-29a is active in hepatocytes; the functions of miR-29a in stellate cells versus hepatocytes will require selective inhibition of the miRNA in a single cell type. Overall, these results provide the first in vivo support for miR-29a function in hepatic fibrosis.
Among the genes up-regulated by ASO-inhibition of miR-29, we have focused on two, Igf1, and Il1RAP, and used reporter assays to demonstrate that they are directly regulated by miR-29. High levels of Igf1 have previously been shown to be associated with cholangiocyte survival in clinical primary biliary cirrhosis samples (49). In experimental BA, increased expression of miR-29, which down-regulates Igf1, would be predicted to increase the likelihood of cholangiocyte cell death, and may contribute to the cholangiopathy in children with BA (50).
Il1RAP is involved in IL-1 signaling through its receptor, IL-1RA, and is believed to promote IL-1 signaling (an alternatively spliced gene product, sIl1RAP, acts as a secreted modulatory sink for IL-1) (51, 52). The regulation of Il1RAP expression by sites within its 3′UTR has previously been described (53). Here we link this regulation in a disease model to over-expression of a specific miRNA, miR-29. In this context, miR-29 over-expression may be an adaptive regulatory mechanism to temper the inflammatory response by down-regulating signaling through the IL-1 receptor.
One limitation of our approach to detecting miR-29 targets was the use of adult mice for the in vivo miR-29 inhibition, because there may be target genes expressed specifically in juvenile animals. Furthermore, confirmation of a functional role for miR-29a in BA will require successful inhibition of the miRNA in the context of the mouse model. To address these limitations, we performed antisense IP injections in RRV-infected and control pups. We utilized a range of doses, dosing schedules, and oligonucleotide chemistries (cholesterol tagged locked nucleic acid, 2-MOE, or 2-MOE-alpha-fluoro; all with phosphothiorylated backbones). Despite this, we were unsuccessful in inhibiting miR-29, as measured by up-regulation of Dnmt3a/b or any other miR-29a target gene identified in the adult mice. This precluded any functional analysis of miR-29a in the RRV model by antisense inhibition. The failure of the ASO to function in the mouse pups may be due to a variety of factors, including variable bioavailability via the intraperitoneal route, decreased liver absorption of the ASO, or rapid growth of liver cells at this age. To circumvent this experimental challenge, we are developing genetic tools to conditionally overexpress or inhibit selected miRNAs in a manner that does not depend on oligonucleotide delivery. Use of this approach will enable us to investigate the role of miR-29 in liver development, growth, function, and disease models, including biliary atresia. Finally, in collaboration with the Childhood Liver Disease Research and Education Network (ChiLDREN; childrennetwork.org), we will test our findings in BA and other cholestatic diseases of infancy using clinical specimens.
Supplementary Material
Acknowledgments
Grant support: NIH R01DK079881 (JRF), the University of Pennsylvania School of Medicine Center for Digestive and Liver Diseases (NIH 5P30DK050306), the American Liver Foundation, and the Fred and Suzanne Biesecker Pediatric Liver Center.
The authors thank all members of the Friedman lab, the Division of Gastroenterology and Nutrition at CHOP, and the Fred and Suzanne Biesecker Pediatric Liver Center for support. We thank H. Fred Clark for assistance with the Rhesus rotavirus.
Footnotes
Financial disclosures: No conflicts of interest exist.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
References
- 1.Bezerra JA. Potential etiologies of biliary atresia. Pediatr Transplant. 2005;9:646–651. doi: 10.1111/j.1399-3046.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 2.Hartley JL, Davenport M, Kelly DA. Biliary atresia. Lancet. 2009;374:1704–1713. doi: 10.1016/S0140-6736(09)60946-6. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber RA, Kleinman RE. Biliary atresia. J Pediatr Gastroenterol Nutr. 2002;35(Suppl 1):S11–16. doi: 10.1097/00005176-200207001-00005. [DOI] [PubMed] [Google Scholar]
- 4.Mieli-Vergani G, Vergani D. Biliary atresia. Semin Immunopathol. 2009;31:371–381. doi: 10.1007/s00281-009-0171-6. [DOI] [PubMed] [Google Scholar]
- 5.Sokol RJ, Shepherd RW, Superina R, et al. Screening and outcomes in biliary atresia: summary of a National Institutes of Health workshop. Hepatology. 2007;46:566–581. doi: 10.1002/hep.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riepenhoff-Talty M, Schaekel K, Clark HF, et al. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr Res. 1993;33:394–399. doi: 10.1203/00006450-199304000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Shivakumar P, Campbell KM, Sabla GE, et al. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest. 2004;114:322–329. doi: 10.1172/JCI21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shivakumar P, Sabla G, Mohanty S, et al. Effector role of neonatal hepatic CD8+ lymphocytes in epithelial injury and autoimmunity in experimental biliary atresia. Gastroenterology. 2007;133:268–277. doi: 10.1053/j.gastro.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho E, Liu C, Shivakumar P, et al. Analysis of the biliary transcriptome in experimental biliary atresia. Gastroenterology. 2005;129:713–717. doi: 10.1016/j.gastro.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 10.Leonhardt J, Stanulla M, von Wasielewski R, et al. Gene expression profile of the infective murine model for biliary atresia. Pediatr Surg Int. 2006;22:84–89. doi: 10.1007/s00383-005-1589-0. [DOI] [PubMed] [Google Scholar]
