The maturation of ribosomal RNAs (rRNAs) is an important but incompletely understood process required for rRNAs to become functional. In order to determine the enzymes responsible for initiating 3′ end maturation of 23S rRNA in Escherichia coli, the authors analyzed a number of strains lacking different combinations of 3′ to 5′ exo-RNases. Through these analyses, they identified Ribonuclease (RNase) PH as a key effector of 3′ end maturation.
Keywords: RNA processing, ribosomal RNA, Escherichia coli, ribonucleases
Abstract
The maturation of ribosomal RNAs (rRNAs) is an important but incompletely understood process required for rRNAs to become functional. In order to determine the enzymes responsible for initiating 3′ end maturation of 23S rRNA in Escherichia coli, we analyzed a number of strains lacking different combinations of 3′ to 5′ exo-RNases. Through these analyses, we identified RNase PH as a key effector of 3′ end maturation. Further analysis of the processing reaction revealed that the 23S rRNA precursor contains a CC dinucleotide sequence that prevents maturation from being performed by RNase T instead. Mutation of this dinucleotide resulted in a growth defect, suggesting a strategic significance for this RNase T stalling sequence to prevent premature processing by RNase T. To further explore the roles of RNase PH and RNase T in RNA processing, we identified a subset of transfer RNAs (tRNAs) that contain an RNase T stall sequence, and showed that RNase PH activity is particularly important to process these tRNAs. Overall, the results obtained point to a key role of RNase PH in 23S rRNA processing and to an interplay between this enzyme and RNase T in the processing of different species of RNA molecules in the cell.
INTRODUCTION
In all organisms, ribosomal RNAs (rRNAs) are synthesized as precursors and require processing by RNases to be converted to mature rRNAs (Srivastava and Schlessinger 1990; Condon 2007; Henras et al. 2008; Deutscher 2009). In Escherichia coli, the three rRNAs, 16S, 23S, and 5S, are initially transcribed as a single ribosomal operon RNA, which is first cleaved at two sites by the double strand–specific endonuclease, RNase III (Ginsburg and Steitz 1975). This cleavage results in the separation of the sequences for the three rRNAs. Each of these products still contains precursor sequences at the 5′ and 3′ ends whose removal requires the action of additional RNases. For 16S rRNA, the endonucleases RNase E and RNase G successively cleave the 115 nucleotide (nt) precursor sequence at the 5′ end to yield a mature end (Li et al. 1999b; Wachi et al. 1999). The 3′ end maturation of this RNA is also likely to be carried out by an endonuclease, but the identity of this enzyme is unknown. For 5S rRNA, the RNase III–generated product is subsequently cleaved at both the 5′ and 3′ ends by RNase E (Roy et al. 1983). The resulting product still retains 3 nt of precursor sequences at both the 5′ and 3′ ends. The 3′ end trailer sequences are digested by the 3′ to 5′ exonuclease, RNase T, whereas the enzyme responsible for 5′ end maturation remains unknown (Li and Deutscher 1995).
For 23S rRNA, the RNase III–cleaved precursor contains predominantly 7 nt of unprocessed sequences at the 5′ end and 8–9 nt at the 3′ end, which engage in long-range base-pairing interactions with each other (Fig. 1A). We have previously provided evidence that the maturation of the 3′ and 5′ ends of 23S rRNA are coordinated events, and have suggested that maturation of the 3′ end occurs first (Gutgsell and Jain 2010). Prior studies have indicated that the 3′ end maturation is a two-step process: In the first step the RNase III–cleaved product is partially digested by one or more unknown RNases, and thereafter, the remaining nucleotides are removed by RNase T (Li et al. 1999a). The starting point for the work presented here is the identification of the exonucleases that are responsible for initiating this process.
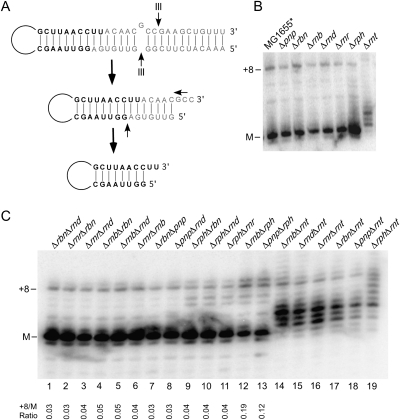
FIGURE 1.
Effect of 3′ to 5′ exonucleases on 23S rRNA 3′ end processing. (A) Schematic description of 23S rRNA processing. The precursor sequences at the 5′ and 3′ ends of 23S rRNA are complementary and form long-range base-pairs. This 23S rRNA precursor is initially cleaved by the double strand–specific RNase, RNase III. The resulting product is subsequently digested by 3′ to 5′ exonucleases to generate a mature 3′ end and by an unknown endonuclease to generate a mature 5′ end. Mature residues are indicated in black lettering; precursor residues, in gray. (B) Effect of deletions in genes encoding 3′ to 5′ exonucleases on 23S rRNA processing. RNA was isolated from a wild-type strain (MGG1655*) or from derivative strains containing Δpnp (PNPase), Δrbn (RNase BN), Δrnb (RNase II), Δrnd (RNase D), Δrnr (RNase R), Δrph (RNase PH), or Δrnt (RNase T) deletions; the encoded enzymes are indicated in brackets. The RNAs were cleaved 50 nt from the mature 3′ end of 23S rRNA using oligonucleotide-directed RNase H cleavage of 23S rRNA, followed by fractionation of the resulting product on a denaturing gel and Northern blotting using a 3′ end probe for 23S rRNA. The migration of mature RNA (M) and a prominent precursor containing eight unprocessed residues (+8) is indicated. (C) RNA was extracted from strains containing 19 different combinations of dual exonuclease gene deletions, as indicated, and analyzed by Northern blotting as described above. The +8 precursor to the mature RNA ratio is indicated below for all strains except those containing Δrnt deletions, which contain negligible levels of fully processed 23S rRNA.
