Although S-adenosylhomocysteine (SAH), a metabolic by-product of S-adenosylmethionine (SAM), differs from SAM only by a single methyl group and an overall positive charge, SAH binds SAM-II riboswitch with more than 1000-fold less affinity than SAM. Here, the authors used atomistic molecular dynamics simulations to show that the positively charged sulfonium moiety on SAM acts as the most crucial anchor point responsible for the formation of key ionic interactions as it fits favorably in the negatively charged binding pocket, regardless of the chirality at the sulfonium center. They show that the unfavorable loss in entropy in SAM-II binding is much greater for SAM than SAH, resulting in a fully formed pseudoknot structure only upon binding SAM, which is compensated by stabilizing electrostatic interactions with the riboswitch.
Keywords: electrostatic interaction, molecular dynamics simulation, S-adenosylhomocysteine, S-adenosylmethionine, SAM-II riboswitch
Abstract
Although S-adenosylhomocysteine (SAH), a metabolic by-product of S-adenosylmethionine (SAM), differs from SAM only by a single methyl group and an overall positive charge, SAH binds the SAM-II riboswitch with more than 1000-fold less affinity than SAM. Using atomistic molecular dynamics simulations, we investigated the molecular basis of such high selectivity in ligand recognition by SAM-II riboswitch. The biosynthesis of SAM exclusively generates the (S,S) stereoisomer, and (S,S)-SAM can spontaneously convert to the (R,S) form. We, therefore, also examined the effects of (R,S)-SAM binding to SAM-II and its potential biological function. We find that the unfavorable loss in entropy in SAM-II binding is greater for (S,S)- and (R,S)-SAM than SAH, which is compensated by stabilizing electrostatic interactions with the riboswitch. The positively charged sulfonium moiety on SAM acts as the crucial anchor point responsible for the formation of key ionic interactions as it fits favorably in the negatively charged binding pocket. In contrast, SAH, with its lone pair of electrons on the sulfur, experiences repulsion in the binding pocket of SAM-II and is enthalpically destabilized. In the presence of SAH, similar to the unbound riboswitch, the pseudoknot structure of SAM-II is not completely formed, thus exposing the Shine-Dalgarno sequence. Unlike SAM, this may further facilitate ribosomal assembly and translation initiation. Our analysis of the conformational ensemble sampled by SAM-II in the absence of ligands and when bound to SAM or SAH reveals that ligand binding follows a combination of conformational selection and induced-fit mechanisms.
INTRODUCTION
Of the various gene regulatory mechanisms that involve either initiation or termination of either transcription or translation, modification of DNA, or stability of mRNAs, the most recently discovered and perhaps the most primitive mechanism is that mediated by riboswitches (Nudler and Mironov 2004; Winkler and Breaker 2005). Riboswitches are self-regulatory elements located in the 5′-untranslated regions of certain bacterial mRNAs that have the remarkable ability to directly bind selective small cellular metabolites without requiring any intermediate protein factors (Nahvi et al. 2002; Winkler and Breaker 2003). Unlike higher eukaryotes, it is natural for prokaryotes, mainly bacteria (Barrick and Breaker 2007), in which genetic information is not compartmentalized within nuclei, to have retained the capability of controlling gene expression by unmediated sensing of small ligands. This functionality is essential for efficient modulation of protein biosynthesis as soon as the changes in the surrounding medium and intracellular concentrations of metabolites are sensed. Typically, riboswitches possess complex sequences and fold into intricate tertiary structures (Mandal and Breaker 2004). The binding of specific ligands takes place in the conserved aptamer domain that is generally known to induce conformational changes in the expression platform, which, in turn, eventually controls the expression of genes located downstream (Soukup and Soukup 2004). Most notably, riboswitches can strongly discriminate between their cognate ligand and their closely related analogs (Lim et al. 2006; Montange et al. 2010). Due to this ability, riboswitches have been recognized as potential anti-microbial drug targets, because designing small organic compounds that bind to them will help inhibition of the activity of selective genes that are important for survival (Zaman et al. 2003; Chen et al. 2010; Deigan and Ferre-D'Amare 2011). Therefore, understanding the physical basis and the underlying interactions that give rise to such specific binding with high affinity is not only of fundamental interest but will also allow an in-depth view of how riboswitches function and how they can be exploited for production of novel antibiotic therapeutics.
