Pluripotent stem cells (PSCs) are defined by their potential to generate all cell types of an organism. The standard assay for pluripotency of murine PSCs is transmission of the cells through the germ line, but for human PSCs, researchers must depend on indirect methods such as differentiation into teratomas in immunodeficient mice. Here we report PluriTest, a robust open-access bioinformatic assay of pluripotency in human cells based on their gene expression profiles.
The current standard for demonstrating that human stem cells are pluripotent is based on their ability to generate a complex variety of tissues in tumors developed in immunodeficient mice. This teratoma assay is widely considered to be the most reliable and informative assay for pluripotency in human cells1 and its use has significantly increased following the report of induction of pluripotency in somatic cells.2 However, the generation of teratomas is technically challenging, resource-intensive and primarily qualitative, is difficult to standardize, and there are conflicting reports about its value as a criterion for pluripotency.3 With the rapid increase in generation of pluripotent human cells, especially induced pluripotent stem cell (iPSC) lines, there is an urgent need for a cost effective, animal-free alternative to the teratoma assay for assessing pluripotency in human cells.4 The low cost and accessibility of microarray-based gene expression data sets makes transcription profiling an attractive alternative. We hypothesized that machine learning methods that are capable of delineating stem cell phenotypes5 based on microarray data could also predict the presence or absence of pluripotent features for unknown samples of cells.
We considerably expanded the gene expression database that we previously used for defining stem cell phenotypes5 to a much larger data set we term ‘Stem Cell Matrix-2’ (SCM2). The SCM2 database contains approximately 450 genome-wide transcriptional profiles from diverse stem cell preparations from multiple laboratories, differentiated cell types, and developing and adult human tissues (Supplementary Table 1). SCM2 contains expression profiles from 223 human embryonic stem cell (hESC) and 41 iPSC lines. We analysed the samples for SCM2 in a highly quality controlled pipeline, using Illumina microarrays. After appropriate transformation and normalization, we used non-negative matrix factorization (NMF) for dimension reduction and to identify unexpected patterns engrained in the datasets.6 NMF provides a systematic, unbiased approach to identify multi-gene features, frequently termed ‘metagenes’ in gene- expression studies7, which can be used to characterize stem cell phenotypes.3
We then use the SCM2 database to assess pluripotency of an unknown, potentially pluripotent sample by comparison of a ‘query gene expression profile’ from the sample to data models derived from SCM2 (see Fig. 1a). Our goals are to not only provide a simple test for pluripotency, but also detailed information on features of the sample that deviate from typical, genomically normal pluripotent stem cell lines. The approach is based on two related classifiers, which use two differently constructed metagene models.
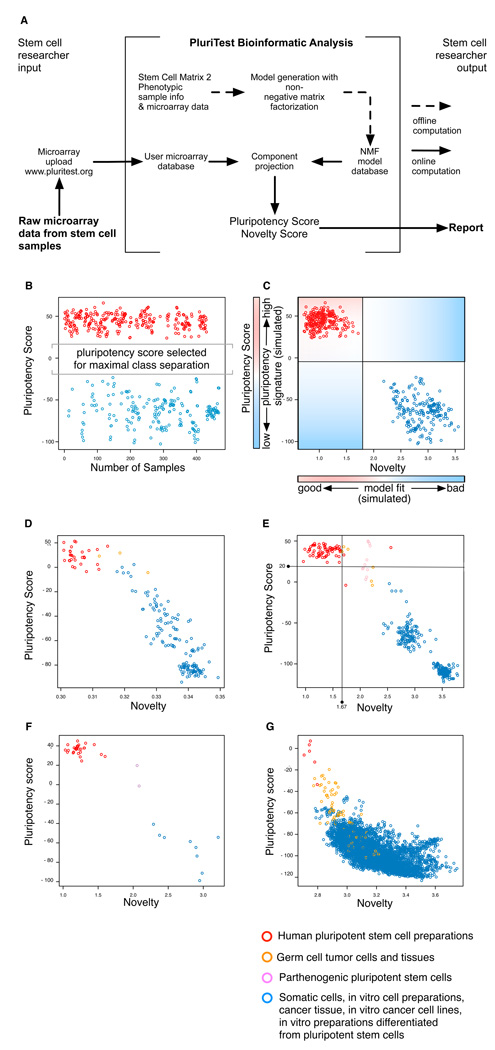
Figure 1. A multidimensional data model for assessing pluripotent stem cells.