RESULTS
23S rRNA processing in strains lacking 3′ to 5′ exonucleases
To identify the enzymes involved in 3′ end processing of 23S rRNA, we first examined strains lacking the known 3′ to 5′ exo-RNases. An expectation was that in strains lacking the exonucleases responsible for processing, an accumulation of unprocessed RNA would be observed. At present, eight such exonucleases are known: RNase II, RNase PH, polynucleotide phosphorylase (PNPase), RNase D, RNase R, RNase BN, RNase T, and oligoribonuclease (Orn), of which the first seven are not essential for viability. Mutations in Orn could not be tested because no mutant allele for this essential enzyme has been described. However, a role for this enzyme in rRNA maturation seemed unlikely given its specialized role in the digestion of small RNAs of 2–5 nt (Ghosh and Deutscher 1999). To determine a possible role for the other seven enzymes in 23S rRNA 3′ end maturation, RNA from strains carrying nonpolar deletion alleles in each of these RNase genes was analyzed by Northern blot. In order to detect changes in rRNA processing at the nucleotide level, 23S rRNA was first cleaved near its 3′ end using RNase H–directed oligonucleotide cleavage of 23S rRNA (Fig. 1B). In the wild-type strain, as well as in six of the deletion strains, mature 23S RNA was observed to be the major product, and a precursor containing eight unprocessed 3′ end nucleotides was a minor product. The precursor to mature rRNA ratio was similar in the wild-type strain and in each of the mutant strains, suggesting that the individual absence of these exonucleases is insufficient to impede 23S rRNA processing. For a strain lacking RNase T, the majority of the RNAs contained 1–4 nt of unprocessed RNA. This profile is consistent with its known role in the removal of the last few nucleotides from immature 23S rRNA (Li et al. 1999a). However, the abundance of precursors containing eight or more unprocessed nucleotides was no greater in this strain than in the other strains, indicating that RNase T has no significant role in initiating 23S rRNA processing.
Many processes involving exonucleases require the removal of multiple such enzymes for significant effects to be observed (Donovan and Kushner 1986; Li and Deutscher 1996). A failure to observe an accumulation of rRNA precursors in the singly deleted strains suggested that 23S rRNA processing might similarly involve multiple exonucleases. To test this possibility, a set of double-deletion strains was constructed. Of the 21 double-deletion combinations possible for the seven nonessential exonucleases, strains lacking PNPase and RNase II or lacking PNPase and RNase R could not be constructed, since these combinations cause synthetic lethality (Donovan and Kushner 1986; Cheng and Deutscher 2003). For the remaining 19 double-deletion combinations, total RNA was extracted from these strains and analyzed as described for the singly deleted strains (Fig. 1C). For two of these combinations, we consistently observed an increased ratio of precursor to mature 23S rRNA. In a strain lacking RNase PH and PNPase, the precursor to mature rRNA level was increased by approximately threefold, whereas in a strain lacking RNase PH and RNase II, the precursor to mature rRNA ratio was increased four- to fivefold. The former effect has been observed previously while examining RNA processing defects in strains lacking PNPase and RNase PH (Jain 2009). These results suggest that RNase II, PNPase, and RNase PH are primarily involved in 23S rRNA processing, with RNase PH playing a prominent role as it was the common factor absent in the two strains that displayed precursor accumulation.
Further evidence for a key role for RNase PH was suggested by an inspection of the processing profiles of the double-deletion strains lacking RNase PH and different combinations of a second exonuclease (Fig. 1C, lanes 9–13). Each of these strains showed an appearance of novel processing intermediates that contained 5–6 nt of unprocessed rRNAs. In addition, while five of the six double-deletion strains lacking RNase T and different combinations of a second exonuclease accumulated precursors containing one to four unprocessed nucleotides (lanes 14–18), a strain lacking both RNase T and RNase PH (lane 19) exhibited products containing up to seven unprocessed nucleotides. These profiles suggest that RNase PH is required to digest the longer set of products, following which the shortened RNAs can be trimmed by RNase T to complete the rRNA maturation process.
The 23S rRNA precursor contains an RNase T stall sequence
Many small RNAs, such as transfer RNAs (tRNAs), 5S rRNA, and small regulatory RNAs, adopt a terminal structure similar to the RNase III–cleaved 23S rRNA precursor, in that the 5′ and 3′ termini of these molecules form intramolecular base-pairs (Fig. 1A). These molecules are usually transcribed with 3′ trailer sequences that are subsequently removed by 3′ to 5′ exonucleases, in particular by RNase T (Li et al. 1998). These findings can be rationalized in terms of the biochemical properties of RNase T, which has shown to be particularly effective at removing the single-stranded residues that abut duplex regions (Zuo and Deutscher 2002). Since the 23S RNase III–cleaved precursor contains three unpaired nucleotides downstream from a duplex stem (Fig. 1A), a failure of RNase T to play a role in the removal of these residues was unexpected.
