Type I collagen is composed of two α1(I) and one α2(I) polypeptides and is the most abundant protein in the human body. Expression of type I collagen is primarily controlled at the level of mRNA stability and translation. Coordinated translation of α(I) and α2(I) mRNAs is necessary for efficient folding of the corresponding peptides into the collagen heterotrimer. In the 5′ untranslated region (5′ UTR), collagen mRNAs have a unique 5′ stem–loop structure (5′ SL). La ribonucleoprotein domain family member 6 (LARP6) is the protein which binds 5′ SL with high affinity and specificity and coordinates their translation. Here the authors show that RNA helicase A (RHA) is tethered to the 5′ SL of collagen mRNAs by interaction with the C-terminal domain of LARP6. Knockdown of RHA prevents formation of polysomes on collagen mRNAs and dramatically reduces the synthesis of collagen protein, without affecting the level of the mRNAs. Accordingly, it appears that RHA is recruited to the 5′ UTR of collagen mRNAs by LARP6 to facilitate their translation.
Keywords: RNA helicase A (RHA), La ribonucleoprotein domain family member 6 (LARP6), type I collagen, post-transcriptional control element (PCE), 5′ stem–loop (5′ SL), translation
Abstract
Type I collagen is composed of two α1(I) polypeptides and one α2(I) polypeptide and is the most abundant protein in the human body. Expression of type I collagen is primarily controlled at the level of mRNA stability and translation. Coordinated translation of α(I) and α2(I) mRNAs is necessary for efficient folding of the corresponding peptides into the collagen heterotrimer. In the 5′ untranslated region (5′ UTR), collagen mRNAs have a unique 5′ stem–loop structure (5′ SL). La ribonucleoprotein domain family member 6 (LARP6) is the protein that binds 5′ SL with high affinity and specificity and coordinates their translation. Here we show that RNA helicase A (RHA) is tethered to the 5′ SL of collagen mRNAs by interaction with the C-terminal domain of LARP6. In vivo, collagen mRNAs immunoprecipitate with RHA in an LARP6-dependent manner. Knockdown of RHA prevents formation of polysomes on collagen mRNAs and dramatically reduces synthesis of collagen protein, without affecting the level of the mRNAs. A reporter mRNA with collagen 5′ SL is translated three times more efficiently in the presence of RHA than the same reporter without the 5′ SL, indicating that the 5′ SL is the cis-acting element conferring the regulation. During activation of quiescent cells into collagen-producing cells, expression of RHA is highly up-regulated. We postulate that RHA is recruited to the 5′ UTR of collagen mRNAs by LARP6 to facilitate their translation. Thus, RHA has been discovered as a critical factor for synthesis of the most abundant protein in the human body.
INTRODUCTION
Type I collagen is a heterotrimer composed of two α1(I) polypeptides and one α2(I) polypeptide and is the most abundant protein in the human body (Kivirikko 1998). It is expressed at high levels in skin, bones, and tendons, with a low expression in parenchymal organs. During fibrosis of soft tissues, type I collagen expression is highly up-regulated at both transcriptional and post-transcriptional levels (Stefanovic et al. 1999, 2002; Lindquist et al. 2000b). The 50–100-fold up-regulation of type I collagen expression and secretion in fibrosis is primarily due to a 16-fold increase in α1(I) mRNA stability and a threefold increase in the transcription rate of collagen genes (Stefanovic et al. 1995, 1999, 2000; Stefanovic and Brenner 2003; Lindquist et al. 2004; Rippe and Stefanovic 2005). mRNAs encoding type I collagen have cis-acting elements controlling mRNA stability in both, the 3′ and the 5′ untranslated regions (UTRs). In the 3′ UTR of α1(I) mRNA, there is a cytosine-rich region that interacts with αCP protein that is shown to stabilize the message (Stefanovic et al. 1997, 2000; Lindquist et al. 2000a,b, 2004). In the 5′ UTR, 75 nt from the 5′-terminal 7-methylguanine cap, there is a discrete 5′ stem–loop structure (5′ SL). This 5′ SL is found only in three collagen mRNAs—α1(I), α2(I), and α1(III); the latter encodes for type III collagen. We have cloned La ribonucleoprotein domain family member 6 (LARP6) as the protein that binds 5′ SL with high affinity and specificity. Binding of LARP6 prevents premature and random translation of type I collagen mRNAs (Cai et al. 2010a). LARP6 also mediates the association of the mRNAs with nonmuscle myosin filaments that is necessary for their coordinated translation at the membrane of the ER (Cai et al. 2009; Cai et al. 2010b). When LARP6 was knocked down or when 5′ SL was mutated in the endogenous α1(I) gene, the cells failed to secrete normal heterotrimeric type I collagen (Stefanovic and Brenner 2003; Cai et al. 2010b). Instead, collagen α1(I) and α2(I) polypeptides were degraded intracellularly, and small amounts of unnatural homotrimer composed of three α1(I) chains were secreted (Uitto 1979; Schwarze et al. 2004; Pace et al. 2008). Disulfide bonding and post-translational modifications of collagen polypeptides take place during the translational elongation phase and before procollagen is being released into the endoplasmic reticulum (ER) lumen (Beck et al. 1996). This led to the hypothesis that the translation of collagen mRNAs occurs in a close proximity in the same ER compartment to allow coordinated synthesis and productive folding of α1(I) and α2(I) polypeptides into the heterotrimer (Stefanovic et al. 2000; Stefanovic and Brenner 2003). Previous work established that the LARP6-dependent mechanism is critical for a high level of collagen synthesis, as seen in wound healing and fibrosis.
DEIH motif DHX9 helicase, also known as RNA helicase A (RHA), is an essential well-conserved RNA binding protein with ATPase and RNA helicase activities (Chester et al. 1998; Zhang and Grosse 2004; Hartman et al. 2006). Recently, it was demonstrated that RHA is a necessary cofactor to promote translation initiation of highly structured mRNAs containing the post-transcriptional control element (PCE) (Fujii et al. 2001; Zhang and Grosse 2004; Hartman et al. 2006). PCE is a highly structured, orientation-dependent motif present in the 5′ UTRs of some retroviral mRNAs (Butsch et al. 1999; Roberts and Boris-Lawrie 2000; Hull and Boris-Lawrie 2002). The 5′PCE activity is positively regulated by RHA and is essential for the translation efficiency of viral mRNAs (Butsch et al. 1999; Hull and Boris-Lawrie 2002). In addition, Hartman et al. (2006) demonstrated that the PCE-like element in the 5′ UTR of human JunD mRNA interacts with RHA and regulates translation. JunD protein regulates cell growth in response to a stress, so tightly controlled translational regulation provides the means to rapidly alter protein expression (van Dam and Castellazzi 2001; Short and Pfarr 2002). During cap-dependant translation initiation, RHA ensures unwinding of the secondary structures in the 5′ UTR to facilitate ribosomal scanning (Kozak 1989; Sonenberg and Gingras 1998; Merrick 2004). Initiation factor 4A (eIF4A) is a global translation DEAD-box factor with helicase activity (Cordin et al. 2006). For the highly structured 5′ UTR, it is likely that eIF4A needs the activity of additional factors with helicase activity for efficient ribosomal scanning (Pelletier and Sonenberg 1985). RHA may be specifically recruited by the PCE elements to provide such activity (Lindquist et al. 2000b).
The collagen 5′ UTR may functionally resemble a PCE-like region (Yamada et al. 1983; Stefanovic et al. 1999, 2000). The start codon of collagen mRNAs is located in the 5′ stem–loop (Stefanovic et al. 1999, 2000), thus the RNA helicase A activity may be crucial in the unwinding of the 5′ UTR and translation initiation. Here we report that RHA interacts with LARP6, which recruits RHA to the 5′ UTR of collagen mRNAs. There, RHA activity is necessary for efficient formation of polysomes and a high level of collagen protein synthesis.