A Schematic for PluriTest. (b–c) We constructed and optimized a multi-class classifier (Pluripotency Score, b, c) and a one-class classifier (Novelty Score, c) to distinguish pluripotent stem cells (PSC) from other cell types and tissues. In (b), we show pluripotent (red) and somatic samples (blue) in the training dataset as assessed with the Pluripotency Score and in (c) with both PluriTest scores. In (d–g), we plot Pluripotency Scores against Novelty Scores for test data set samples. The classifiers were tested against datasets generated on four different microarray platforms: Illumina WG6v1 (c, 177 samples)5, HT12v3 (d, 498 samples), HT12v4 (e, 38 samples) and Affymetrix U133A (f, 5372 samples)10). Samples for these datasets were independently generated (c and d) or curated from published studies (c, d, f). In d, the lines in the plot indicate empirically determined thresholds for defining normal pluripotent lines (see also Supplementary Fig. 2).
For the first classifier, termed the ‘Pluripotency Score’, we used all samples, pluripotent and non-pluripotent, to identify the metagenes that have the capability to separate pluripotent from non-pluripotent samples in SCM2 (Fig. 1b, Supplementary Figs. 2 and 3).5 The rank and number of metagenes were selected by identifying those that provided the largest distance between margins of known pluripotent and non-pluripotent samples in the training set (Fig. 1; Online Methods and Supplementary Fig. 4). The Pluripotency Score is a logistic regression model, thus enabling a probability-based choice between the two phenotypic classes.
The second classifier, termed the ‘Novelty Score’, measures the ability of an NMF model to approximate a given query gene expression profile (Online Methods).8 We compare the query sample to an NMF-reconstructed sample based on the well-characterized pluripotent stem cells in the SCM2 dataset and determine model fit and identify deviations from the expected gene expression patterns (Fig 1c–g).8 The Novelty Score detects technical as well as biological variations in the data; to deemphasize the technical variation, we applied an exponential transformation to empirically weight biological over technical deviations from our model (see Online Methods.).
The combination of the Pluripotency Score and the Novelty Score enables the open-ended assessment of pluripotent features in a query sample when that sample is a novel kind of pluripotent stem cell. The first classifier reports to what degree a query sample contains a pluripotent signature, and the second reports on how much of the signal measured in a query sample can be explained by the normal PSC lines contained in the SCM2 (Supplementary Note 1 and Supplementary Fig. 1). The utility of the two-classifier approach is exemplified in a test analysis of germ cell tumor cell lines. These cells are pluripotent and resemble normal PSCs, but have genetic and epigenetic abnormalities.9 These cells have high Pluripotency Scores, as expected, but the Novelty Score indicates that they deviate from the normal PSCs in the SCM2 (Fig. 1 and Supplementary Fig. 2).
We tested the combined classification approach and communication framework, which we term ‘PluriTest’, using several independently generated test datasets containing pluripotent and non-pluripotent samples: Illumina WG6v15 (Fig. 1d), HT12v3 (Fig. 1e), and HT12v4 (Fig. 1f) datasets generated in- house on our own microarray scanner and datasets that were generated in six different core facilities (Online Methods and Supplementary Table). We also used PluriTest to examine a recently published human transcriptome atlas based on Affymetrix U133A arrays (Fig. 1g).10
PluriTest predicted pluripotency with excellent sensitivity and specificity. We could set thresholds that could separate pluripotent from non-pluripotent samples in a HT12v3 test data sets with 98% sensitivity and 100% specificity (Fig. 1e and Supplementary Fig. 2) and could also distinguish germ cell tumor cell lines (orange, Fig. 1 d, e and g) and parthenogenetic stem cell lines (Fig 1e and f) from the bulk of pluripotent stem cells. A few pluripotent samples displayed unusually high novelty scores (Fig. 1e), indicating that these test samples should be further evaluated for epigenetic or genetic abnormalities or unwanted differentiation (Supplementary Fig. 1). For the most informative analysis, the query sample should be analyzed on the same platform as the training dataset (Illumina HT12), but acceptable results can be obtained with data from other platforms (Fig. 1f and Supplementary Fig. 3, Supplementary Note 2).