To verify the above-mentioned property of RNase T and compare it with different exonucleases, we performed digestion reactions using a substrate composed of an RNA duplex with 3′ single-stranded residues on one strand (Fig. 2A). Treatment of the substrate with purified RNase T, RNase PH, RNase II, or PNPase resulted in the digestion of most of the single-stranded residues in each case. However, only RNase T was able to digest all of the single-stranded residues. RNase PH was slightly less effective at removing single-stranded residues, whereas both PNPase and RNase II left multiple single-stranded residues undigested. Similar results using RNase II or PNPase on this substrate have been observed previously (Cheng and Deutscher 2005). Thus, among the RNases tested, RNase T was the most effective at digesting single-stranded residues downstream from a duplex, and hence, it would have been expected to be capable of digesting the 3′ single-stranded residues in the 23S rRNA precursor.
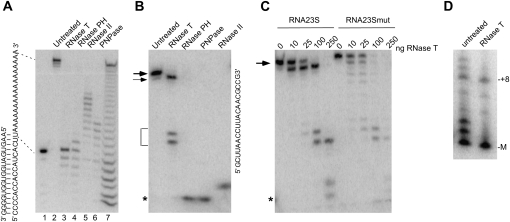
FIGURE 2.
Biochemical analysis of an RNase T stall sequence in the 23S rRNA 3′ trailer. (A) Digestion of single-stranded regions by 3′ to 5′ exonucleases. A labeled RNA substrate containing both duplex and single-stranded regions, with the longer strand labeled at its 5′ end, is shown at the left. This substrate was treated with 100 ng of purified RNase T, RNase PH, RNase II, or PNPase. The RNase-treated reactions (lanes 3–6) and a control untreated reaction (lane 2) were electrophoresed on a denaturing gel, along with a labeled RNA oligonucleotide that corresponds to the product derived from the digestion of all single-stranded residues in this complex (lane 1). An RNA ladder, generated by alkaline hydrolysis of the labeled single-stranded RNA, was also included (lane 7). The positions of the undigested RNA and of the product derived from the digestion of all single-stranded residues are indicated by dotted lines. (B) Digestion of a 23S substrate by 3′ to 5′ exonucleases. An RNA substrate, RNA23S (shown on the right), which corresponds to the unprocessed 3′ end of 23S rRNA, was radiolabeled at its 5′ end and treated with 100 ng of PNPase, RNase T, RNase PH, or RNase II, followed by the fractionation of the treated or untreated reactions on a denaturing gel. The positions of the undigested substrate or a product generated by removal of one 3′ nucleotide are indicated by thick or thin arrows, respectively. Products generated by stalling of RNase T at an internal CC dinucleotide are indicated by brackets, and limit digestion products are indicated by an asterisk. (C) Effect of mutation of the CC dinucleotide in the 23S rRNA leader sequence. Oligonucleotides RNA23S or RNA23Smut (5′-GCUUAACCUUACAACGUUG-3′), a RNA23S variant that has the 3′ CC dinucleotide of RNA23S replaced with U residues (underlined), were treated with the indicated amounts of RNase T and fractionated on a denaturing gel. The migration of the undigested substrates is indicated by an arrow. (D) Digestion of ribosomal particles with RNase T. 50S large ribosomal subunit particles were isolated from a Δrnt strain and treated with 1 μg of RNase T. RNA was extracted from RNase T–treated or –untreated particles and analyzed by Northern blot, as described in Figure 1. The positions of the mature end and the +8 precursor are indicated.
An examination of the 23S rRNA precursor sequence revealed that the two positions on the 3′ trailer immediately upstream of the RNase III cleavage site are both cytosine (Fig. 1A). Earlier work on RNase T has indicated that a CC dinucleotide sequence is particularly inhibitory to RNase T digestion and can reduce kcat/KM by up to 100-fold (Zuo and Deutscher 2002). We speculated that the ineffectiveness of RNase T to act on the 23S rRNA precursor could be due to the presence of the CC dinucleotide at its 3′ terminus. To test this hypothesis, we examined the action of several different exonucleases on a single-stranded RNA substrate (RNA23S) that corresponds to the 3′ region of 23S rRNA. This substrate contained nine residues corresponding to the mature 23S rRNA 3′ end and nine precursor residues, one more than what we have observed to be the major unprocessed product in wild-type cells (Fig. 1). Cleavage of RNA23S with PNPase, RNase II, or RNase PH resulted in the complete digestion of the RNA (Fig. 2B). With RNase T, however, only the first guanine residue preceding the CC dinucleotide sequence could be removed effectively. These observations indicate that the CC dinucleotide sequence in the 23S rRNA precursor, in fact, inhibits digestion by RNase T. Interestingly, additional bands were observed with the RNase T digestion reaction, consistent with RNase T stalling at a second, internal CC dinucleotide present within the 23S rRNA precursor.
To further confirm a role of the 3′ CC dinucleotide in inhibiting RNase T digestion, we tested an oligonucleotide substrate (RNA23Smut) similar to RNA23S but with the CC dinucleotide replaced with two uridine residues. When treated with different amounts of RNase T, RNA23Smut was digested to completion with 100 ng of RNase T, unlike RNA23S, which exhibited retarded digestion past the first guanine residue at the 3′ end (Fig. 2C). Finally, to assess the role of RNase T digestion on a biologically relevant substrate, we isolated 50S ribosomal particles from a Δrnt strain and performed digestion assays with RNase T. As shown in Figure 2D, 23S rRNA from an Δrnt strain contains elevated levels of precursors that have undergone partial processing as well as precursors that contain eight or more unprocessed residues at the 3′ end. When treated with RNase T, two different effects were observed. For a subset of the substrates containing one to two unprocessed residues, treatment with RNase T resulted in the substantial digestion of these residues to generate a mature 3′ terminus, as has been observed before (Li et al. 1999a). For the population of substrates that contained eight or more unprocessed residues, treatment with RNase T led to the trimming of these residues up the CC dinucleotide, with little evidence for digestion beyond this point. Thus, as was observed on synthetic RNA substrates, the action of RNase T on a physiologically relevant substrate is also impeded by the presence of the CC dinucleotide sequence in the 23S rRNA trailer.