RESULTS
LARP6 interacts with RNA helicase A
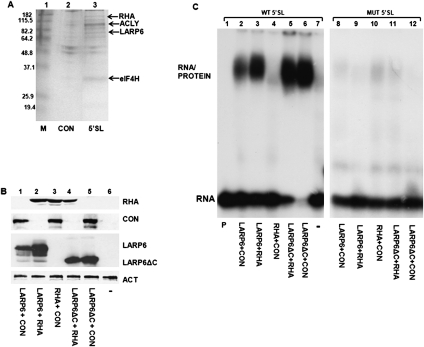
LARP6 was cloned as the protein that directly binds the 5′ SL of collagen mRNAs (Cai et al. 2010a). To identify additional proteins that may be associated in a complex with LARP6 and 5′ SL, we performed pull-down experiments using biotin-labeled collagen α1(I) 5′ SL RNA (Fig. 1A). Inverted 5′ SL RNA was used for control pull-down. Several proteins were pulled down with 5′ SL RNA (indicated in Fig. 1A). RNA helicase A (RHA) was identified by LC/MS/MS as the 140-kDa protein band (indicated in Fig. 1A). This result was further confirmed by MALDI/TOF in a separately repeated experiment.
FIGURE 1.
RHA interaction with collagen 5′ SL RNA. (A) Pull-down of RHA with biotinylated 5′ SL RNA. Biotin-tagged 5′ SL RNA (5′ SL, lane 3) and inverted 5′ SL RNA (CON, lane 2) were used to pull down proteins from cytosolic extracts of HLF. Proteins were analyzed by SDS-PAGE and Coomassie staining. The protein bands identified by MALDI-TOF are indicated: RNA helicase A (RHA), ATP citrate lysate isoform 1 (ACLY1), ATP-dependent DNA helicase 2 subunit 1(XRCC6), eukaryotic initiation factor 4H (eIF4H). (B) Expression of proteins (immunoblot) used in gel mobility shift experiments. HA-tagged LARP6 and LARP6ΔC, His-tagged RHA and Flag-tagged control protein TRIM45 (CON) were transfected into HEK293 cells, and their expression in cytosolic extract was analyzed by Western blot. (–) Nontransfected cells. Actin (ACT) is shown as the loading control. (C) RHA does not bind 5′ SL in the gel mobility shift assay with high affinity. Gel mobility shift assay with wild-type (wt) 5′ SL RNA (lanes 1–7) and mutant 5′ SL RNA with a single point mutation that abolishes interaction with LARP6 (lanes 8–12) and extracts shown in B. Migration of RNA/protein complex and free probe is indicated. (Lane 1) The wt probe alone (P).
To test if RHA interacts with the 5′ SL of collagen mRNAs with high affinity, we performed a gel mobility shift assay using 5′ SL RNA as a probe. This assay is very stringent because only RNA/protein complexes that are stable and formed by high-affinity interactions can endure gel electrophoresis. To increase the readout of these experiments, we overexpressed RHA together with LARP6 or with LARP6 mutant lacking the C-terminal domain (LARP6ΔC) as our positive control or with a control protein (TRIM45) as a negative control in HEK293 cells. Figure 1B shows the expression of the transfected proteins. Overexpression of LARP6 resulted in formation of the 5′ SL/protein complex in a gel mobility shift assay (Fig. 1C, lane 2). Cotransfection of LARP6 and RHA yielded twofold higher expression of LARP6 (Fig. 1B, cf. lane 2 to lane 1). Therefore, the RNA/protein complex in a gel mobility shift experiment using this extract is stronger (Fig. 1C, lane 3). However, the electrophoretic mobility of this complex was identical to that of LARP6 alone, suggesting that RHA has no effect on binding of LARP6 to 5′ SL. Overexpression of RHA with control protein (Fig. 1C, lane 4) failed to produce the RNA/protein complex, only the background binding was seen, which was identical to the background in the nontransfected cells (lane 7). This suggests that binding of RHA to 5′ SL cannot be detected by gel mobility shift experiments. LARP6ΔC was used as an additional control; this protein cannot interact with RHA (see below), but can bind 5′ SL (Cai et al. 2010a). When LARP6ΔC was coexpressed with RHA or control protein (Fig. 1C, lanes 5,6), the identical shift was seen.
Binding of LARP6 to 5′ SL is strictly sequence-specific and when a single U nucleotide in the bulge of the 5′ SL is changed into an A, LARP6 cannot bind (Cai et al. 2010a). When this mutant 5′ SL probe was used in gel mobility shift experiments (Fig. 1C, lanes 8–12), no binding of LARP6 was detected, while binding of RHA was at the background level seen in the nontransfected cells. This verified that LARP6 binds 5′ SL in the sequence-specific manner and that RHA cannot bind 5′ SL with high affinity to withstand the electropheresis. Therefore, it is likely that RHA had been recruited to 5′ SL in the pull-down experiments by a protein–protein interaction with LARP6.
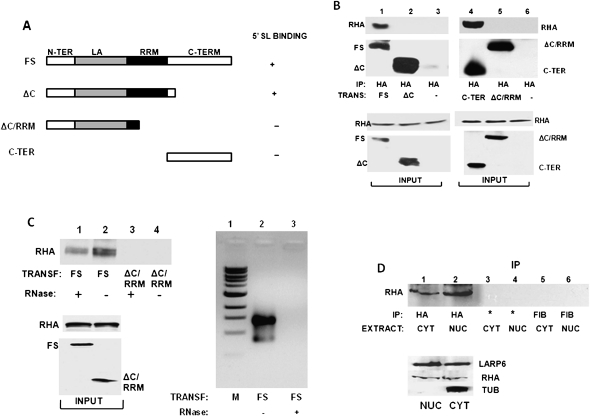
Since it likely that LARP6 and RHA interact by protein–protein interactions, we tested the RHA–LARP6 interaction by coimmunoprecipitation experiments in the presence and absence of intact RNA. We have designed adenoviruses expressing different mutants of the HA-tagged LARP6 protein, as shown in Figure 2A, and analyzed these constructs for their ability to interact with the endogenous RHA. After expression of HA-tagged LARP6 constructs and immunoprecipitation with anti-HA antibody, RHA was analyzed by Western blot. Figure 2B shows that RHA precipitated with the full-size LARP6 (Fig. 2B, lane 1), while the LARP6 lacking the C terminus failed to pull down RHA (lane 2). To assess if the C-terminal domain of LARP6 is sufficient to pull down RHA, we expressed the HA-tagged C-terminal domain (LARP6ΔC) and performed immunoprecipitation (Fig. 2B, lane 4). The C-terminal domain (C-TER) pulled down RHA as efficiently as full-size LARP6 (Fig. 2B, cf. lanes 1 and 4). As the control in this experiment, the LARP6(ΔC/RRM) construct showed no interaction with RHA. These results suggested that the C-terminal domain of LARP6 is necessary and sufficient for the interaction of LARP6 with RHA.
FIGURE 2.
LARP6 interacts with RHA. (A) Schematic representation of LARP6 constructs used in immunoprecipitations (IP). All constructs had an HA-tag at the N terminus. (FS) Full-size LARP6 with the domains indicated. The ability of the constructs to bind 5′ SL RNA is indicated as + or −. (B) IP of RHA with LARP6. The constructs shown in A were transfected into HEK293 cells, and IP was performed using anti-HA antibody. IP material was analyzed by Western blot with anti-RHA antibody (RHA) and anti-HA antibody (FS, ΔC, ΔC/RRM, and C-TER). (Bottom panels) Expression of proteins in the input material. (–) Nontransfected cells. (C) Intact RNA is not required for LARP6/RHA interaction. (Left top panel) IP of RHA with FS and ΔC/RRM LARP6 constructs after digestion of the samples with RNase A (lanes 1,3) or without RNase A digestion (lanes 2,4). (Left bottom panel) Expression of proteins in the input material. (Right panel) RNA from untreated extracts (lane 2) and extracts treated with RNase A (lane 3) was analyzed by agarose gel electrophoresis. (Lane 1) Size marker. (D) Interaction of LARP6 and RHA in the nuclear extract. HEK293 cells were transfected with full-size HA-tagged LARP6, and cytosolic (CYT) and nuclear (NUC) extracts were used for IP with anti-HA antibody (lanes 1,2), protein A/G beads without antibody (*, lanes 3,4), and anti-fibronectin antibody (lanes 5,6). (Top panel) Analysis of RHA in the IP samples. (Bottom panel) Analysis of the proteins in the input material.
It is still possible that LARP6/RHA interaction requires intact collagen mRNA. To test this, we treated the lysate with RNase A prior to immunoprecipitation. Figure 2C shows that RHA is able to bind to LARP6 even after digestion of the total RNA (Fig. 2C, lane 1), although the interaction was stronger without RNase A treatment (Fig. 2C, cf. lanes 1 and 2). The C-terminus-truncated LARP6 (ΔC/RRM) did not show any interaction with RHA, as before (Fig. 2C, lanes 3,4). The right panel in Figure 2C shows that the RNase A treatment completely degraded RNA in the lysate (lane 3). This indicated that the interaction of RHA and LARP6 is not mRNA-dependent.