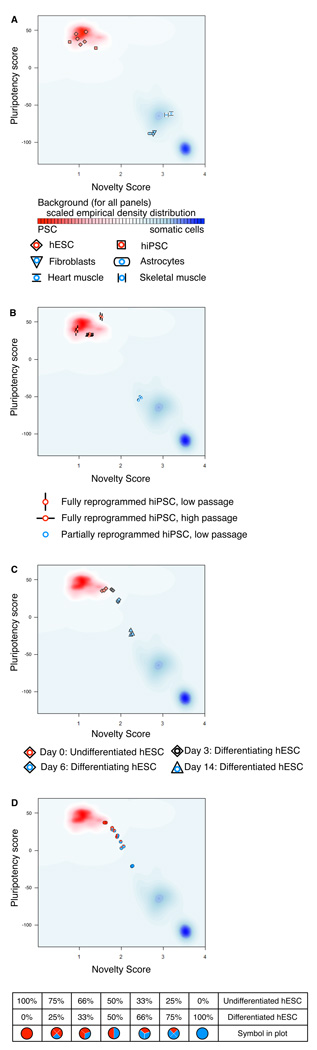
We demonstrated the performance of PluriTest on sets of query samples. hESC (SIVF014, SIVF011, SIVF042, Fisher42, WA01) and hiPSC (HDF51IPS12, HDF51IPS1) lines, which were part of the training dataset, group together and are separated from somatic samples (Fig. 2a). PluriTest also separates fully and partially reprogrammed iPSC lines (samples that were not included in the training datatset, Fig. 2b); partially reprogrammed cell lines cluster with non-pluripotent cells. We then applied PluriTest to samples from a neural differentiation time course that was also not used in the training dataset (Fig. 2c, d). WA09 cells were differentiated into neural precursors and three biological replicates sampled at day 0, day 3, day 6 and day 14 after neural induction. We observed that the Novelty Score changed after 3 days of differentiation, while the Pluripotency Score was still high at this time-point, whereas samples from later time points dropped out of the pluripotency space and scored increasingly higher on the Novelty Score (Fig. 2c). In a mixing experiment in which we combined RNA samples from different time points (day 0 and day 14) at varying ratios, PluriTest could separate the differentially mixed samples (Fig. 2d).
Figure 2. Application of PluriTest.
The graphs show the actual output of PluriTest. Pluripotency score is plotted against novelty score for the indicated samples. The background encodes an empirical density map indicating pluripotency (red) and novelty (blue). (a–c) PluriTest results for known pluripotent cells and somatic cells and tissues (a), for fully and partially reprogrammed iPSC lines (b), and for an hESC line (WA09) being differentiated into neural precursors, at the indicated time points. In (d) PluriTest was run on mixed samples of hESC and hESC-derived neural precursor RNA (day 0 and day 14 from c) at the indicated ratios. hESC, human embryonic stem cell, hiPSC, human induced pluripotent stem cell.
The PluriTest is contained within a single R/Bioconductor open-source open-access workspace11 (Supplementary Data 1 and Supplementary Note 3) that also includes the SCM2 database-derived NMF models. To enable easy access to PluriTest, we programmed a Rich Internet Application (RIA) using Microsoft Silverlight4 and C# (accessible under: www.pluritest.org). The RIA automatically performs all data extraction and preprocessing steps after the upload of an unmodified microarray scanner output file. All data and results are stored securely in an MS-SQL database. We chose to use the binary microarray scanner output file (*.idat-file) as the most basic ‘stem cell query term’. After upload, the results of our PSC-prediction algorithm are reported back to the user via a web interface (Fig. 2 and Supplementary Fig. 5). PluriTest runs on every recent Apple and Windows computer and requires internet access and a local installation of the Silverlight4 plug-in. A typical online analysis with 12 samples takes less than 10 minutes including data upload (Supplementary Note 2).
In summary, we have demonstrated the general feasibility of a web-based prediction of stem cell properties.12 PluriTest breaks from the conventional marker-based approaches to assess pluripotency of human cells, which typically assay a small number of markers by methods such as RT-PCR. With the lowered cost of whole genome analysis, reduction of a gene expression profile to a few markers is no longer necessary. Using all of the expression information available provides much higher discriminatory power and the ability to identify deviations from known patterns that may lead to further insights into cellular phenotypes.
The PluriTest framework could be applied to any unbiased high-content dataset, such as global DNA methylation analysis or RNA-seq data, provided that there is sufficient representation of a defined target phenotype in the training data set. Our work suggests that it will be relatively straightforward to construct similar models of developmental pathways such as differentiation along the neural, endodermal or hematopoietic lineages. Such databases will inform further experimentation and may be applicable as a rapid method to quality control PSC-derived preparations for experimental and pre-clinical investigations.