The biochemical experiments described above provide a rationalization for the observation that processing of the 23S rRNA 3′ end occurs via a two-step mechanism (Li et al. 1999a). A first step, performed primarily by RNase PH, would be required to remove the residues that inhibit RNase T action. Thereafter, based on its ability to digest close to a duplex stem, RNase T would be expected to remove additional precursor residues. Collectively, both steps would be required to generate the mature 23S rRNA 3′ end.
Physiological role of a CC dinucleotide in the 23S 3′ trailer sequence
Although CC dinucleotides are not uncommon, we hypothesized that the presence of a CC dinucleotide at a strategic location in the 23S rRNA trailer could be important for the optimal fitness of cells, possibly by preventing premature digestion by RNase T. In that event, replacement of this sequence with one that allows RNase T digestion would be expected to cause a measurable cellular defect. To test this prediction, we made use of a strain, SQ2518/p19Cr, which has all chromosomal copies of the rRNA operons deleted. In this strain, rRNAs are transcribed from p19Cr, a multicopy plasmid, which encodes the rRNA operon.
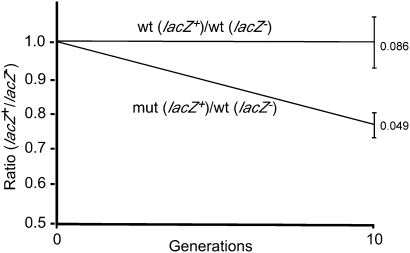
To examine the role of the CC dinucleotide in the 23S rRNA trailer sequence, plasmid p19Cr was mutated to change the CC dinucleotide to a UU sequence, resulting in a strain designated as SQ2518/p19Crmut. Strains SQ2518/p19Cr and SQ2518/p19Crmut were each grown in competition with a reference strain, SQ2518lacZ/p19Cr, which is derived from SQ2518/p19Cr but contains a mutated lacZ gene, and cells were plated on indicator X-gal plates both before and after competitive growth. By counting the numbers of blue and white colonies in each case, the fitness of each of the strains relative to the reference strain could be determined. For the competition experiments involving SQ2518/p19Cr and SQ2518lacZ/p19Cr, no change in the relative ratios of the two strains was observed after competitive growth for 10 generations, indicating that the lacZ mutation in the reference strain had a negligible effect on fitness (Fig. 3). However, when SQ2518/p19Crmut was grown in competition with SQ2518lacZ/p19Cr, the abundance of the former strain was reduced by a significant margin of 24 ± 5%. Based on the extent of the changes observed, the growth rate of SQ2518/p19Crmut is calculated to be 2% less than that of SQ2518/p19Cr. Hence, the replacement of the CC dinucleotide within the 3′ trailer of the 23S rRNA confers a small but significant fitness defect, indicating that its presence within the 23S rRNA trailer confers an evolutionary benefit to E. coli. The significance of these results is elaborated upon in the Discussion section.
FIGURE 3.
Competition assays. Strains SQ2518/p19Cr (wt) and SQ2518/p19Crmut (mut) were grown in competition with a reference strain, SQ2518lacZ/p19Cr, over 10 generations of growth in LB medium at 37°C. The ratios of the strains after competitive growth were normalized to the ratio of the strains prior to competitive growth. Standard deviations, as indicated, are derived from 10 experiments for each set of competing strains. P < 0.05 using the matched-pair test.
Revisiting the role of RNase PH in tRNA maturation
All tRNAs in E. coli are synthesized with 3′ trailer sequences and require exonucleases to perform 3′ end maturation, the most effective being RNase T and RNase PH (Li et al. 1998). It has been observed that the effectiveness of the processing RNases is sequence-dependent (Li and Deutscher 1994). We speculated that a role for RNase PH in tRNA maturation could be similar to its role in 23S rRNA maturation, i.e., to remove precursor sequences that inhibit digestion by RNase T. To determine whether this could be the case, first, we surveyed the 3′ trailer sequences on all E. coli tRNAs. For two tRNAs (tRNAAsp1 and tRNAVal1), the dinucleotide sequence immediately downstream from the mature 3′ ends was a mixture of CC and other dinucleotide sequences. This was so because each of these tRNAs is encoded by multiple gene copies with different trailer sequences present downstream. For purposes of our analysis, we denote these tRNAs as Class I tRNAs. We next looked for examples in which the dinucleotide sequence in the trailer sequences was either CU or UC. The presence of these dinucleotides has also been shown to be inhibitory to RNase T digestion, though less so than a CC dinucleotide (Zuo and Deutscher 2002). The six tRNAs that meet these criteria were denoted as Class II tRNAs. Finally, tRNAs that did not contain CC, CU, or UC dinucleotides downstream from the mature 3′ end were denoted as Class III.