LARP6 is found in the nucleus and cytoplasm of collagen-producing cells (Cai et al. 2010a). To test if RHA and LARP6 can form a complex in nuclear extracts, we fractionated cellular extracts into nuclear and cytosolic fractions and performed the immunoprecipitation experiments (Fig. 2D). The association between LARP6 and RHA was found in both nuclear and cytosolic extracts (Fig. 2D, lanes 1,2). Control immunoprecipitations using protein A/G beads only (Fig. 2D, lanes 3,4) or anti-fibronectin antibody (lanes 5,6) did not pull down RHA, suggesting the specific interaction between LARP6 and RHA. As a control to show that there was no cross-contamination of the fractions, we probed the cytosolic and nuclear extracts for the presence of tubulin, which is a strictly cytosolic protein. The absence of tubulin in nuclear fractions suggested that there was no contamination of the nuclear extract with cytosolic proteins (Fig. 2D, bottom panel). These results show that LARP6 can associate with RHA in the nucleus and, thus, prior to the onset of translation initiation on collagen mRNAs.
LARP6 mediates RHA association with collagen α1(I) and α2(I) mRNAs in vivo
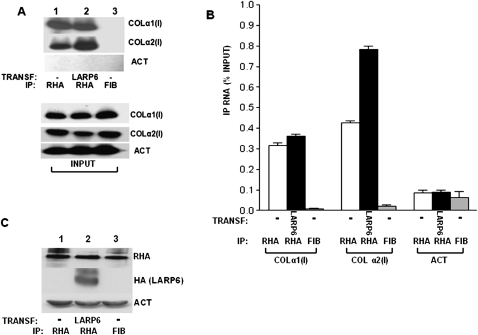
Next, we wanted to test if RHA is associated with collagen mRNAs in vivo. LARP6 interacts with 5′ SL of collagen α1(I) and α2(I) mRNAs with high affinity, and immunoprecipitation experiments using LARP6 always pull down both collagen mRNAs from the cytosolic extracts (Cai et al. 2010a). Since LARP6 binds RHA, we tested if antibodies against RHA can also pull down both collagen mRNAs in human lung fibroblasts (HLFs), which express detectable level of endogenous LARP6. After immunoprecipitation with anti-RHA antibody, we analyzed the immunoprecipitated material by RT-PCR using primers specific for collagen α1(I) and α2(I) mRNAs and with radiolabeling of the PCR products (radiolabeled RT-PCR) (Fig. 3A). Collagen α1(I) and α2(I) mRNAs were strongly immunoprecipitated with anti-RHA antibody (Fig. 3A, lane 1), while the immunoprecipitation reaction with the nonspecific antibody (anti-fibronectin antibody) was negative (Fig. 3A, lane 3). Actin mRNA was not pulled down with any antibody, suggesting that the interaction of collagen mRNAs with RHA is specific. When LARP6 was overexpressed in HLFs, there was no increase in the amount of collagen α1(I) mRNAs coimmunoprecipitated with RHA, but collagen α2(I) mRNA was coimmunoprecipitated more efficiently (Fig. 3A, cf. lanes 1 and 2). Overexpression of LARP6 did not change the input levels of collagen mRNAs, which are shown in the bottom panel of Figure 3A. Figure 3B shows more quantitative determination of collagen mRNAs in the immunoprecipitate by real-time PCR. This analysis verified the specific immunoprecipitation of collagen mRNAs with RHA and that overexpression of LARP6 increased the pull-down of collagen α2(I) mRNA twofold. The input levels of proteins are shown in Figure 3C.
FIGURE 3.
Association of collagen mRNAs with RHA in human lung fibroblasts (HLFs). (A) HLFs (lanes 1,3) and HLFs transduced with adenovirus expressing LARP6 (lane 2) were used for IP with anti-RHA antibody (lanes 1,2) or anti-fibronectin antibody (lane 3). The IP material was analyzed for the presence of collagen α1(I), collagen α2(I), and actin (ACT) mRNAs by radiolabeled RT-PCR. (Bottom panel) Expression of the mRNAs in the input material. (B) Analysis of collagen mRNAs in the IP by real-time PCR. The amount of immunoprecipitated mRNAs is presented as the percentage of the amount in the input. The bars representing the standard error of the mean are indicated. (C) Protein expression of endogenous RHA and transduced LARP6 from the input material. Loading control: actin (ACT).
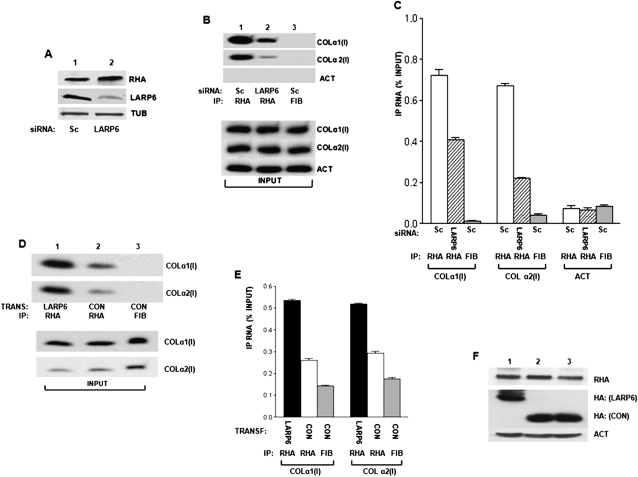
To further determine if LARP6 is mediating the interaction of RHA with collagen mRNAs, we knocked down LARP6 by siRNA in human lung fibroblasts (HLFs) using LARP6-specific siRNA, which was expressed from an adenovirus as shRNA (Cai et al. 2010a). Adenoviral delivery of LARP6 shRNA reduced LARP6 protein expression to <50% and did not affect RHA expression or tubulin expression (Fig. 4A, cf. lanes 1 and 2). Using the LARP6 knockdown cells, we performed the pull-down with antibodies against RHA and analyzed the immunoprecipitation of collagen α1(I) and α2(I) mRNAs by radiolabeled RT-PCR. The amount of collagen mRNAs in the input was similar in cells expressing LARP6 shRNA and scrambled shRNA and is shown at the bottom of Figure 4B. Figure 4B, top panel, shows that the pull-down of RHA with both, collagen α1(I) and α2(I) mRNAs, was reduced when LARP6 was knocked down, compared to the cells expressing scrambled shRNA (Fig. 4B, cf. lanes 1 and 2). The control immunopercipitation reaction with anti-fibronectin antibody was negative (Fig. 4B, lane 3), and actin mRNA was not immunoprecipitated with any antibody. To provide more quantitative assessment of the amount of collagen mRNAs in the immunoprecipitate, we analyzed the samples by real-time PCR (Fig. 4C). This analysis showed that knockdown of LARP6 decreased the fraction of collagen α1(I) mRNA associated with RHA twofold and that of α2(I) mRNA 2.5-fold. Thus, our data suggest that RHA binding to the type I collagen mRNAs may be LARP6-dependent.
FIGURE 4.
Association of RHA with collagen mRNAs is LARP6-dependent. (A) Knockdown of LARP6 in HLFs. HLFs were transduced with adenovirus expressing control shRNA (SC, lane 1) or LARP6-specific shRNA (LARP6, lane 2). Expression of endogenous RHA, LARP6, and tubulin was analyzed by Western blot. (B) Knockdown of LARP6 decreases association of RHA with collagen mRNAs. (Top panel) Control shRNA (Sc, lanes 1,3) or LARP6-specific shRNA (LARP6, lane 2) was expressed in HLFs, and IP was done with anti-RHA antibody (lanes 1,2) or anti-fibronectin antibody (lane 3). The IP samples were analyzed for pull-down of collagen α1(I), collagen α2(I), and actin mRNA by radiolabeled RT-PCR. (Bottom panel) Analysis of mRNAs in the input material. (C) Analysis of collagen mRNA in the IP by real-time PCR. The data are presented as means and standard errors of the mean from three independent experiments. (D) LARP6 enhances association of RHA with collagen mRNAs. HEK293 cells were transfected with LARP6 (lane 1) or control protein RBMS3 (lanes 2,3). IP was performed with anti-RHA antibody, and the IP material was analyzed by RT-PCR for presence of collagen α1(I) and collagen α2(I) mRNAs by radiolabeled RT-PCR. (Bottom panel) Analysis of mRNAs in the input material. (E) Analysis of collagen mRNA in the IP by real-time PCR. Error bars, ±1 standard error of the mean (SEM). (F) Analysis of proteins in the input material. Endogenous RHA and loading control actin (ACT), HA blot with transfected LARP6 and RBMS3 (CON).