- 11.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hand NJ, Master ZR, Eauclaire SF, et al. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, Irizarry R, Gentleman R, et al. A Model Based Background Adjustment for Oligonucleotide Expression Arrays. Johns Hopkins University, Dept. of Biostatistics Working Papers; 2004. Working Paper 1. [Google Scholar]
- 14.Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008;33:474–481. doi: 10.1016/j.tibs.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkerova M, Vasikova A, Belickova M, et al. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem cells and development. 2010;19:17–26. doi: 10.1089/scd.2009.0071. [DOI] [PubMed] [Google Scholar]
- 19.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay S, Friedman RC, Marquez RT, et al. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J Infect Dis. 2011;203:1753–1762. doi: 10.1093/infdis/jir186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cushing L, Kuang PP, Qian J, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roderburg C, Urban GW, Bettermann K, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y, Fang JH, Yun JP, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer metastasis reviews. 2010;29:613–639. doi: 10.1007/s10555-010-9257-9. [DOI] [PubMed] [Google Scholar]
- 27.Mott JL, Kurita S, Cazanave SC, et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. Journal of cellular biochemistry. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onori P, Alvaro D, Floreani AR, et al. Activation of the IGF1 system characterizes cholangiocyte survival during progression of primary biliary cirrhosis. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2007;55:327–334. doi: 10.1369/jhc.6R7125.2006. [DOI] [PubMed] [Google Scholar]
- 29.Jensen LE, Muzio M, Mantovani A, et al. IL-1 signaling cascade in liver cells and the involvement of a soluble form of the IL-1 receptor accessory protein. Journal of immunology. 2000;164:5277–5286. doi: 10.4049/jimmunol.164.10.5277. [DOI] [PubMed] [Google Scholar]
- 30.Zahm AM, Thayu M, Hand NJ, et al. Circulating microRNA is a biomarker of pediatric Crohn disease. Journal of pediatric gastroenterology and nutrition. 2011;53:26–33. doi: 10.1097/MPG.0b013e31822200cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meenhuis A, van Veelen PA, de Looper H, et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, Luna C, Qiu J, et al. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130:731–741. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 139:1654–1664. 1664, e1651. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roderburg C, Urban GW, Bettermann K, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 37.Rogler CE, Levoci L, Ader T, et al. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 38.Sekiya Y, Ogawa T, Yoshizato K, et al. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem Biophys Res Commun. doi: 10.1016/j.bbrc.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 39.Song G, Sharma AD, Roll GR, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 51:1735–1743. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lou Y, Yang X, Wang F, et al. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med. 26:819–827. doi: 10.3892/ijmm_00000530. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen S, Baker A, Moller S, et al. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Harris TA, Yamakuchi M, Ferlito M, et al. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews RP, Eauclaire SF, Mugnier M, et al. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53:905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwiecinski M, Noetel A, Elfimova N, et al. Hepatocyte Growth Factor (HGF) Inhibits Collagen I and IV Synthesis in Hepatic Stellate Cells by miRNA-29 Induction. PLoS One. 2011;6:e24568. doi: 10.1371/journal.pone.0024568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekiya Y, Ogawa T, Yoshizato K, et al. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem Biophys Res Commun. 2011;412:74–79. doi: 10.1016/j.bbrc.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 48.Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011 doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onori P, Alvaro D, Floreani AR, et al. Activation of the IGF1 system characterizes cholangiocyte survival during progression of primary biliary cirrhosis. J Histochem Cytochem. 2007;55:327–334. doi: 10.1369/jhc.6R7125.2006. [DOI] [PubMed] [Google Scholar]
- 50.Gatto M, Drudi-Metalli V, Torrice A, et al. Insulin-like growth factor-1 isoforms in rat hepatocytes and cholangiocytes and their involvement in protection against cholestatic injury. Lab Invest. 2008;88:986–994. doi: 10.1038/labinvest.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen LE, Whitehead AS. Expression of alternatively spliced interleukin-1 receptor accessory protein mRNAs is differentially regulated during inflammation and apoptosis. Cell Signal. 2003;15:793–802. doi: 10.1016/s0898-6568(03)00039-1. [DOI] [PubMed] [Google Scholar]
- 52.Jensen LE, Muzio M, Mantovani A, et al. IL-1 signaling cascade in liver cells and the involvement of a soluble form of the IL-1 receptor accessory protein. J Immunol. 2000;164:5277–5286. doi: 10.4049/jimmunol.164.10.5277. [DOI] [PubMed] [Google Scholar]
- 53.Jensen LE, Whitehead AS. The 3′ untranslated region of the membrane-bound IL-1R accessory protein mRNA confers tissue-specific destabilization. J Immunol. 2004;173:6248–6258. doi: 10.4049/jimmunol.173.10.6248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.