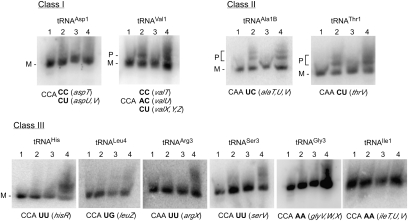
To determine the consequences of RNase PH depletion on tRNA maturation, we performed Northern blotting using probes to each of the two Class I tRNAs, to two Class II tRNAs, and to six Class III tRNAs. In each experiment, RNA was analyzed from a wild-type strain, from strains containing individual Δrph or Δrnt mutations, or from a doubly mutated ΔrphΔrnt strain. The question we wanted to address was whether an absence of RNase PH, by itself, could lead to precursor accumulation. An expectation was that if sequences inhibitory to RNase T action are present in the 3′ trailer, there should be significant precursor accumulation in a Δrph strain because, without RNase PH, RNase T would be unable to perform processing in this strain background. An examination of the Northern blotting results indicated that in some cases a visible accumulation of precursors in the Δrph strain could be observed (Fig. 4). One of these tRNAs corresponds to a Class I tRNA and two correspond to Class II tRNAs. Accumulation of tRNA precursors in an RNase PH–deficient strain has also been previously observed for the class I tRNA, tRNAVal1 (Li and Deutscher 1996). None of the Class III tRNAs accumulated precursors in a Δrph strain, even though this was the largest group tested. Thus, precursor accumulation in an Δrph strain background appears to be correlated with the presence of CC, CU, or UC sequences downstream from mature tRNA sequences. As an exception to this rule, no significant accumulation of precursors in a Δrph strain was observed for one of the Class I tRNAs (tRNAAsp1), possibly because this tRNA undergoes processing by a mechanism that does not involve RNase T or RNase PH. Interestingly, in each case where precursors were observed in the Δrph strain, the levels of precursors were nearly as high as those observed in the ΔrphΔrnt double-mutant strain. This result would be expected if the trailer sequences were to inhibit digestion by RNase T completely or nearly so. Overall, these results help to clarify the role of RNase T and RNase PH in the maturation process; for most of the tRNAs, either enzyme is sufficient to promote maturation, but for tRNAs that contain an RNase T stall sequence in the 3′ trailer, RNase PH generally assumes a major role in 3′ end maturation.
FIGURE 4.
Analysis of tRNA maturation. RNAs extracted from strains that were wild-type (lane 1) or contained deletions in rph (lane 2), in rnt (lane 3), or in both rph and rnt (lane 4) were analyzed by Northern blot using probes to specific tRNAs. Below each panel, the genes that encode each tRNA as well as the corresponding dinucleotide sequence following the mature tRNA CCA end are shown. The precursors (P) and mature tRNAs (M) that accumulate for the Class I and Class II tRNAs are indicated.
DISCUSSION
Among the different classes of RNAs in the cell, rRNAs are the most abundant class in prokaryotes and constitute up to 80% of total RNA in the cell. Investigating the rRNA processing pathway therefore becomes important not only to understand how rRNAs are rendered competent for incorporation into ribosomes but also to define a prominent set of metabolic processes in the cell. Additionally, rRNAs maturation is impeded under a variety of circumstances, including conditions of slow growth, the addition of antibiotics, and mutations in ribosome assembly factors, through mechanisms that remain unclear (Michaels 1972; Schlessinger et al. 1974; Maguire 2009). Despite its significance, several details concerning rRNA processing remain unknown. In particular, in E. coli, the most thoroughly characterized prokaryotic organism, the overall pathway of rRNA maturation still remains to be delineated.
We have been interested in understanding the steps that lead to the maturation of the largest of the three rRNAs in E. coli, the 23S rRNA. Our interest in this molecule derives partly from the observation that mutations in two RNA helicases, SrmB and DeaD, two factors being investigated in the laboratory, cause defects in 23S rRNA processing (Charollais et al. 2003, 2004). Earlier, we provided evidence that processing of the 3′ end of 23S rRNA precedes 5′ end processing and therefore is likely to be the rate-limiting step for 23S rRNA maturation (Gutgsell and Jain 2010). This order of events is likely to be a consequence of base-pairing interactions between the 5′ and 3′ regions of 23S rRNA precursor; removal of the 3′ end trailer would be a requisite for 5′ end maturation, since the latter is probably performed by a single strand–specific endonuclease. To identify the enzymes involved in 3′ processing, we systematically examined strains lacking different combinations of 3′ to 5′ exonucleases. The most dramatic effects were observed in strains lacking RNase PH in combination with either PNPase or RNase II. On the basis of these observations, we conclude that RNase PH is a key ribonuclease involved in initiating 23S rRNA maturation. Further support for this proposal stems from the observation that in cells lacking RNase PH, novel intermediates containing five to six unprocessed nucleotides were observed (Fig. 1C).
In theory, any of the 3′ to 5′ exonucleases in the cell should be capable of initiating the maturation process, particularly RNase II, PNPase, RNase T, or RNase PH, the exonucleases that are primarily involved in digesting single-stranded RNA residues inside the cell (Deutscher and Li 2001). That RNase PH should have a prominent role appears to derive from two features of the unprocessed 23S rRNA end. First, the 23S rRNA precursor contains three single-stranded residues at its 3′ end that follow five unprocessed residues that are base-paired with 5′ end sequences (Fig. 1A). A short 3′ tail is likely to limit the accessibility of RNases to this substrate. As shown in Figure 2A, RNase T and RNase PH are more effective at digesting sequences close to duplex regions compared with RNase II or PNPase. By this criterion, the former set of enzymes would be expected to be more effective at digesting the 23S 3′ precursor. A second feature is the presence of a CC dinucleotide sequence in the 3′ terminus sequence, one that is known to inhibit digestion by RNase T. In vitro experiments performed using either synthetic RNA substrates or 50S particles confirmed that its presence, in fact, impedes digestion by RNase T. Consequently, the best candidate to initiate the maturation of 23S rRNA would be RNase PH, which is what was observed. An examination of tRNAs revealed that a similar role is also performed by RNase PH during tRNA maturation. Thus, in the absence of RNase PH, noticeable levels of precursors were observed only for those tRNAs that contained dinucleotide sequences in the 3′ trailer that retard RNase T digestion (Fig. 4). Efficient processing of these tRNAs therefore requires RNase PH function, whereas for tRNAs that do not contain RNase T stall sequences, either RNase T or RNase PH is sufficient for maturation.