To assess if overexpression of LARP6 could increase association of collagen mRNAs with RHA, we used HEK293 cells, which express low levels of LARP6 (Cai et al. 2010a). These cells also express much less collagen mRNA. We overexpressed control RNA binding protein, RBMS3 (Fig. 2D, lane 2; Fritz and Stefanovic 2007), and LARP6 (Fig. 2D, lane 1) and compared how much collagen mRNAs can be pulled down with anti-RHA antibody. RHA was able to pull down a small amount of collagen α1(I) and α2(I) mRNAs in the presence of the control protein. However, more collagen mRNAs were found in complex with RHA when LARP6 was overexpressed (Fig. 2D, cf. lanes 2 and 3). As a control, anti-fibrinogen antibody failed to immunoprecipitate collagen mRNAs (Fig. 2D, lane 3). The input levels of collagen mRNAs are shown in the bottom panel of Figure 4D. Real-time PCR analysis of the collagen mRNAs in the immunoprecipitate verified that overexpression of LARP6 can increase the association of collagen mRNAs with RHA twofold (Fig. 4E). The levels of the relevant proteins in the input material are shown in Figure 4F. These results further supported our hypothesis that LARP6 is involved in tethering RHA to collagen mRNAs.
RHA is required for efficient synthesis of type I collagen
The interaction of RHA with LARP6 and collagen mRNAs suggests that there is a functional role of RHA in expression of type I collagen. To assess this role, we knocked down RHA by transfecting two RHA-specific siRNAs and used scrambled siRNA as a control (for sequences, see Table 1) into HEK293 cells. HEK293 cells were used because they express low but detectable levels of type I collagen and can be efficiently transfected with siRNAs. By using a mixture of two RHA-specific siRNAs, its level was constantly reduced by ∼80%, as presented by Western blot (Fig. 5A, lane 2). The samples were then analyzed for expression of collagen α1(I) polypeptide. Depletion of RHA resulted in a severalfold decrease in the collagen α1(I) polypeptide steady-state level, while the control protein tubulin (TUB) was unaffected (Fig. 5A, cf. lanes 1 and 2). Collagen α2(I) polypeptide could not be detected in HEK293 cells with the available antibodies. To verify that this phenotype was specific to the depletion of RHA, we rescued RHA expression by transfecting the cells with an RHA construct that could not be targeted by the siRNAs used (RHA*). In cells in which RHA was knocked down, collagen α1(I) polypeptide was again greatly reduced (Fig. 5B, lane1). When RHA* was transfected into the RHA knockdown cells, the expression of RHA was restored to a level twofold higher than in control cells, and collagen expression was rescued to the level seen in the control cells (Fig. 5B, cf. lanes 2 and 3). Tubulin expression was not affected by any treatment. This indicates that the effect of RHA knockdown on collagen expression is specific and that RHA is required to maintain high expression of type I collagen.
TABLE 1.
Primers used for RT-PCR and sequence of siRNAs used
FIGURE 5.
RHA is required for efficient synthesis of collagen protein. (A) Knockdown of RHA decreases collagen expression. HEK293 cells were transfected with control siRNA (Sc, lane 1) or with two RHA-specific siRNAs (RHA, lane 2), and the expression of RHA, collagen α1(I) polypeptide, and tubulin was analyzed by Western blot. (B) Rescue of collagen expression by the siRNA-resistant RHA gene (RHA*). RHA was knocked down by RHA-specific siRNAs (RHA, lanes 1,2) or the cells were transfected with control siRNA (Sc, lane 3). The siRNA-resistant RHA construct (RHA*) was then transfected (lane 2), or the cells were untransfected (lanes 1,3). Collagen α1(I), RHA, and tubulin expression was measured by Western blot. (C) Knockdown of RHA does not affect expression of collagen mRNAs. Total RNA from the experiment in A was analyzed for expression of collagen α1(I), collagen α2(I), and GAPDH mRNA by quantitative real-time RT-PCR. Relative expression of collagen α1(I) and collagen α2(I) mRNAs was standardized by the internal control GAPDH. Error bars, ±1 SEM.
We also analyzed the expression of collagen mRNAs in RHA-depleted cells. Real-time RT-PCR showed no change in the steady-state level of collagen α1(I) and α2(I) mRNAs in the RHA knockdown samples compared with the scrambled (Sc)–treated control samples (Fig. 5C). This result suggests that RHA regulates translation and not the stability of collagen mRNAs or transcription of collagen genes.
Polysomal distribution of RHA
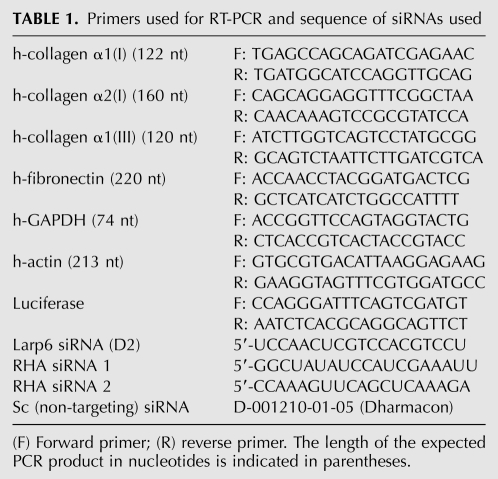
The experiments shown in Figure 5 suggested that RHA may facilitate translation of collagen mRNAs. To assess in which step of translation of collagen mRNAs, RHA is involved, we first analyzed if RHA is found on polysomes. We examined the distribution of RHA in polysomal fractions prepared from HEK293 cells by Western blot. Figure 6A, top panel, shows the OD260 profile of the sucrose fractions of HEK293 cells, and the distribution of ribosomal RNA in the fractions is shown in the bottom panel. From this analysis, we estimated that fractions 1–9 contained polysomes, fractions 10–14 contained ribosomal subunits, and fractions 16 and17 were ribosome-free cytosolic fractions. This overall polysomal profile did not significantly change when RHA was knocked down by siRNAs, suggesting that the overall distribution of polysomes was not greatly affected. Figure 6B shows the knockdown of RHA protein in these experiments.
FIGURE 6.
Collagen mRNAs are inefficiently translated in the absence of RHA. (A) Polysomal profile of HEK293 cells with and without RHA. (Top panel) Cells were transfected with RHA-specific siRNA or control siRNA and polysomes fractionated on linear sucrose gradients (fraction 1, 45% sucrose; fraction 16, 15% sucrose; fractions 16 and 17, postpolysomal supernatant). (Top panel) The OD260 of the fractions from cells transfected with siRNAs (RHA) (dotted line) and control siRNA(Sc) (full line) is shown. (Bottom panel) Distribution of ribosomal RNA in the fractions. Fractions containing polysomes are indicated. (B) Knockdown of RHA by siRNA. Control siRNA (Sc, lane 1) and RHA-specific siRNA (RHA, lane 2) were transfected into HEK293 cells, and expression of RHA was analyzed by Western blot. Loading control: tubulin (TUB). (C) RHA distribution in polysomal fractions of HEK293 cells. (Top panel) Sucrose fractions as in A were probed for the presence of RHA by Western blot. (Bottom panel) The fractions were analyzed by Western blot for LARP6. (D) Polysomal loading of collagen α1(I) mRNA is RHA-dependent. Polysomes were fractionated from HEK293 cells transfected with control siRNAs (Sc, top panel) or RHA-specific siRNA (RHA, bottom panel), and the fractions were analyzed for collagen α1(I) mRNA by radiolabeled RT-PCR. (E) Polysomal loading of collagen α2(I) mRNA is RHA-dependent. Same experiment as in C, except collagen α2(I) was analyzed. (F) Knockdown of RHA does not affect polysomal loading of GAPDH mRNA. Same experiment as in C, except GAPDH mRNA was analyzed.