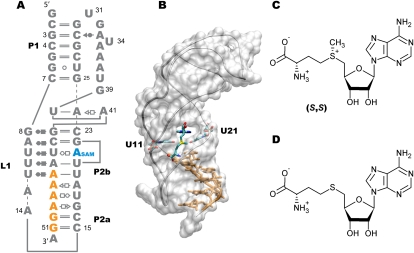
S-Adenosyl-L-methionine (SAM) is a major biological methyl donor in all living organisms and is such a commonly used enzyme substrate that it ranks second in importance to ATP (Cantoni 1952). All the chemical moieties and functional groups of SAM are used as a source of a wide range of biochemical derivatives that are involved in sulfur metabolism; biosynthesis of amino acids, biotin, phospholipids, and plant hormones; post-transcriptional modification of tRNAs; DNA methylation; and iron–sulfur enzymes (Fontecave et al. 2004). Several genes involved in biosynthetic pathways of SAM, sulfur-containing amino acids, and general sulfur metabolism are themselves under the control of SAM-binding riboswitches (Grundy and Henkin 1998; Epshtein et al. 2003; Winkler et al. 2003). Five riboswitches (SAM-I to SAM-V) (McDaniel et al. 2003; Corbino et al. 2005; Fuchs et al. 2006; Weinberg et al. 2008; Poiata et al. 2009) that recognize SAM have been identified to date, each with their exclusive presence in a particular type of organism (Barrick and Breaker 2007). While SAM-I riboswitches are found in Gram-positive bacteria (Winkler et al. 2003), representatives of the SAM-II class are predominantly distributed in proteobacteria (Corbino et al. 2005). Despite the fact that the first two classes of SAM riboswitches share SAM as their natural ligand, both SAM-I and SAM-II have no similarity in sequence and structure (Corbino et al. 2005) (i.e., SAM-I has a pseudoknot structure with a four-way junction architecture [Montange and Batey 2006], while SAM-II has a much simpler H-type pseudoknot [Gilbert et al. 2008]) (Fig. 1A,B). Interestingly, for the same stereoisomer of SAM, SAM-I exhibits about 200 times higher affinity than SAM-II (Lim et al. 2006). From the solved X-ray crystal structures of SAM in complex with SAM-I and SAM-II, it is noted that SAM binds to SAM-II in an extended configuration, while SAM is observed in a bent conformation in the binding pocket of SAM-I (Gilbert et al. 2008). When the methyl group of SAM is transferred, the by-product S-adenosylhomocysteine (SAH) is produced, which has toxic effects if present in higher concentrations in cells (Ueland 1982). SAH too binds both SAM-I and SAM-II but almost about 2500 times better to SAM-I than SAM-II (Lim et al. 2006; Wang and Breaker 2008). Moreover, while SAH binds SAM-I with an affinity reduced by 80 times (reported from in-line probing assays) (Lim et al. 2006) or 550 times (reported from isothermal titration calorimetric studies) (Montange et al. 2010) as compared to that of SAM, the binding of SAH to SAM-II is weaker than that of SAM by more than 1000-fold (Lim et al. 2006). Similarly, the SAH riboswitch can discriminate SAH over SAM by more than three orders of magnitude (Wang et al. 2008). Such discrepancies between binding affinities are quite remarkable given the chemical similarity between SAM and SAH with the only difference being the presence of methyl group and an overall positive charge on SAM (Fig. 1C,D). Investigation of the binding affinities of SAM-I and SAM-II for many analogs of SAM with substitution of the thioether group clearly demonstrated that SAM-II has a greater discriminatory ability for SAM than SAM-I, i.e., SAM-II exclusively binds SAM and rejects even those SAM analogs for which SAM-I has high affinity (Lim et al. 2006). It seems that the lack of a methyl group on the analogs does not weaken the binding to SAM-I as much as it does when the net positive charge on the sulfur atom is missing. However, both the methyl group and the positive charge density seem to be equally essential determinants for binding to SAM-II (Lim et al. 2006). While the functional consequence of binding of SAM to SAM riboswitches is either the termination of mRNA transcription or inhibition of translation initiation of downstream genes, SAH fails to exert any gene-repressing effects (Epshtein et al. 2003; McDaniel et al. 2003; Fuchs et al. 2006). SAM is produced from methionine and ATP in a stereospecific biochemical reaction with the sulfur atom and the amino group purely in the (S,S)-configuration. The (S,S) isomer can spontaneously convert to the (R,S) form, but the latter does not participate in the methyl transferase or other metabolic reactions involving SAM (Haba et al. 1959; Wu et al. 1983). Although the binding affinity of the racemic mixture (S,S and R,S) of SAM is reduced by only half as compared to the chirally pure (S,S)-SAM (Corbino et al. 2005), we still do not know how the binding of (R,S)-SAM modulates the structure and function of SAM-II.
FIGURE 1.
Structures of ligand-bound SAM-II riboswitch and its ligands. (A) Secondary structure representation of SAM-II riboswitch showing the helices P1, P2a/b, and the loop L1. The standard base-pairing interactions seen in the crystal structure (Gilbert et al. 2008) of the 52-nt SAM-II are shown here using the Leontis-Westhof notation (Leontis and Westhof 2001). (Orange) The sequence of the expression platform; (blue) the adenine moiety of SAM. (B) Three-dimensional (3D) structure of the SAM-II riboswitch bound to its cognate metabolite SAM, which is seen in an extended configuration and perpendicular to the bases U11 and U21 of the riboswitch that closely interacts with the sulfur (yellow) moiety of SAM. The expression platform (orange) that contains the Shine-Dalgarno sequence is part of the P2a/b helix and follows its curvature. Chemical structures of (C) SAM and (D) SAH ligands. SAM contains a methyl group attached to the sulfur giving the ligand a +1 overall charge, while SAH is neutral. SAM is shown in the S configuration at both the sulfur atom and the Cα of the methionyl moiety.