After the first nucleotides of the 23S rRNA precursor have been removed, the removal of the remaining precursor residues involves RNase T, as suggested by the accumulation of partially processed intermediates in a Δrnt strain (Fig. 1B,C; Li et al. 1999a). Thus, the key role of RNase PH in 23S rRNA maturation may be to remove the outermost 3′ residues, including the CC dinucleotide sequence, to allow RNase T to digest the subsequent residues. Such a mechanism ensures that 23S rRNA processing requires multiple enzymes to proceed to completion. Interestingly, among the other maturation pathways that have been elucidated for E. coli rRNAs, specifically 5′ end maturation of 16S rRNA and 3′ end maturation of 5S rRNA, each also requires two different enzymes: RNase E and RNase G for the former process and RNase E and RNase T for the latter. This contrasts with the maturation of the 3′ ends of small regulatory RNAs and of both the 3′ and 5′ ends of tRNAs, each of which can be performed by a single enzyme. The question as to why multiple enzymes are required for rRNA processing remains to be answered, but hints at a regulatory role in the processing of these more complex RNA molecules.
A remaining question regarding 3′ end maturation of 23S rRNA concerns the mechanism by which RNase T can remove the five precursor nucleotides that are base-paired with 5′ precursor sequences (Fig. 1A). Earlier, it has been shown that RNase T can digest single-stranded nucleotides but digestion does not proceed significantly into regions of duplex RNA, a result that we have confirmed (Fig. 2A). However, it has also been demonstrated that RNase T can remove the base-paired nucleotides from partially processed 23S rRNA precursors in vitro, especially when 23S rRNA is part of a 50S subunit particle (Li et al. 1999a). These observations suggest that the increased activity of RNase T on ribosomal substrates could be facilitated by an interaction with proteins present on the ribosomal subunit, which could help to recruit RNase T. Alternatively, the 3′ unprocessed nucleotides within the 50S subunits might adopt a conformation that allows RNase T to digest these residues more effectively. Further work on this topic will be required to clarify how RNase T is able to digest the multiple base-paired precursor residues on the 23S rRNA precursor that would normally have been expected to be resistant to its action.
Although CC dinucleotides are expected to occur relatively frequently, its presence at the extreme terminus of the 23S rRNA precursor led us to speculate that its location could have some strategic significance. In that event, a replacement of this dinucleotide with one that does not inhibit digestion by RNase T might be expected to confer a cellular defect. Indeed, through competition assays, we were able to detect a measurable fitness defect when the CC dinucleotide was altered to a UU dinucleotide. That this defect should be caused by a small change in a precursor region indicates that the CC dinucleotide in the 23S trailer could be serving a regulatory function. If that were the case, what might that regulatory role be? As shown in Figure 1A, the 5′ and 3′ ends of the 23S rRNA precursor engage in long-range base-pairing interactions, which ultimately helps to define the processing pathway. However, because these sequences are separated by ∼3000 nt, a delay before the two ends find each other would not be unexpected. If, during this time, RNase T was able to initiate digestion of the sequences downstream from the 3′ end of mature 23S rRNA, in the absence of a stalling sequence, digestion could progress into the mature regions and result in degraded 23S rRNA. Instead, the presence of a strategically located CC dinucleotide sequence ensures that any such premature digestion, if it were to happen, would remain limited to the precursor sequence. Moreover, such RNAs would still remain capable of base-pairing with the 5′ precursor and of going through a normal maturation process. By this model, the RNase T stall sequence helps to ensure that rRNAs are protected from inadvertent digestion and thereby contributes to the overall fitness of cells. Interestingly, an inspection of the sequence of 16S rRNA, one other rRNA that forms long-range base-pairs between 5′ and 3′ precursors sequences, shows that the 2 nt immediately downstream from the mature 16S rRNA 3′ end are both cytosine. The maturation of the 16S rRNA 3′ end is known to be dissimilar to that of 23S rRNA and is likely to involve endonucleolytic cleavage (Deutscher 2009). Given such a mechanism, the optimal location of an RNase T stall sequence to prevent unwarranted digestion proceeding into mature rRNA would be just downstream from the mature end, which is indeed observed.
A large fraction of the cell's resources is devoted toward rRNA transcription and its assembly into ribosomes, and therefore, rRNA sequences have evolved in a manner to optimize function. That the precursor sequences could also have evolved to ensure that rRNAs are processed and folded efficiently would not be unexpected. The formation of base-pairing interactions between the 5′ and 3′ end precursor sequences and an RNase T stall sequence in the 3′ precursor each provides an example of how precursor sequences help to ensure that rRNA processing will proceed in an orderly fashion. Other examples of sequences in the precursor whose mutation causes phenotypic defects, such as cold-sensitive growth, have also been documented (Schaferkordt and Wagner 2001). Although our current understanding of the role of precursor sequences is still limited, we can expect that further work on this topic will provide further examples of how precursor sequences contribute to the efficacy of rRNA maturation and help to ensure that the incorporation of rRNAs into ribosomes is ultimately carried out with a high degree of effectiveness.