When RHA was analyzed in the polysomal fractions of HEK293 cells (Fig. 6C, top panel), it was found in fractions representing polysomes, but also in fractions 10–14, representing free ribosomes and ribosomal subunits, as well as in postpolysomal supernatant (fractions 16 and 17). LARP6 was analyzed in the same fractions and was found predominantly in the lightest fractions (fractions 14 and 15) and in the postpolysomal supernatant (Fig. 6C, bottom panel). The long exposure of the Western blot, as shown in Figure 6C, indicated that only tracing amounts of LARP6 can be seen in polysomal fractions. Such distribution of LARP6 is consistent with its role in translation initiation, rather than in elongation, but the latter cannot be completely excluded. Based on the distribution of LARP6 and RHA in sucrose gradients, it is likely that LARP6 and RHA interact freely in the cytosol and before formation of polysomes. This is further supported by the fact that RHA and LARP6 can also interact in nuclear extracts (Fig. 2D).
RHA is necessary for polysomal loading of collagen mRNAs
Since knockdown of RHA does not appear to dramatically change general formation of polysomes (Fig. 6A), we tested if RHA is specifically needed for formation of polysomes on collagen mRNAs. To test this, we knocked down RHA by siRNAs and compared the polysomal distribution of collagen mRNAs with that of control siRNA. As a control, we analyzed the distribution of GAPDH mRNA. In control cells, collagen α(I)1 (Fig. 6D, top panel) and α2(I) mRNAs (Fig. 6E, top panel) were found in the polysomal fractions (fractions 1–9). Compared with the amount found in the non-polysomal fractions, it appears that ∼50% of these mRNAs were actively translated. When RHA was knocked down, collagen α1(I) and α2(I) mRNAs were not found on polysomes, except in fraction 9, representing the smallest polysomes, and accumulated in fractions containing ribosomal subunits (fractions 10–13) (Fig. 6D,E, bottom panels). The polysomal loading of GAPDH mRNA was not affected by knockdown of RHA (Fig. 6F). This clearly indicated that RHA is needed for efficient formation of polysomes on collagen mRNAs, suggesting that RHA is an essential and specific factor for translation of collagen mRNAs.
RHA stimulates collagen translation in a 5′ SL-dependent manner
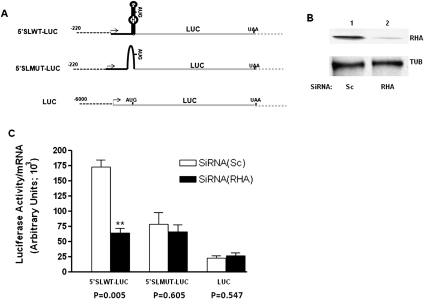
It was reported that RHA stimulates translation of mRNAs containing a PCE (Fujii et al. 2001; Zhang and Grosse 2004; Hartman et al. 2006; Bolinger et al. 2007). We hypothesized that the 5′ stem–loop may serve as the collagen mRNA–specific PCE. To test if collagen's 5′ UTR can confer translational regulation onto a heterologous mRNA and thus act as a PCE, we created a series of reporter genes encoding luciferase protein (Fig. 7A). One reporter gene had the 5′ UTR of human collagen α1(I) mRNA with the intact 5′ stem–loop (5′ SLWT-LUC), while the other had collagen α1(I) 5′ UTR but the 5′ stem–loop was abolished (5′ SLMUT-LUC). Otherwise, these genes were identical and were driven by the same promoter. The third construct had the 5′ UTR of luciferase and was driven by a different promoter (Stefanovic et al. 2000, 2005; Jiang and Stefanovic 2008); this gene (LUC) did not have any collagen sequences in the 5′ UTR. The constructs were transfected into HEK293 cells, which had either a normal level of RHA (pre-transfected with scrambled siRNA) or had the RHA knocked down by RHA-specific siRNAs. Figure 7B shows the level of RHA in control and knocked down cells. The luciferase protein was then measured by a standard assay and normalized to the total protein in the extract. Luciferase mRNA was analyzed by real-time RT-PCR using primers specific for the luciferase sequence and normalized to the endogenous GAPDH mRNA, measured in the same experiment. Thus, the expression of both, luciferase protein and luciferase mRNA, was normalized to the endogenous protein or mRNA, and the ratio of LUCprot/LUCmRNA was plotted in Figure 7C. The reporter mRNA with wild-type (WT) collagen 5′ SL produced a high level of luciferase protein, but when RHA was knocked down, there was a threefold reduction in luciferase protein synthesis per mRNA (p = 0.005) (Fig. 7C). There was no significant difference in luciferase expression with or without RHA from the reporter mRNA with the 5′ SL mutated (p = 0.605). Likewise, the reporter mRNA with no collagen sequences produced luciferase protein in an RHA-independent manner (p = 0.547). This indicates that RHA stimulates translation of a heterologous mRNA having a collagen 5′ stem–loop and that the other sequences in the collagen 5′ UTR cannot substitute for the lack of the 5′ stem–loop.
FIGURE 7.
Stimulation of translation by RHA depends on the presence of collagen 5′ SL. (A) Schematic representation of the collagen/luciferase reporter genes. (Dotted line) Promoters; (black line) collagen 5′ UTR; (gray line) luciferase sequence. The transcription start site is marked by an arrow, and positions of the start and stop codons are indicated. (B) Immunoblot showing knockdown of RHA by siRNA. (C) Knockdown of RHA affects protein expression from the mRNA with 5′ SL. RHA-specific siRNA (RHA, black bars) and control siRNA (Sc, open bars) were transfected into HEK293 cells, followed by transfection of the reporter genes. (The two stars represent the significance p < 0.01.) Expression of luciferase protein was measured as the enzymatic activity and was normalized to the total protein in the extract. Luciferase mRNA was measured by real-time RT-PCR and was normalized to GAPDH mRNA. The ratio of luciferase protein to mRNA was plotted on the y-axis. The results of five independent experiments with error bars of ±1 SEM and the P-values are shown.
RHA is up-regulated during hepatic stellate cell (HSC) activation
Hepatic stellate cells (HSCs) are liver cells responsible for excessive collagen synthesis in fibrosis (Friedman 1996, 1999, 2008). When isolated from normal rat livers, HSCs are quiescent and do not synthesize large amounts of type I collagen. However, when cultured in vitro, they spontaneously activate into myofibroblasts and up-regulate collagen synthesis 50–100-fold (Friedman 1996, 2008). The activation starts at Day 4 in culture and is maximal by Day 8, and the collagen level gradually increases during this time period. Thus, HSCs are an excellent model to follow the changes in gene expression from collagen-nonproducing to collagen-producing cells. We used these cells to assess if there is a temporal association between the RHA expression and type I collagen expression.
HSCs were isolated from rat livers and cultured in vitro (Weiskirchen and Gressner 2005). The samples were collected at Days 2, 4, 6, and 8 in culture and analyzed by Western blot for collagen α1(I) and RHA protein expression (Fig. 8A). As controls, we analyzed expression of tubulin, which does not change during activation, and expression of α-smooth muscle actin (αSMA), which is a marker of activation and expression of which gradually increases (Mak and Lieber 1988). At Day 2, when the cells are still quiescent, there was no detectable expression of RHA by Western blot (Fig. 8A, lane 1). However, more sensitive detection of RHA mRNA by RT-PCR showed a low level of expression (Fig. 8B, lane 1), suggesting that RHA is expressed, but at low levels. Collagen and αSMA were also not expressed at Day 2 (Fig. 8A, lane 1). At Day 4, RHA protein was clearly detected by Western blot (Fig. 8A, lane 2), which correlated with the increased level of its mRNA (Fig. 8B, lane 2). Type I collagen protein was still undetectable at this time point (Fig. 8A, lane 2), suggesting that up-regulation of RHA precedes the onset of collagen expression. Expression of RHA slightly increases at Days 6 and 8, which coincides with a large increase in collagen synthesis and an increase in expression of the marker of myofibroblasts, αSMA. These results indicate that, in the cells that undergo transformation from collagen-nonproducing into collagen-producing cells, RHA is temporally expressed before type I collagen and that later its levels closely parallel that of collagen. This is consistent with the role of RHA in high levels of collagen synthesis.
FIGURE 8.
Temporal expression of RHA and type I collagen in activation of hepatic stellate cells (HSCs). (A) Temporal expression of RHA protein. HSCs were isolated from rat livers and plated into uncoated plastic dishes. The cells were cultured for the indicated time periods, collected, and analyzed for expression of RHA, collagen α1(I), α-smooth muscle actin, and tubulin by Western blot. (B) Temporal expression of RHA mRNA. Same experiment as in A, except total RNA was analyzed by radiolabeled RT-PCR for RHA and actin (ACT) mRNAs.