We have only begun to understand the exact mechanistic details of how SAM riboswitches control transcription attenuation or translation initiation upon metabolite-binding, since SAM riboswitches have been discovered only within the last decade, and not until recently were their three-dimensional (3D) structures available. The X-ray crystal structure of SAM-II is the first available structure of an entire riboswitch comprising the ligand-binding aptamer domain as well as the expression platform (EP), a purine-rich region located few nucleotides upstream of the start codon (Gilbert et al. 2008). Therefore, it serves as a perfect model to understand in atomic detail using molecular dynamics (MD) simulations how SAM exerts regulation of SAM-II by transmitting the effects of its presence to the conformational changes in the structure of the EP.
As shown in Figure 1, SAM-II is a continuous single-nucleotide chain that forms two major helices P1 and P2a/b and consists of a loop L1 that forms a triple helix with P2b. The EP is located at the 3′ end and interacts with P2a/b when SAM positions itself in the binding pocket formed in the center of the P2b helix. Our recent computational investigation of the SAM-bound and unbound forms of the SAM-II riboswitch revealed that in the presence of (S,S)-SAM the curvature as well as the base-pairing in the EP increases, resulting in a fully formed pseudoknot (Kelley and Hamelberg 2010). In the absence of SAM, the EP has significantly reduced curvature with a very low probability of forming key base-pairing interactions with the P2a/b helix. As a result, the pseudoknot unzips below the binding pocket. For SAM-II, the most likely explanation for the down-regulation of expression of downstream genes seems to rest on the unavailability of the ribosome binding site (RBS). The tertiary interactions between the EP and the P2a/b helix lock the RBS that reside on the EP, preventing the binding of the ribosome and thereby translation initiation. The questions we address in the current study are what are the atomistic bases for the difference in the specificity and the free energy of binding between SAM and SAH and why binding of SAH is incapable of turning off gene expression. Similar to our previous study in which we performed 200-nsec-long MD simulations of the SAM–SAM-II complex, we investigated the SAH-bound complex of SAM-II during an equally long MD simulation. Additionally, we simulated the complex of SAM-II with the (R,S) form of SAM, which has so far not been characterized experimentally.
RESULTS AND DISCUSSION
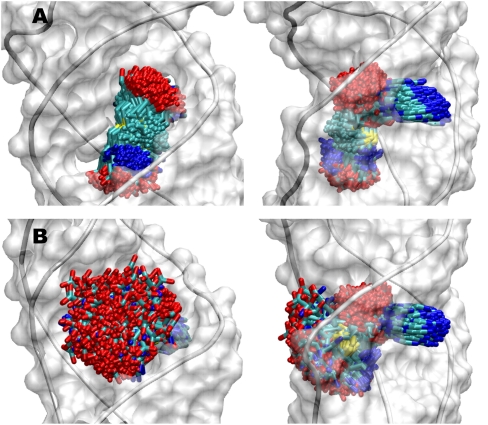
Comparison of the MD trajectories of the SAM-II riboswitch bound to the (S,S)-SAM and SAH ligands revealed that just as in the case of SAM, the adenine moiety in SAH also intercalates between U21 and G22 and is involved in triple helix base-pairing interactions. Also, the ribose sugar in both cases is lodged in the major groove of the triple helix. However, there are marked differences in the flexibility of the methionyl and homocystyl tails of SAM and SAH, respectively. As seen in Figure 2, the methionyl tail of SAM is relatively much more localized than the homocystyl tail of SAH, which exhibits much larger flexibility, thereby suggesting that the loss in entropy for the SAM ligand upon moving from the unbound state into the binding pocket of SAM-II is greater than that for SAH. Since the SAM–SAM-II complex is stabilized by more than ∼17 kJ/mol (at room temperature) than the SAH–SAM-II complex, i.e., KdSAM/KdSAH < 1000, the binding of SAM should be all the more enthalpically driven to compensate for the higher unfavorable entropy of binding.
FIGURE 2.
Dynamics of SAM and SAH ligands in their binding pockets in SAM-II riboswitch. Snapshots from 200-nsec MD trajectory are shown every 1 nsec in two different views (left and right panels) for the SAM-II riboswitch bound to (A) SAM and (B) SAH. The ligands follow the color code: carbon (cyan), nitrogen (blue), oxygen (red), and sulfur (yellow).