MATERIALS AND METHODS
Strains
MG1655* is a derivative of the sequenced strain MG1655 (Blattner et al. 1997), which contains an engineered point mutation that converts a defective rph gene to wild-type. Derivatives of MG1655* containing nonpolar Δpnp, Δrbn, Δrnb, Δrnd, Δrnr, Δrph, and Δrnt alleles were constructed by transduction of deletion alleles that are interrupted by a kanamycin-resistance (kanR) marker (Baba et al. 2006). The kanR marker was removed from these strains by trans-formation with plasmid pCP20 (Datsenko and Wanner 2000), which expresses the FRT recombinase and harbors a temperature-sensitive replicon, followed by recombination of frt sites that flank the kanR gene during growth at 30°C, and the removal of pCP20 by restreaking at 42°C. Double-deletion strains were constructed as described above, resulting in 19 strains containing combinations of the aforementioned deletion alleles. For these strains, the kanR markers were again removed, as described above, with the exception of ΔpnpΔrph::kan, which we were unable to grow at 30°C. A successful deletion of alleles was confirmed by PCR in each case.
To test the effect of mutations in 23S rRNA trailer sequences on growth fitness, we used a strain, SQ2518, which is a derivative of strain MG1655 and contains deletions in each of its seven rRNA operons. SQ2518 contains two plasmids, a kanR multicopy plasmid, pK4-16, that expresses the rRNA operon, and ptRNA67 (Asai et al. 1999), which expresses tRNAs that are encoded only within chromosomal rRNA operons. Another multicopy plasmid, p19Cr, which expresses the rRNA operon and confers chloramphenicol resistance (camR), was used to mutate the CC dinucleotide in the 23S rRNA trailer gene sequence to TT, as described below. Both the wild-type and the mutant p19Cr plasmid (p19Crmut) were introduced into SQ2518, and in each case, pK4-16 was displaced through repeated rounds of selection for camR and loss of the kanR phenotype. In this manner, strains expressing rRNA from p19Cr or p19Crmut were constructed.
Mutation of the CC dinucleotide in the 23S rRNA trailer sequence was performed by PCR of a part of the 23S rRNA gene with primers 5′-TATGCGTTGTTGGGTAGGGGAGC-3′ and 5′-GGCTGAAAATCTTCTCTCATCCGCCAAAACAGCTTCAACGTTG-3′, with the mutated nucleotides underlined and an XmnI restriction site in italics. The 2 kb PCR product was digested with XmnI and SphI, with the latter restriction site being internal of the primer binding sites, and cloned between the unique XmnI and SphI sites of p19Cr to generate p19Crmut. Sequence analysis was performed to confirm that no inadvertent mutations had been incorporated into the cloned amplicon in p19Crmut prior to use.
A lacZ− allele of SQ2518/p19Cr was constructed through several steps. First, the Tn5 transposon kanR gene was amplified using primers (5′-CGTTTTACAACGTCGTGACTGGGAAAACCCTGGCGTTACCATTGAACAAGATGGATTG-3′ and 5′-CGCGAAATACGGGCAGACATGGCCTGCCCGGTTATTATTATCAGAAGAACTCGTCAAG-3′) that contain flanking homology with lacZ. The PCR product was electroporated into MG1655 cells expressing λRed protein from plasmid pKD46 (Datsenko and Wanner 2000). The transformed cells were plated on agar plates supplemented with kanamycin, X-gal, and IPTG, and the recombinant strain was identified based on its lacZ− phenotype. The lacZ− allele was transferred to SQ2518/p19Cr by P1 transduction, yielding strain SQ2518lacZ/p19Cr.
RNA isolation and analysis
RNA was prepared from cultures growing exponentially in LB medium using the hot-phenol method, with variations, as described previously (Gutgsell and Jain 2010). Analysis of 3′ end processing of 23S rRNA was performed using oligonucleotide directed RNase H cleavage of 23S rRNA 50 nt from the mature 3′ end and Northern blot analysis, as described previously (Gutgsell and Jain 2010). For tRNA analysis, RNA was isolated from MG1655*, MG1655*Δrph, MG1655*Δrnt, or MG1655*ΔrphΔrnt strains, fractionated on 6% polyacrylamide–8 M urea sequencing gel, and transferred to positively charged nylon membranes (Nytran, Whatman Inc.). The tRNAs were probed using radiolabeled oligonucleotides at 50°C. The oligonucleotides sequences used were 5′-GGATTCGAACCCACGAC-3′ [tRNAHis]; 5′-CTACCGATTCCACCATCC-3′ [tRNALeu4]; 5′-TGAGACCTCTGCCTCCGGAGGG-3′ [tRNAArg3]; 5′-GAACCGCAGACCTCCTGCGTGC-3′ [tRNAAla1B]; 5′-GAACTCGCGACCCCGACCTTGGC-3′ [tRNAGly3]; 5′-GAACCACCGACCTCACCCTTATC-3′ [tRNAIleu1]; 5′-CGAACTGGGGACCCCACCCCTACC-3′ [tRNAThr1]; 5′-GATGCAGCTTTTGACCGCA-3′ [tRNASer3]; 5′-CGGGACTCGAACCCGCGACC-3′ [tRNAAsp1]; 5′-CCCAGCTGAGCTAATCACCC-3′ [tRNAVal1].