DISCUSSION
Previous studies described the role of RNA helicase A in transcription (Aratani et al. 2001; Fujii et al. 2001; Zhang and Grosse 2004), mRNA processing (Lee and Hurwitz 1992; Nakajima et al. 1997; Lee et al. 1998; Li et al. 1999), and translation of viral and cellular mRNAs with highly structured 5′ UTRs containing a PCE (Zhang and Grosse 2004; Hartman et al. 2006). This study describes the role of RHA in translation of collagen mRNAs, which have only one clearly defined structural element in the 5′ UTR, the 5′ SL (Stefanovic et al. 1999, 2000; Stefanovic and Brenner 2003). We showed that (1) RHA interacts with LARP6 and is tethered to the collagen mRNAs in vivo. (2) RHA is required for high collagen synthesis; knockdown of RHA results in poor loading of polysomes on collagen mRNAs. (3) The effects of RHA on translation of collagen mRNAs are mediated by the 5′ SL. (4) Expression of RHA is coregulated with the expression of type I collagen in cells that differentiate from collagen-nonproducing into collagen-producing cells. These results suggest that RHA is a critical factor for high levels of synthesis of type I collagen.
Although it has been described that RHA can bind RNA (Hartman et al. 2006; Bolinger et al. 2007), we could not demonstrate high affinity of binding of RHA to 5′ SL using gel mobility shift experiments. This does not exclude the possibility that RHA can interact with the 5′ SL with low affinity or with some other sequence in collagen α1(I) and α2(I) mRNAs. However, since LARP6 and RHA interact in an RNA-independent manner (Fig. 2C), it is likely that RHA is tethered to collagen mRNAs by interaction with LARP6. LARP6 is the only protein that binds the 5′ SL of collagen mRNAs with high affinity, and it was proposed that it regulates translation (Cai et al. 2010a). Collagen α1(I) and α2(I) mRNAs are designed to be poorly translated, because in their 5′ UTR there are two short uORFs and the start codon is buried within the 5′ stem–loop and not in the optimal sequence context for the initiation codon (Stefanovic et al. 1999, 2000, 2002; Stefanovic and Brenner 2003). The recruitment of RHA may be necessary to increase their translational competitiveness. This is best evidenced by the fact that knockdown of RHA results in poor formation of polysomes on collagen mRNAs (Fig. 6) and low cellular levels of collagen protein (Fig. 5). We found that RHA is associated with polysomes in HEK293 cells (Fig. 6C), and a similar finding was reported before (Jin et al. 2011). Knockdown of RHA does not significantly change the polysomal profile of HEK293 cells (Fig. 6A), so it is not likely that RHA is required for translation elongation of the majority of mRNAs. Its association with polysomes suggests a role in translation elongation of a selected subset of mRNAs, possibly including collagen mRNAs. Significant amounts of RHA were found associated with ribosomal subunits and in postpolysomal supernatant. LARP6 is also found predominately in the postpolysomal supernatant (Fig. 6C; Cai et al. 2010a), suggesting that LARP6 and RHA form a complex prior to translation initiation on collagen mRNAs. In this way, RHA may unwind the 5′ stem–loop during the translation initiation phase and promote ribosomal recognition of the collagen main reading frame.
RHA is also found in the nucleus and can interact with LARP6 in the nucleus (Fig. 2D), again indicating that it may be recruited to collagen mRNAs at early stages of their metabolism.
Since LARP6 interacts with RHA in the absence of intact RNA (Fig. 2), it is probably tethered to the 5′ stem–loop of collagen mRNAs by interaction with LARP6. LARP6 interacts with RHA with its C-terminal domain, and LARP6 lacking the C-terminal domain strongly suppresses collagen synthesis (Cai et al. 2010b). That this tethering is required for high levels of collagen synthesis is also evidenced by using reporter genes (Fig. 7). When 5′ stem–loop was introduced into a reporter gene, the high protein expression from this gene was observed when RHA was present and was threefold lower when RHA was knocked down. Two reporter mRNAs without the 5′ stem–loop were translated in an RHA-independent manner. This again suggests that RHA is not a general stimulator of translation and that it must associate with a particular mRNA to exert its effect. RHA and other RNA helicases, such as DDX3 and Ded1, complement RNA helicase eIF4A and, if recruited, can enhance translation of the specific mRNAs (Lee and Hurwitz 1992; Chuang et al. 1997; Lee et al. 2008). The PCE elements were proposed to tether RHA to some viral and cellular mRNAs (Hartman et al. 2006), while our results indicate that 5′ SL and LARP6 serve this function for collagen mRNAs. Eif4H was pulled down as one of the proteins using 5′ SL RNA (Fig. 1). Although its function in translation of collagen mRNAs was not studied, it is possible that ei4H also contributes to efficient translation of collagen mRNAs.
In conditions of high collagen synthesis, like in wound healing or fibrosis, there is activation of quiescent fibroblasts and stellate cells, which produce low levels of type I collagen, into myofibroblasts, which up-regulate expression of type I collagen 50–100-fold (Stefanovic et al. 1995, 1997, 1999; Stefanovic and Brenner 2003; Lindquist et al. 2004; Rippe and Brenner 2004). Cultivation of HSC in vitro mimics this process (Friedman 1996, 1999) and enabled us to compare the temporal profiles of RHA and collagen expression (Fig. 8). In activation of HSCs, RHA is expressed at low levels in quiescent HSCs. Its expression was undetectable by Western blot (Fig. 8A), but RT-PCR analysis showed low expression (Fig. 8B). During activation, an increase in RHA expression is observed before an increase of type I collagen expression, consistent with the hypothesis that RHA activity is a prerequisite for high collagen synthesis. Activation of HSCs is associated with dramatic up-regulation and down-regulation of many genes (Friedman 1996). Although RHA is required for type I collagen synthesis, it is possible that during this process, other mRNAs may also require activity of RHA. However, so far, only junD mRNA was reported to require RHA for translation (Hartman et al. 2006).
In conclusion, we have identified one of the key interactions necessary for high levels of collagen synthesis, the interaction of LARP6 and RHA. This interaction is needed to tether RHA to the 5′ UTR of collagen mRNAs, which have the 5′ SL as the high-affinity docking site of LARP6. When tethered to collagen mRNAs, RHA may help in unwinding the secondary structures and assembly of the ribosomes competent for translational elongation of the main reading frame. This is the first description of the role of RHA in synthesis of extracellular matrix proteins and may have profound implications for future development of antifibrotic drugs.
MATERIALS AND METHODS
Plasmid and adenovirus construction
Full-size Larp6 was cloned into a pCDNA3 vector (Stratagene) with an HA-tag at the N terminus. Deletion mutants were constructed using conventional restriction enzyme HindIII (ΔC/RRM) and XcmI (ΔC) (Cai et al. 2010a), and by cloning of the PCR amplified C-terminal region. The identity of the mutants was confirmed by Western blot and sequencing.
Adenoviruses were constructed by recloning the constructs into a pAdCMV-TRACK vector, followed by recombination with a pAd-Easy vector, as described (He et al. 1998). All adenoviruses also expressed GFP from a separate transcription unit to monitor transduction efficiency. Adenoviruses were amplified and purified as described (He et al. 1998).
A luciferase reporter gene containing the 5′ UTR of the mouse collagen α1(I) gene (5′ SLWT-LUC) was constructed by cloning 220 nt of the promoter of the mouse collagen α1(I) gene together with the complete 5′-UTR sequence and 5′ stem–loop into the Basic Luciferase Vector (Promega). The reporter gene with the mutated 5′ stem–loop (5′ SLMUT-LUC) was made from the above construct by mutating sequences involved in formation of the 5′ SL. The reporter gene containing no collagen 5′ UTR was described before (Stefanovic et al. 2005; Jiang and Stefanovic 2008). It was driven by a hybrid promoter consisting of the upstream collagen promoter and the proximal SV40 promoter, followed by the luciferase open reading frame of the Basic Lucifirase Vector (Promega) with 6000 nt of the mouse collagen α1(I) promoter.
Cells and transfections
Human lung fibroblasts (HLFs), hepatic stellate cells (HSCs), and HEK293 cells were grown under standard conditions (Graham et al. 1977). HEK293 cells and human fibroblasts were transfected with 1 μg of plasmid per 35-mm culture dish using 293TransIT reagent (Mirus). The cells were harvested 48–72 h after the transfection. SiRNAs were transfected using Lipofectamine 2000 reagent (Invitrogen) at the final concentration of 125 nM per 100,000 cells. The sequence of the siRNA is shown in Table 1. Adenoviruses were used at an MOI of 500, which resulted in >95% transfection of HLFs (Cai et al. 2010a).