Methods
Methods, supplementary information and any associated references are available in the Online Methods section of this paper.
Supplementary Material
Acknowledgements
F.J.M. is supported by an Else-Kröner Fresenius Stiftung fellowship, and J.F.L. by grants from the California Institute for Regenerative Medicine (RT1-01108, TR1-01250, and CL1-00502), the NIH (R43 GM085981 and R21 MH087925), the Bill and Melinda Gates Foundation, and the Esther O’Keeffe Foundation. B.M.S. is supported by Bayer Technology Services GmbH and the Deutsche Forschungsgemeinschaft through grant GSC 111. L.C.L. is supported by an NIH/NICHD K12 Career Development Award. We thank Andreas Schuppert, Suzanne Peterson and Kit Nazor for insightful comments, valuable criticisms, and for reading the manuscript for clarity. We thank Michel Sadelain (Memorial Sloan Kettering Cancer Center, New York, USA) for providing us with samples and data. M. S. and E. P. were supported the New York State Stem Cell Science grant N08T-060. We thank Kirt Haden and Ivan Mikoulitch from Illumina Inc. (San Diego, CA, USA) for help with handling Illumina BeadArray data formats and letting us use the idat.reader.dll program module in PluriTest RIA. We thank Corina Becker, David Barker and Anja Fritz for helpful discussions.
Footnotes
Author contributions
F.J.M. conceived and designed the study. F.J.M. and B.M.S. developed the PluriTest algorithm. F.J.M., J.F.L., L.C.L., and J.B.A. oversaw the sample collection, microarray analysis and coordinated biological and bioinformatic experiments. R.W., D.M., B.M.S., and A.H. implemented the bioinformatic online platform. R.W., D.M., F.J.M., B.M.S., and G.A. provided bioinformatic analyses. E.P., S.D., J.E.G., and N.O.S. prepared important biological samples for the study. F.J.M., B.M.S. and J.F.L. wrote the manuscript with input from all co-authors.
References Main Text
- 1.Daley GQ, Lensch MW, Jaenisch R, et al. Cell Stem Cell. 2009;4(3):200. doi: 10.1016/j.stem.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Cell. 2006;126(4):663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Müller FJ, Goldmann J, Loser P, et al. Cell Stem Cell. 2010;6(5):412. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Russell WMS, Burch RL. The Principles of Humane Experimental Technique. London: Methuen; 1959. [Google Scholar]
- 5.Müller FJ, Laurent LC, Kostka D, et al. Nature. 2008;455(7211):401. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DD, Seung HS. Nature. 1999;401(6755):788. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 7.Brunet JP, Tamayo P, Golub TR, et al. Proc Natl Acad Sci U S A. 2004;101(12):4164. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David M, Tax J, Muller Klaus-Robert. Proceedings of the Pattern Recognition, 17th International Conference on (ICPR'04) Volume 3 - Volume 03; IEEE Computer Society.2004. [Google Scholar]
- 9.Josephson R, Ording CJ, Liu Y, et al. Stem Cells. 2007;25(2):437. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- 10.Lukk M, Kapushesky M, Nikkila J, et al. Nat Biotechnol. 28(4):322. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R_Development_Core_Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 12.Gray Jim. In: Data-Intensive Scientific Discovery. Hey Toney, Tansley Stewart, Tolle Kristin., editors. Redmond, WA: Microsoft Research; 2009. [Google Scholar]
- 13.Du P, Kibbe WA, Lin SM. Bioinformatics. 2008;24(13):1547. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 14.Doulatov S, Notta F, Eppert K, et al. Nat Immunol. 2010;11(7):585. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 15.Parris TZ, Danielsson A, Nemes S, et al. Clin Cancer Res. 2010;16(15):3860. doi: 10.1158/1078-0432.CCR-10-0889. [DOI] [PubMed] [Google Scholar]
- 16.Gandhi KS, McKay FC, Cox M, et al. Hum Mol Genet. 2010;19(11):2134. doi: 10.1093/hmg/ddq090. [DOI] [PubMed] [Google Scholar]
- 17.Kunarso G, Chia NY, Jeyakani J, et al. Nat Genet. 2010;42(7):631. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 18.Fuentes-Duculan J, Suarez-Farinas M, Zaba LC, et al. J Invest Dermatol. 2010;130(10):2412. doi: 10.1038/jid.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan EM, Ratanasirintrawoot S, Park IH, et al. Nat Biotechnol. 2009;27(11):1033. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 20.Gaujoux R, Seoighe C. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.