Ribonuclease digestion assays
RNA oligonucleotides were synthesized by Sigma-Genosys or obtained from the laboratory of Dr. Murray Deutscher (University of Miami Miller School of Medicine). For enzymatic digestion assays, oligoribonucleotides were kinased at the 5′ end using polynucleotide kinase and γ-32P-ATP. Duplex substrates were made by annealing labeled strands with an excess of unlabeled complementary RNA. In some cases, RNAs were gel purified prior to use. RNA substrates were digested with E. coli exoribonucleases using about 100 counts of labeled substrate and indicated amounts of enzyme in CP buffer (50 mM MOPS at pH 7.0, 3 mM MgCl2, 5 mM NaPO4, and 1 mM DTT) for 1 h at 37°C. The reactions were terminated by adding 0.6 volume of gel loading buffer (96% formamide, 4% 0.5 M EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol) and fractionated through a denaturing 22.5% polyacrylamide gel. The RNA digestion products were visualized using a GE Healthcare Storm 840 Phosphorimager. Digestion of 50S particles with RNase T was carried after fractionating cell extracts derived from strain MG1655Δrnt by sucrose density gradient ultracentrifugation and collecting the 50S fraction. The ribosomal particles were passed over a Sephadex spin column (5% Sephadex in buffer CP), and the resulting 50S particles were treated with RNase T as described above. To analyze 23S RNAs present in 50S particles, the RNA was first extracted by phenol extraction and ethanol precipitation, followed by RNase H cleavage and Northern analysis.
Competition experiments
Strains SQ2518/p19Cr, SQ2518/p19Crmut, and SQ2518lacZ/p19Cr were grown to saturation, diluted 1:50, and regrown to mid-log phase at 37°C. Equal volumes of the SQ2518lacZ/p19Cr were combined with either wild-type or mutant lacZ+ cells. A sample of these mixtures was removed, diluted 1000-fold into prewarmed LB, and grown until they had reached the density of the mixture before dilution (10 generations of growth). Different dilutions of the mixtures were plated on LB-agar plates supplemented with X-gal either at the time of dilution or after competitive growth, and the plates were incubated overnight at 37°C. The next day, blue and white colonies were counted for dilutions that yielded 150–300 colonies per plate. The ratio of blue to white colonies was determined in each case and was used to derive the fitness of the competing strains with respect to each other.
ACKNOWLEDGMENTS
We thank Dr. Selwyn Quan (Stanford University) for strain SQ2518 and plasmids, Mr. Brad Schmier and Dr. Arun Malhotra (University of Miami Miller School of Medicine) for providing purified enzymes, and Dr. Murray Deutscher for oligonucleotides and comments on the manuscript. This work was supported by a RO1 grant from the National Institutes of Health (GM81735).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.027854.111.
REFERENCES
- Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol 181: 3803–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474 [DOI] [PubMed] [Google Scholar]
- Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol 48: 1253–1265 [DOI] [PubMed] [Google Scholar]
- Charollais J, Dreyfus M, Iost I 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res 32: 2751–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci 100: 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP 2005. An important role for RNase R in mRNA decay. Mol Cell 17: 313–318 [DOI] [PubMed] [Google Scholar]
- Condon C 2007. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol 10: 271–278 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP 2009. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci 85: 369–391 [DOI] [PubMed] [Google Scholar]
- Deutscher MP, Li Z 2001. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol 66: 67–105 [DOI] [PubMed] [Google Scholar]
- Donovan WP, Kushner SR 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci 83: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Deutscher MP 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci 96: 4372–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D, Steitz JA 1975. The 30 S ribosomal precursor RNA from Escherichia coli. A primary transcript containing 23 S, 16 S, and 5 S sequences. J Biol Chem 250: 5647–5654 [PubMed] [Google Scholar]
- Gutgsell NS, Jain C 2010. Coordinated regulation of 23S rRNA maturation in Escherichia coli. J Bacteriol 192: 1405–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C 2009. Identification and characterization of growth suppressors of Escherichia coli strains lacking phosphorolytic ribonucleases. J Bacteriol 191: 5622–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP 1994. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J Biol Chem 269: 6064–6071 [PubMed] [Google Scholar]
- Li Z, Deutscher MP 1995. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc Natl Acad Sci 92: 6883–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP 1996. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86: 503–512 [DOI] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP 1998. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc Natl Acad Sci 95: 2856–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP 1999a. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA 5: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP 1999b. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J 18: 2878–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire BA 2009. Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol Mol Biol Rev 73: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels GA 1972. Ribosome maturation of Escherichia coli growing at different growth rates. J Bacteriol 110: 889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy MK, Singh B, Ray BK, Apirion D 1983. Maturation of 5-S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur J Biochem 131: 119–127 [DOI] [PubMed] [Google Scholar]
- Schaferkordt J, Wagner R 2001. Effects of base change mutations within an Escherichia coli ribosomal RNA leader region on rRNA maturation and ribosome formation. Nucleic Acids Res 29: 3394–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger D, Ono M, Nikolaev N, Silengo L 1974. Accumulation of 30S preribosomal ribonucleic acid in an Escherichia coli mutant treated with chloramphenicol. Biochemistry 13: 4268–4271 [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol 44: 105–129 [DOI] [PubMed] [Google Scholar]
- Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K 1999. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem Biophys Res Commun 259: 483–488 [DOI] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP 2002. The physiological role of RNase T can be explained by its unusual substrate specificity. J Biol Chem 277: 29654–29661 [DOI] [PubMed] [Google Scholar]