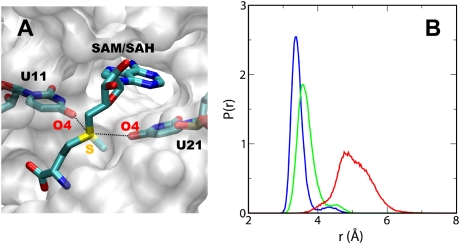
The question that now arises is which interactions between SAM and the SAM-II riboswitch are responsible for the enthalpic stabilization of the complex. SAM-I and SAM-II riboswitches have not only evolved to have dissimilar sequences and divergent folds but recognize also two different configurations of SAM. Despite this fact, it is noteworthy that in both these riboswitches the sulfonium group of SAM is positioned next to the carbonyl oxygens of two uridine bases. As observed in the crystal structure (Fig. 1B), the O4 of the uridine bases, U11 and U21, are in close proximity to the sulfur of the methionyl/homocystyl chain. The distance between the O4 of U11 and U21 and the sulfur atom is approximately equal to the sum of the van der Waals radii for these atoms. We therefore calculated the average distance between these oxygens and the sulfur atoms on the individual ligands and used this as a metric for the interaction between the tail of the ligands and the riboswitch. For the length of the trajectory, the distribution of this average distance in case of (S,S)-SAM exhibited a narrow peak centered at ∼3.25 Å, which suggested that the sulfur atom provided an anchor point that allowed SAM to remain more or less ordered in the binding pocket (Fig. 3). When SAM-II was bound to the (R,S) configuration of SAM, the average distance slightly increased but still showed a predominantly narrow distribution as in the case of the (S,S) isomer. In contrast, the average distance between the sulfur atom of SAH and the O4s of the bases fluctuated over a much wider range with the distribution shifted to larger distances, thus implying weaker interaction. As a consequence, SAH was allowed to sample larger conformational space and would be relatively less stabilized enthalpically.
FIGURE 3.
The sulfur atom acts as the major anchor point and is involved in the key interactions. (A) Detailed view of the binding pocket of SAM-II with the ligand bound showing the distance (dotted lines) between the sulfur atom on SAM/SAH and O4 oxygens on the bases U11 and U21. (B) Probability distributions of the average distance between the sulfur atom and the two O4s when (S,S)-SAM (blue), (R,S)-SAM (green), or SAH (red) is bound to SAM-II.
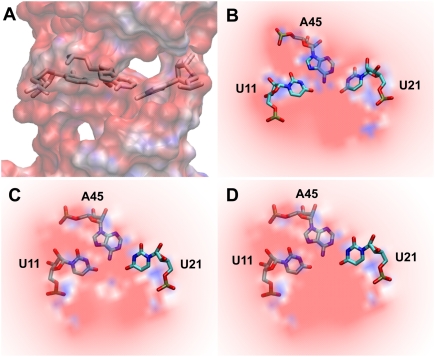
We further explored the electrostatic potential maps of the binding pocket of SAM-II bound to either (S,S)-SAM or SAH. SAM possesses the positively charged sulfonium ion involved in three single bonds with methylene/methyl groups that render an overall positive charge to SAM. The highly electronegative binding pocket generated by the bases A45, U11, and U21 (Fig. 4) thus favorably accommodated the electropositive ligand, SAM. In the case of SAH, however, sulfur is involved in two single bonds and has two lone pairs of electrons that render an electronegative character and a neutral formal charge to sulfur. As a result, the homocystyl chain experienced repulsion in the electronegative binding pocket that led to large fluctuations (Fig. 2B) and an inability to form key interactions with the sulfur atom of SAH (Fig. 3).
FIGURE 4.
Electrostatic basis for the binding specificity of SAM. Electrostatic potential map of the binding pocket of the SAM-II riboswitch. Shown are the cross-sectional (A) and the detailed (B) views of the binding pocket of free SAM-II riboswitch formed by U11, U21, and A45. Red (−10 kT/e) and blue (+10 kT/e) areas depict electronegative and electropositive regions, respectively. Also shown are the detailed views of the binding pocket when (S,S)-SAM (C) with the electropositive sulfur or SAH (D) with the electronegative sulfur atom is bound to the riboswitch. The blue dot in the center of the electronegative pocket in C depicts the electropositive sulfur atom. Hydrogens are not shown in the above electrostatic potential map; however, they were included in the calculation of the map.
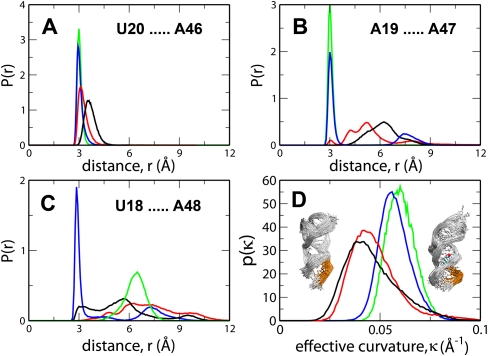
As shown previously in the comparative study of SAM-bound and unbound forms of the SAM-II riboswitch, it was only in the presence of SAM that the pseudoknot structure was fully formed (Kelley and Hamelberg 2010). Binding of SAM not only stabilized the loop L1 but also allowed for favorable base-pairing interactions in the EP and increased the effective curvature in the EP. Here, to investigate how different the effects of SAH-binding were on the EP, we compared the distribution of the distances between the three key base pairs obtained from the MD trajectories of SAM-II in complex with two different ligands and the free riboswitch (Fig. 5A–C). These base pairs, which are located on the EP along the P2a/b helix on one strand and the P2a/b helix on the opposite strand, monitor the level of sequestration of the ribosome binding site on the EP. During the length of the simulations, these base-pairing distances were quite stable in the case of the SAM-bound complexes, as suggested from their very narrow distributions. For both, the unbound and SAH-bound riboswitches, the base-pairing interactions between the EP and the P2a/b helix were poorly formed and therefore