HSCs were purified by perfusing rat livers with 0.5 mg of pronase and 0.04 mg of collagenase per gram of animal weight and centrifugation of the cell suspension over a 20% Nycodenz gradient (Weiskirchen and Gressner 2005). The purity of the cells was assessed by immunostaining with anti-desmin antibody. The cells were cultured on uncoated plastic dishes in DMEM with 10% FCS for the indicated time periods.
RT-PCR analysis
Total RNA was isolated using an RNA isolation kit (Sigma-Aldrich). The RNA was treated with DNase I to remove contaminating DNA and analyzed by two RT-PCR techniques. RT-PCR with radiolabeling of the PCR products was performed as previously described (Stefanovic et al. 2000; Fritz and Stefanovic 2007; Cai et al. 2010a; Challa and Stefanovic 2011). Briefly, 50 ng of total RNA was reverse-transcribed using rTth reverse transcriptase (Boca Scientific) and the primer specific for the mRNA under analysis. The PCR reactions were performed in the presence of [α-32P]dATP. PCR products were visualized by autoradiography. The identity of the PCR bands was verified by sequencing and by their expected size.
Quantitative real-time RT-PCR analysis
Total RNA was isolated using an RNA isolation kit (Sigma-Aldrich). The RNA was treated with DNase I to remove contaminating DNA. The cDNA for the quantitative real-time polymerase chain reaction (qRT-PCR) was synthesized using SuperScript II RT (Invitrogen) following the manufacturer's protocol. Five percent of the cDNA was used in qRT-PCR (BioRad-IQ5 Thermocycler) using specific primers with the sequences shown in Table 1. The qRT-PCR was performed in triplicates of each RNA immunoprecipitation experiment. The threshold cycle (CT) was computed using IQ-5 software (Bio-Rad) with standard curves constructed for each primer set with a stepwise dilution of input DNA with an efficiency of 100% (±15%). The analysis of the dissociation curve was performed at the end of 40 cycles. The enrichment values presented were acquired by the data calculated as percent input of IP-RNA as a percent of total immunopercipitated RNA to input fraction (Figs. 3B, 4C,E). The total collagen mRNA (Fig. 5C) and the luciferase mRNA expression (Fig. 7C) were standardized to the GAPDH and the negative control (empty vector). The equation for the determination of error propagation (Standard Error) for normalized expression and the graphics were computed using the biostatistics comprehensive program GraphPad Prism 3.02. The statistical data were analyzed on Microsoft Excel and GraphPad Prism 3.02. Values are obtained from three independent experiments ± standard error of the mean. One-way ANOVA followed by a Tukey's multiple comparison test, p < 0.05, was used to determine significance of enrichment. The P-values in Figure 7C were determined by the Student's t-test.
Western blotting
Cytosolic proteins were prepared by lysis in isotonic buffer containing 0.5% NP-40. The protein concentration was estimated using a Bradford assay. The following antibodies were used: anti-collagen α1(I) polypeptide antibody from Rockland, anti-collagen α2(I) polypeptide antibody from Santa Cruz Biotechnology, anti-fibronectin antibody from BD Transduction Laboratories, anti-tubulin antibody from Cell Signaling, anti-Larp6 antibody from Abnova, anti-RHA antibody from Abcam, and anti-HA and anti-Flag antibodies from Sigma-Aldrich.
Cytosolic cell extracts were prepared in isotonic buffer (140 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl at pH 7.6, 0.5% NP-40), and after removal of the nuclei by centrifugation, the supernatant was used as a cytosolic extract. The nuclei pellet was washed three times in the same lysis buffer, and nuclear extracts were prepared as described by Dignam et al. (1983).
Biotin-RNA pull-down
For identification of 5′ SL RNA interacting proteins, biotinylated 5′ SL RNA or inverted 5′ SL RNA (CON) was incubated in cytosolyc extracts prepared from 3 × 107 human lung fibroblasts and pulled down with streptavidin agarose. After washing five times with PBS, the pull-down material was analyzed by SDS-PAGE and Coomassie staining. Proteins specifically pulled down with 5′ SL RNA were excised from the gel and identified by MALDI-TOF (FSU) or LC/MS/MS (Tufts University Core Facility).
Immunoprecipitation (IP)
Cells were lysed as for Western blots, and the nuclei were removed by centrifugation. Lysate containing 1 mg of total protein was incubated with 1 μg of the antibody for 1 h at 4°C. Twenty microliters of equilibrated protein A/G beads (Santa Cruz Biotechnology) was added to the samples and incubated for 4 h at 4°C. The beads were washed three times with PBS supplemented with 0.5% NP-40, and the samples were analyzed by Western blotting or by RT-PCR when coprecipitation of collagen mRNAs was analyzed. In some experiments, the lysate was treated with 1 μg of RNase A prior to the IP.
Fractionation of polysomes
Cells were treated with cycloheximide for 1 h to stabilize the polysomes prior to harvesting. Cell lysates were prepared from 3 × 107 cells, as described for immunoprecipitation. The lysate was laid on the top of a linear 15%–45% sucrose gradient and centrifuged for 2 h at 38,000g at 4°C (Veyrune et al. 1996; Cai et al. 2010a). Five hundred-microliter fractions were collected and the OD260's of the fractions were measured. Total RNA was extracted from the fractions by phenol:chloroform followed by isopropanol precipitation. The RNA was analyzed on a 1% agarose gel to determine the distribution of ribosomal RNA, and expression of the collagen mRNAs and GAPDH mRNA in the fractions was estimated by RT-PCR. Proteins were extracted from the fractions by precipitation with 6.5% TCA and 0.05% DOC; the protein pellets were dissolved in 0.1% SDS, 10 mM Tris (pH 6.8) and analyzed by Western blot.
Gel mobility assay
The RNA probe described in Cai et al. (2010a) was prepared by in vitro transcription from the template cloned by the pGEM3 vector (Promega). The mobility shift assay was done using 4 ng of labeled 5′ SL RNA and 20 μg (total protein) of cytosolic extracts of cells transfected with various constructs by standard laboratory protocol as described (Cai et al. 2010a). The input of transfected proteins (Fig. 2B) was estimated by theWestern blot. Quantification of the intensity of bands was done using a phosphorimager.
Luciferase assay
Lucifarese reporter constructs were transfected into HEK293 cells, and luciferase activity was measured by the standard assay 2 d after transfection (Alam and Cook 1990; Jiang and Stefanovic 2008). The activity was normalized to total protein in the extract, as loading control. Luciferase mRNA was analyzed by real time RT-PCR using primers specific for the luciferase sequence (Table 1). The mRNA expression was normalized to the expression of GAPDH, as loading control. The normalized activity of luciferase protein was divided to the normalized expression of luciferase mRNA, and the ratio is shown as arbitrary units on the y-axis. The results plotted were from three independent experiments. Student's t-test was used to assess the statistical significance. The statistical significance was set at p < 0.05, and the error bars represent ±1 SD.
ACKNOWLEDGMENTS
Special thanks to Dr. Chee-Gun Lee for the generous gift of the RHA clone; to Dr. Kathleen Boris-Lawrie for generous information in regard to siRNAs (RHA); to Sergio Iniguez for statistical analysis and help with GraphPad Prism; to Dr. Yoichi and Akiko Kato for advice on qRT-PCR; to Janel Rodriguez and Brett Mulvey for help on IQ5 Optical Software; to Lela Stefanovic for laboratory help; and to Azariyas Challa, Dr. Milica Vukmirovic, and Dr. Carlos A. Bolaños for valuable comments on this manuscript. This work was supported by the National Institutes of Health (grant number 5R01DK059466-08 to B.S.; grant number 1F31AA019845-01A1 to Z.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030288.111.