exhibited much wider distributions. It should also be noted from Figure 5A–C that, similar to free SAM-II, not only the distances between the base pairs in the EP in SAH-bound SAM-II exhibited progressively broader distributions but the average distances also increased going from 5′ to 3′, i.e., d(U20…A46) = ∼3.5 Å (Fig. 5A), d(A19…A47) = ∼6 Å (Fig. 5B), and d(U18…A48) > 6 Å. The distance between U18–A48, which, of the three base pairs, is closest to the 3′ terminal, showed a relatively narrow distribution with much larger fluctuations when the riboswitch was bound to the (R,S)-SAM. This suggested that the base pair was stabilized, and, as noticed from the 130-nsec trajectory, the base pair was indeed buckled with increased stacking. Furthermore, we calculated the effective curvature of the EP (Fig. 5D) from each snapshot of these individual trajectories by defining an osculating circle that best approximates the curvature of every three consecutive backbone phosphorus atoms of the EP (for details, see Materials and Methods). Regardless of the stereochemistry at the sulfur atom, the effective curvature of the EP was found to be >0.05 Å−1. Our previous studies have shown that the effective curvature of the EP was significantly reduced in the free riboswitch as compared with that in the SAM-bound form (Kelley and Hamelberg 2010). Notably, we found that the probability distribution of the effective curvature of the EP in the SAH-bound riboswitch was closely comparable to that in the free riboswitch (Fig. 5D). Just as in the case of the free riboswitch, lack of stabilizing interactions and reduced curvature in the EP resulted in the local melting of the pseudoknot structure, downstream from the ligand-binding region to the 3′ end in the SAH-bound riboswitch (Fig. 5D, inset).
FIGURE 5.
Effects of ligand binding to SAM-II riboswitch on its EP. Probability distributions of the distances between the key base pairs in the expression platform along the P2a/b helix, i.e., N3 of U20 and N1 of A46 (A), N6 of A19 and N3 of A47 (B), and O2 of U18 and N6 of A48 (C) when the SAM-II riboswitch is unbound (black) or bound either to (S,S)-SAM (blue), (R,S)-SAM (green), or SAH (red). The distributions of the effective curvature of the expression platform are shown in D for all four cases: the unbound riboswitch (black) and that bound to (S,S)-SAM (blue), (R,S)-SAM (green), or SAH (red). (Inset to D) Ensemble of structures of the free (left inset) and the SAM-bound (right inset) SAM-II with curvatures corresponding to the peak of the respective distributions.
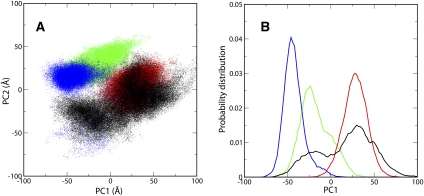
To further differentiate the effects of binding of closely related but distinct ligands to the SAM-II riboswitch on its EP, we analyzed the conformations of the EP from the individual trajectories of SAM- and SAH-bound complexes of SAM-II as well as the free riboswitch. The complexity underlying the 3D atomic coordinates of the EP obscures the relevant information about the differences in its conformations. Therefore, to reduce the multidimensional coordinate system to a few dimensions, we carried out Principal Component Analysis (see Methods), which brings about linear transformation of correlated variables to yield uncorrelated or Principal Components, which accounts for the largest variability. Shown in Figure 6A is the two-dimensional (2D) projection of the conformational space of the EP along the two main Principal Components. Obviously, the EP in the free riboswitch sampled more conformational space than in the ligand-bound ones. The free riboswitch also already visited those conformations that were sampled by the riboswitch when the ligand was bound, suggesting that SAM-II followed conformational selection in ligand recognition. Figure 6, A and B, clearly indicated that the EP in the SAH-bound riboswitch sampled conformational space that showed considerable overlap with that sampled by the free SAM-II. This confirmed that the structure of the EP in the SAH-bound riboswitch was melted as in free SAM-II. In contrast, the overlap between the conformations of the EP in the (S,S)- or (R,S)-SAM-bound riboswitch and the free SAM-II was to a much smaller degree (Fig. 6B). Moreover, in the presence of (S,S)- or (R,S)-SAM, the EP of the SAM-II riboswitch occupied exclusive conformational space, indicating an induced-fit mechanism underlying the high specificity of SAM-II for SAM. The presence of SAM induced those conformations in SAM-II that facilitated the formation of favorable electrostatic interactions with the ligand and more than compensated for the loss in conformational entropy.
FIGURE 6.
Conformational space sampled by the expression platform of the SAM-II riboswitch in the free as well as SAM- and SAH-bound forms. (A) Projection of the configurations of the expression platform (nucleotides 46–51) on the first two Principal Components when the SAM-II riboswitch is free (black) and bound to either (S,S)-SAM (blue), (R,S)-SAM (green), or SAH (red). (B) Probability distribution of the configurations of the expression platform along the first Principal Component. The color code is the same as in A.
The details provided by the above atomistic MD simulations clearly indicated that in the presence of SAH, the ribosome binding site was exposed, while the presence of SAM sequestered the bases in the EP in very stable interactions such that the resulting pseudoknot structure was completely formed. Our results are consistent with the very recent biophysical studies including NMR, fluorescence spectroscopy, and single-molecule FRET that showed that it was only upon addition of SAM that the stem–loop element of the riboswitch got involved in the base-pairing with the EP, resulting in the formation of the pseudoknot fold (Haller et al. 2011). It should, however, be noted that in our studies, apart from the local melting in the EP region in free or SAH-bound SAM-II, the integrity of the pseudoknot structure was maintained in the L1 loop and the P1 region of the riboswitch in both the free and the ligand-bound SAM-II (Fig. 5D, inset). We did not observe any global unfolding/folding of the riboswitch in our nanosecond-timescale simulations. Our results further suggested that the conformational ensemble sampled predominantly by the free and the SAH-bound riboswitch with partially unfolded structure and uncurled EP might facilitate ribosomal assembly and thereby maintain the riboswitch in the ON state, eventually allowing for translation initiation. In a scenario when SAM (either of the chiral forms) was bound, the riboswitch mostly sampled conformations in which the pseudoknot is fully formed and the RBS would be inaccessible due to base-pairing interactions in the EP. As a result, ribosomal binding would be prevented and gene expression would be down-regulated. Cryo-electron microscopic studies on different stages of translation initiation have suggested that it is possible for the ribosome to bind the pseudoknot structure even in the presence of the ligand (Marzi et al. 2007). However, the 30S ribosomal protein S15 mRNA (rpsO) investigated in such studies has a 5′-pseudoknot structure of different architecture from that of SAM-II and is not a riboswitch since it binds the S15 protein and not a small ligand. Moreover, unlike SAM-II in which the Shine-Dalgarno sequence forms an integral part of the P2a/b helix only in the context of SAM, the location of the Shine-Dalgarno sequence is on loop 2 on the rpsO mRNA, which keeps it exposed irrespective of the presence or absence of its ligand protein S15. In the case of rpsO, despite the assembly of ribosome on the mRNA, translation is repressed because the presence of S15 does not allow the pseudoknot structure to unfold and the start codon to be accessible to tRNA. Our results suggest that translation of genes under the control of SAM-II is repressed in the presence of SAM due possibly to the lack of base-pairing between the Shine-Dalgarno sequence on the mRNA and its complementary anti-Shine-Dalgarno sequence on the 16S ribosomal rRNA resulting from the complete sequestration of the ribosome recognition site.
CONCLUSIONS
The chemical structures of SAM and SAH are very similar and differ only in the presence of positively charged sulfonium moiety on SAM versus a neutral sulfo-ether group on SAH. Despite this small difference, the SAM-riboswitches exhibit remarkable discrimination between the two ligands, binding SAM with high selectivity and affinity. Since there has been a growing interest in riboswitches as potential antibiotic drug targets, it is essential to understand the molecular recognition principles of SAM riboswitches that will help in the design of new drugs. Our atomistically detailed MD simulations on the free and SAM- and SAH-bound SAM-II riboswitch show that SAM is entropically more destabilized than SAH in the binding pocket. To achieve a 1000-fold higher affinity than SAH and compensate for the unfavorable loss in entropy, SAM forms stabilizing electrostatic interactions as it is favorably accommodated in the electronegative binding pocket of SAM-II. Remarkably, the single sulfur atom acts as the predominant anchor point for forming key interactions with the riboswitch. SAH, on the other hand, with the lone pair of electrons is electronegative and experiences repulsion in the electronegative binding pocket. SAH thus fails to form electrostatic interactions with the carbonyl oxygens of the two conserved uridine bases. Moreover, our results demonstrate that SAH-bound SAM-II samples conformations that, most of the time, are very similar to those sampled by the free riboswitch, with the pseudoknot structure that is melted from below the binding pocket and the EP that has significantly reduced curvature in the presence of SAH. Unlike SAM-bound SAM-II, the ribosome binding site on the SAH-bound riboswitch is exposed and available for base-pairing with the ribosome, thus allowing for ribosomal assembly and translational machinery to synthesize nascent protein from the mRNA. Furthermore, since the unbound riboswitch appears to share a portion of the conformational space sampled by SAM-bound SAM-II and a considerable amount of that sampled by SAH-bound riboswitch, the binding of ligands seems to predominantly follow the conformational selection mechanism. Moreover, the fact that the presence of SAM allows SAM-II to populate exclusive conformational space suggests that induced fit is at play for achieving much higher binding specificity for SAM as compared with SAH. Our studies also show that apart from the buckling of the terminal base pairs, the (R,S) configuration of SAM has no effects that are significantly dissimilar to those of the (S,S) isomer when bound to SAM-II.