REFERENCES
- Alam J, Cook JL 1990. Reporter genes: Application to the study of mammalian gene transcription. Anal Biochem 188: 245–254 [DOI] [PubMed] [Google Scholar]
- Aratani S, Fujii R, Oishi T, Fujita H, Amano T, Ohshima T, Hagiwara M, Fukamizu A, Nakajima T 2001. Dual roles of RNA helicase A in CREB-dependent transcription. Mol Cell Biol 21: 4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K, Boswell BA, Ridgway CC, Bachinger HP 1996. Triple helix formation of procollagen type I can occur at the rough endoplasmic reticulum membrane. J Biol Chem 271: 21566–21573 [DOI] [PubMed] [Google Scholar]
- Bolinger C, Yilmaz A, Hartman TR, Kovacic MB, Fernandez S, Ye J, Forget M, Green PL, Boris-Lawrie K 2007. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res 35: 2629–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Hull S, Wang Y, Roberts TM, Boris-Lawrie K 1999. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J Virol 73: 4847–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Fritz D, Stefanovic L, Stefanovic B 2009. Coming together: Liver fibrosis, collagen mRNAs and the RNA binding protein. Expert Rev Gastroenterol Hepatol 3: 1–3 [DOI] [PubMed] [Google Scholar]
- Cai L, Fritz D, Stefanovic L, Stefanovic B 2010a. Binding of LARP6 to the conserved 5′ stem–loop regulates translation of mRNAs encoding type I collagen. J Mol Biol 395: 309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Fritz D, Stefanovic L, Stefanovic B 2010b. Nonmuscle myosin-dependent synthesis of type I collagen. J Mol Biol 401: 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa AA, Stefanovic B 2011. A novel role of vimentin filaments: Binding and stabilization of collagen mRNAs. Mol Cell Biol 31: 3773–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester N, Kuo F, Kozak C, O'Hara CD, Leder P 1998. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev 12: 3382–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH 1997. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275: 1468–1471 [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL 1996. Hepatic stellate cells. Prog Liver Dis 14: 101–130 [PubMed] [Google Scholar]
- Friedman SL 1999. Stellate cell activation in alcoholic fibrosis–an overview. Alcohol Clin Exp Res 23: 904–910 [PubMed] [Google Scholar]
- Friedman SL 2008. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 88: 125–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz D, Stefanovic B 2007. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J Mol Biol 371: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T 2001. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J Biol Chem 276: 5445–5451 [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36: 59–74 [DOI] [PubMed] [Google Scholar]
- Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K 2006. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol 13: 509–516 [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B 1998. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci 95: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull S, Boris-Lawrie K 2002. RU5 of Mason-Pfizer monkey virus 5′ long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J Virol 76: 10211–10218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Stefanovic B 2008. Homeobox gene Prx1 is expressed in activated hepatic stellate cells and transactivates collagen α1(I) promoter. Exp Biol Med (Maywood) 233: 286–296 [DOI] [PubMed] [Google Scholar]
- Jin J, Jing W, Lei XX, Feng C, Peng S, Boris-Lawrie K, Huang Y 2011. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res 39: 3724–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivirikko KI 1998. Collagen biosynthesis: A mini-review cluster. Matrix Biol 16: 355–356 [DOI] [PubMed] [Google Scholar]
- Kozak M 1989. The scanning model for translation: An update. J Cell Biol 108: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Hurwitz J 1992. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J Biol Chem 267: 4398–4407 [PubMed] [Google Scholar]
- Lee CG, da Costa Soares V, Newberger C, Manova K, Lacy E, Hurwitz J 1998. RNA helicase A is essential for normal gastrulation. Proc Natl Acad Sci 95: 13709–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R 2008. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36: 4708–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, Wong-Staal F 1999. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc Natl Acad Sci 96: 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist JN, Kauschke SG, Stefanovic B, Burchardt ER, Brenner DA 2000a. Characterization of the interaction between αCP(2) and the 3′-untranslated region of collagen α1(I) mRNA. Nucleic Acids Res 28: 4306–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist JN, Marzluff WF, Stefanovic B 2000b. Fibrogenesis. III. Posttranscriptional regulation of type I collagen. Am J Physiol Gastrointest Liver Physiol 279: G471–G476 [DOI] [PubMed] [Google Scholar]
- Lindquist JN, Parsons CJ, Stefanovic B, Brenner DA 2004. Regulation of α1(I) collagen messenger RNA decay by interactions with αCP at the 3′-untranslated region. J Biol Chem 279: 23822–23829 [DOI] [PubMed] [Google Scholar]
- Mak KM, Lieber CS 1988. Lipocytes and transitional cells in alcoholic liver disease: A morphometric study. Hepatology 8: 1027–1033 [DOI] [PubMed] [Google Scholar]
- Merrick WC 2004. Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332: 1–11 [DOI] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90: 1107–1112 [DOI] [PubMed] [Google Scholar]
- Pace JM, Wiese M, Drenguis AS, Kuznetsova N, Leikin S, Schwarze U, Chen D, Mooney SH, Unger S, Byers PH 2008. Defective C-propeptides of the proα2(I) chain of type I procollagen impede molecular assembly and result in osteogenesis imperfecta. J Biol Chem 283: 16061–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40: 515–526 [DOI] [PubMed] [Google Scholar]
- Rippe RA, Brenner DA 2004. From quiescence to activation: Gene regulation in hepatic stellate cells. Gastroenterology 127: 1260–1262 [DOI] [PubMed] [Google Scholar]
- Rippe RA, Stefanovic B 2005. Methods for assessing the molecular mechanisms controlling gene regulation. Methods Mol Med 117: 141–160 [DOI] [PubMed] [Google Scholar]
- Roberts TM, Boris-Lawrie K 2000. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J Virol 74: 8111–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze U, Hata R, McKusick VA, Shinkai H, Hoyme HE, Pyeritz RE, Byers PH 2004. Rare autosomal recessive cardiac valvular form of Ehlers-Danlos syndrome results from mutations in the COL1A2 gene that activate the nonsense-mediated RNA decay pathway. Am J Hum Genet 74: 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short JD, Pfarr CM 2002. Translational regulation of the JunD messenger RNA. J Biol Chem 277: 32697–32705 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Gingras AC 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol 10: 268–275 [DOI] [PubMed] [Google Scholar]
- Stefanovic B, Brenner DA 2003. 5′ stem-loop of collagen α1(I) mRNA inhibits translation in vitro but is required for triple helical collagen synthesis in vivo. J Biol Chem 278: 927–933 [DOI] [PubMed] [Google Scholar]
- Stefanovic B, Hellerbrand C, Brenner DA 1995. Post-transcriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Nucleic Acids Symp Ser 1995: 212–214 [PubMed] [Google Scholar]
- Stefanovic B, Hellerbrand C, Holcik M, Briendl M, Aliebhaber S, Brenner DA 1997. Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol Cell Biol 17: 5201–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Hellerbrand C, Brenner DA 1999. Regulatory role of the conserved stem-loop structure at the 5′ end of collagen α1(I) mRNA. Mol Cell Biol 19: 4334–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Lindquist J, Brenner DA 2000. The 5′ stem–loop regulates expression of collagen α1(I) mRNA in mouse fibroblasts cultured in a three-dimensional matrix. Nucleic Acids Res 28: 641–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Schnabl B, Brenner DA 2002. Inhibition of collagen α1(I) expression by the 5′ stem–loop as a molecular decoy. J Biol Chem 277: 18229–18237 [DOI] [PubMed] [Google Scholar]
- Stefanovic L, Stephens CE, Boykin D, Stefanovic B 2005. Inhibitory effect of dicationic diphenylfurans on production of type I collagen by human fibroblasts and activated hepatic stellate cells. Life Sci 76: 2011–2026 [DOI] [PubMed] [Google Scholar]
- Uitto J 1979. Collagen polymorphism: Isolation and partial characterization of α1(I)-trimer molecules in normal human skin. Arch Biochem Biophys 192: 371–379 [DOI] [PubMed] [Google Scholar]
- van Dam H, Castellazzi M 2001. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20: 2453–2464 [DOI] [PubMed] [Google Scholar]
- Veyrune JL, Campbell GP, Wiseman J, Blanchard JM, Hesketh JE 1996. A localisation signal in the 3′ untranslated region of c-myc mRNA targets c-myc mRNA and β-globin reporter sequences to the perinuclear cytoplasm and cytoskeletal-bound polysomes. J Cell Sci 109: 1185–1194 [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, Gressner AM 2005. Isolation and culture of hepatic stellate cells. Methods Mol Med 117: 99–113 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Mudryj M, de Crombrugghe B 1983. A uniquely conserved regulatory signal is found around the translation initiation site in three different collagen genes. J Biol Chem 258: 14914–14919 [PubMed] [Google Scholar]
- Zhang S, Grosse F 2004. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim Biophys Sin (Shanghai) 36: 177–183 [DOI] [PubMed] [Google Scholar]