MATERIALS AND METHODS
Preparation of SAM-II–SAH/SAM complex and molecular dynamics simulations
We used the AMBER 10 suite of programs (Case et al. 2008) to carry out all molecular dynamics (MD) simulations on the SAH-bound SAM-II. The crystal structure of SAM-II in complex with (S,S)-SAM (PDB id: 2qwy; chain A) (Gilbert et al. 2008) was used as the starting structure for the simulations. To generate the SAH–SAM-II complex, the coordinates of the methyl group on SAM were deleted from this pdb file, while the water molecules and ions in the crystal structure were left unmodified. For SAM-II bound to the (R,S) configuration of SAM, the chirality at the sulfur atom was simply modified in the XLEAP program. As described earlier (Kelley and Hamelberg 2010), starting with the SAM ligand removed from the X-ray structure of the complex, the methyl group was deleted to generate SAH, and the missing hydrogens on SAH were added using the GaussView program. Similarly, the (R,S) form of SAM was generated from its (S,S) isomer with GaussView. This was followed by quantum mechanical optimization of both SAH and (R,S)-SAM at the HF/6-31-g(d) level of theory in the Gaussian program (Gaussian 03, Revision E.01. Gaussian Inc.). Using the same level of theory, electrostatic potential was calculated with the Gaussian program, which was then used to obtain partial charges using the RESP method (Bayly et al. 1993). The force field parameters for SAM or SAH were obtained from the ANTECHAMBER module of AMBER, while for the riboswitch, we used the reoptimized nucleic acid parameters (Perez et al. 2007) of the Cornell et al. force field (Cornell et al. 1995). The initial structure of the SAH– or (R,S)-SAM–SAM-II complex was then generated by loading the charges and the force field parameters in the XLEAP program. The complexes were further solvated in an octahedral box filled with about 11,000 TIP3P (Jorgensen et al. 1983) water molecules and brought to neutral charge by addition of 50 Na+ ions. The systems were minimized and then subjected to 10 nsec of equilibration in which the temperature was brought to 300 K using a Langevin thermostat (collision frequency = 1.0/psec). After the equilibration, we carried out the MD simulations at the NPT ensemble with a constant temperature of 300 K and constant pressure of 1 bar. While the SAH–SAM-II complex was simulated for 200 nsec, the production run for the (R,S)-SAM-bound SAM-II was performed for 130 nsec. Long-range electrostatics was treated with Particle mesh Ewald summation (Essmann et al. 1995), and a cutoff of 9 Å was used for the calculation of short-range non-bonded interactions. The SHAKE (Ryckaert et al. 1977) algorithm was applied for constraining all bonds in which hydrogen atoms were participants. Newton's equations of motion were solved using a time step of 2 fsec.
Calculation of the effective curvature
The expression platform comprises 6 nt in the region of the P2a/b helix. At first, we calculated the vectors joining every two consecutive backbone phosphorus atoms of the expression platform using their x, y, z coordinates, i.e., 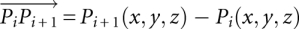 , where Pi is the phosphorus atom of nucleotides 46–50. We next defined four osculating circles that best approximated the individual curvature of the backbone of the expression platform for four sets of three consecutive phosphorus atoms, i.e., set 1: P46, P47, P48; set 2: P47, P48, P49; and so on. We therefore assumed the formation of an isosceles triangle by the two sides equal to the radii of each such circle since they joined the center of each such circle to two consecutive phosphorus atoms. The third side was simply the distance connecting the two consecutive phosphorus atoms,
, where Pi is the phosphorus atom of nucleotides 46–50. We next defined four osculating circles that best approximated the individual curvature of the backbone of the expression platform for four sets of three consecutive phosphorus atoms, i.e., set 1: P46, P47, P48; set 2: P47, P48, P49; and so on. We therefore assumed the formation of an isosceles triangle by the two sides equal to the radii of each such circle since they joined the center of each such circle to two consecutive phosphorus atoms. The third side was simply the distance connecting the two consecutive phosphorus atoms,
Using the properties of isosceles triangles and right triangles, the radius of each osculating circle was given by
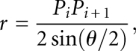 |
where θ was the angle made at that vertex of each isosceles triangle that was also the center of each osculating circle. This θ was also equal to the smaller angle between the vectors 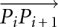 and
and 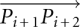 and therefore could be obtained from their cross-product, i.e.,
and therefore could be obtained from their cross-product, i.e.,
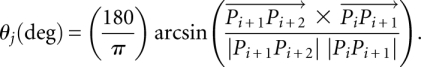 |
θj could be positive or negative. In this manner, four values of θ between four pairs of vectors formed by six phosphorus atoms were calculated. The effective curvature of the expression platform was then given by 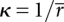 , where
, where
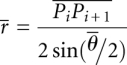 |
and 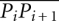 is the average distance between every two consecutive phosphorus atoms and
is the average distance between every two consecutive phosphorus atoms and 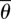 is the average angle. If all of the phosphorus atoms fell on a straight line or if the average angle
is the average angle. If all of the phosphorus atoms fell on a straight line or if the average angle 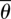 was zero, the effective curvature would be zero.
was zero, the effective curvature would be zero.
Principal Component Analysis
Using the ptraj module of AMBER 10, the Principal Component Analysis (Levy et al. 1984; Jolliffe 2002) was performed on all the atoms of the nucleotides of the expression platform, i.e., from 46 to 51, after aligning the phosphorus atoms of the entire SAM-II structure from each snapshot. The covariance matrix of the x, y, and z coordinates of all the atoms of the expression platform obtained from each snapshot of the combined trajectories of the free SAM-II, SAM-bound [both the (S,S) and the (R,S) forms], and SAH-bound SAM-II was calculated. The covariance matrix was further diagonalized to produce orthonormal eigenvectors and their corresponding eigenvalues, ranked on the basis of their corresponding variances. The first two eigenvectors, the Principal Components, which contributed ∼50% of all atomic fluctuations, were used to project the conformational space onto them, i.e., along two dimensions.
ACKNOWLEDGMENTS
This work was supported in part by research initiation grants from Georgia State University, the Department of Chemistry, the Georgia Cancer Coalition (GCC) scholar award, and a National Science Foundation CAREER Award (MCB-0953061). This work was also supported by Georgia State's IBM System p5 supercomputer, acquired through a partnership of the Southeastern Universities Research Association and IBM supporting the SURAgrid initiative. We thank Dr. W.D. Wilson for helpful discussions during the preparation of this manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028779.111.
REFERENCES
- Barrick JE, Breaker RR 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239 doi: 10.1186/gb-2007-8-11-r239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly CI, Cieplak P, Cornell W, Kollman PA 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J Phys Chem 97: 10269–10280 [Google Scholar]
- Cantoni GL 1952. The nature of the active methyl donor formed enzymatically from L-methionine and adenosinetriophosphate. J Am Chem Soc 74: 2942–2943 [Google Scholar]
- Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. 2008. AMBER 10. University of California, San Francisco, CA [Google Scholar]
- Chen L, Cressina E, Leeper FJ, Smith AG, Abell C 2010. A fragment-based approach to identifying ligands for riboswitches. ACS Chem Biol 5: 355–358 [DOI] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR 2005. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol 6: R70 doi: 10.1186/gb-2005-6-8-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA 1995. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J Am Chem Soc 117: 5179–5197 [Google Scholar]
- Deigan KE, Ferre-D'Amare AR 2011. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. doi: 10.1021/ar200039b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E 2003. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci 100: 5052–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG 1995. A smooth particle mesh Ewald method. J Chem Phys 103: 8577–8593 [Google Scholar]
- Fontecave M, Atta M, Mulliez E 2004. S-Adenosylmethionine: Nothing goes to waste. Trends Biochem Sci 29: 243–249 [DOI] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM 2006. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol 13: 226–233 [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182 [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM 1998. The S box regulon: A new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol Microbiol 30: 737–749 [DOI] [PubMed] [Google Scholar]
- Haba G, Jamieson GA, Mudd AH, Richard HH 1959. S-Adenosylmethionine: The relation of configuration at the sulfonium center to enzymatic reactivity. J Am Chem Soc 81: 3975–3980 [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R 2011. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7: 393–400 [DOI] [PubMed] [Google Scholar]
- Jolliffe IT 2002. Principal Component Analysis. Springer, New York [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935 [Google Scholar]
- Kelley JM, Hamelberg D 2010. Atomistic basis for the on-off signaling mechanism in SAM-II riboswitch. Nucleic Acids Res 38: 1392–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy RM, Srinivasan AR, Olson WK, McCammon JA 1984. Quasi-harmonic method for studying very low frequency modes in proteins. Biopolymers 23: 1099–1112 [DOI] [PubMed] [Google Scholar]
- Lim J, Winkler WC, Nakamura S, Scott V, Breaker RR 2006. Molecular-recognition characteristics of SAM-binding riboswitches. Angew Chem Int Ed Engl 45: 964–968 [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR 2004. Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5: 451–463 [DOI] [PubMed] [Google Scholar]
- Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP 2007. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell 130: 1019–1031 [DOI] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM 2003. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci 100: 3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT 2010. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol 396: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS 2004. The riboswitch control of bacterial metabolism. Trends Biochem Sci 29: 11–17 [DOI] [PubMed] [Google Scholar]
- Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE III, Laughton CA, Orozco M 2007. Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophys J 92: 3817–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiata E, Meyer MM, Ames TD, Breaker RR 2009. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 15: 2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J, Cicotti G, Berendsen H 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys 23: 327–341 [Google Scholar]
- Soukup JK, Soukup GA 2004. Riboswitches exert genetic control through metabolite-induced conformational change. Curr Opin Struct Biol 14: 344–349 [DOI] [PubMed] [Google Scholar]
- Ueland PM 1982. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacol Rev 34: 223–253 [PubMed] [Google Scholar]
- Wang JX, Breaker RR 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168 [DOI] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR 2008. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell 29: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR 2008. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA 14: 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR 2003. Genetic control by metabolite-binding riboswitches. ChemBioChem 4: 1024–1032 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR 2005. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59: 487–517 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707 [DOI] [PubMed] [Google Scholar]
- Wu SE, Huskey WP, Borchardt RT, Schowen RL 1983. Chiral instability at sulfur of S-adenosylmethionine. Biochemistry 22: 2828–2832 [DOI] [PubMed] [Google Scholar]
- Zaman GJ, Michiels PJ, van Boeckel CA 2003. Targeting RNA: New opportunities to address drugless targets. Drug Discov Today 8: 297–306 [DOI] [PubMed] [Google Scholar]