Abstract
Resveratrol has demonstrated cancer chemopreventive activity in animal models and some clinical trials are underway. In addition, resveratrol was shown to promote cell survival, increase lifespan and mimic caloric restriction, thereby improving health and survival of mice on high-calorie diet. All of these effects are potentially mediated by the pleiotropic interactions of resveratrol with different enzyme targets including COX-1 (cyclo-oxygenase-1) and COX-2, NAD+-dependent histone deacetylase SIRT1 (sirtuin 1) and QR2 (quinone reductase 2). Nonetheless, the health benefits elicited by resveratrol as a direct result of these interactions with molecular targets have been questioned, since it is rapidly and extensively metabolized to sulfate and glucuronide conjugates, resulting in low plasma concentrations. To help resolve these issues, we tested the ability of resveratrol and its metabolites to modulate the function of some known targets in vitro. In the present study, we have shown that COX-1, COX-2 and QR2 are potently inhibited by resveratrol, and that COX-1 and COX-2 are also inhibited by the resveratrol 4′-O-sulfate metabolite. We determined the X-ray structure of resveratrol bound to COX-1 and demonstrate that it occupies the COX active site similar to other NSAIDs (non-steroidal anti-inflammatory drugs). Finally, we have observed that resveratrol 3- and 4′-O-sulfate metabolites activate SIRT1 equipotently to resveratrol, but that activation is probably a substrate-dependent phenomenon with little in vivo relevance. Overall, the results of this study suggest that in vivo an interplay between resveratrol and its metabolites with different molecular targets may be responsible for the overall beneficial health effects previously attributed only to resveratrol itself.
Keywords: cancer chemoprevention, cyclo-oxygenase (COX), molecular docking, quinone reductase 2, resveratrol metabolite, SIRT1
INTRODUCTION
The discovery and use of natural medicines and remedies to cure diseases, improve human health and promote longevity has been a quest of mankind for thousands of years. Over the past three centuries, numerous natural products have been isolated and modified for use as important therapeutic agents. For example, morphine from the opium poppy has been used for over two centuries as an analgesic, quinine from Cinchona bark has been used to treat malaria, and salicin from willow bark has been formulated into aspirin, which is now being widely used as chemopreventive agent for cardiovascular disease.
A modern example of a plant-derived natural product, which is receiving a substantial amount of attention worldwide, is resveratrol (3, 5, 4′-trans-trihydroxystilbene, Figure 1A). This naturally abundant, polyphenolic compound is found in grape skins, red wine, grape and cranberry juices, and peanuts. Resveratrol is touted as being the principle agent responsible for numerous health benefits associated with the consumption of these dietary foods. For instance, the results of several epidemiological studies have linked the moderate consumption of red wine with increased longevity and reduction of the incidence of cardiovascular diseases and certain cancers, a phenomenon known as the ‘French paradox’ [1,2]. In 1992, it was postulated that resveratrol may be an explanation for the French paradox [3], but it was not until 1997 when Pezzuto and colleagues demonstrated the ability of resveratrol to inhibit carcinogenesis at multiple stages that it received worldwide attention [4].
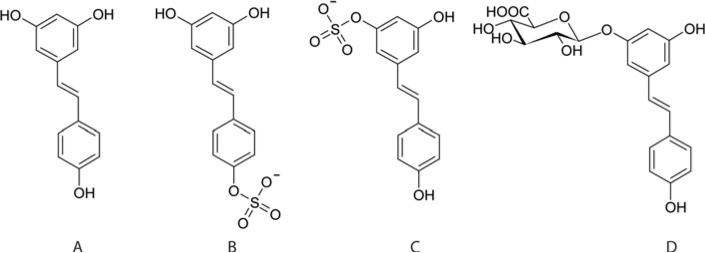
Figure 1. Structures of trans-resveratrol and its three main human metabolites.
Resveratrol (A), resveratrol-4′-O-sulfate (B), resveratrol-3-O-sulfate (C) and resveratrol-3-O-glucuronide (D).
Over the last 10 years, over 2000 research studies on the positive health benefits of resveratrol have been reported, including studies of its ability to promote longevity and to prevent cancer, cardiovascular disease, neurodegenerative disease and brain damage caused by stroke [5,6]. Resveratrol has also been shown to improve the health and survival of mice on high-calorie diets [5]. Nonetheless, despite all of the impressive health benefits being attributed to resveratrol, considerable controversy exists as to whether resveratrol is the active molecule in vivo [7]. The controversy stems from the fact that resveratrol is rapidly metabolized to its 3- and 4′-O-sulfate, and 3-O-glucuronide conjugates (Figures 1B–1D) [8–12]. For instance, it has been reported that after a dietarily relevant 25 mg oral dose of resveratrol, the plasma concentration of resveratrol is only in the nanomolar range compared with the micromolar range of its metabolites [7,10,13]. Similar amounts of resveratrol and its metabolites have been measured in human plasma after a 1 g oral dose of pure resveratrol [8]. These reoccurring observations suggest that in vivo resveratrol concentrations may be too low to elicit an effective biological response. Therefore the question is raised as to whether or not resveratrol and its metabolites are capable of interacting with and eliciting a biological response in identified molecular targets. To help resolve this controversy, we investigated the biological activities of resveratrol metabolites with known biological targets including COX-1 (cyclo-oxygenase 1), COX-2, SIRT1 and QR2 (quinone reductase 2).
EXPERIMENTAL
Synthesis of resveratrol metabolites and NMeH (N-methyldihydronicotinamide)
The resveratrol metabolites in Figure 1 were each synthesized and purified according to the procedures of Yu et al. [14]. Resveratrol and its metabolites were analysed by LC-MS (liquid chromatography MS) before and after each experiment to ensure purity and to assure the compounds had not decomposed. The substrate NMeH was prepared by reduction of N-methylnicotinamide iodide (Sigma) following the procedure described by Ortiz-Maldonado et al. [15].
Expression and purification of COX-1, COX-2, SIRT1 and QR2
COX-1 and COX-2 were expressed and purified as previously described [16,17]. Recombinant hSIRT (human SIRT1) encoded in the SIRT1-pHex was expressed and purified from Rosetta pLysS Escherichia coli according to the procedures described in the Supplementary Methods (see http://www.BiochemJ.org/bj/429/bj4290273add.htm). Human QR2 was cloned, expressed and purified according to published procedures [18] with modifications described in the Supplementary Methods.
Crystallization, X-ray data collection and structural refinement of the COX-1–resveratrol complex
Purified ovine COX-1 was co-crystallized with resveratrol according to published procedures [16], except that 1 mM resveratrol was included in the crystallization solution. Resveratrol (10 mM), dissolved in DMSO, was added directly to COX-1 at 15 mg/ml to give a final resveratrol concentration of 1 mM and a final protein concentration of 0.23 mM (~4:1 inhibitor/protein ratio). Crystals were grown in sitting drops by combining the COX-1–resveratrol complex with 0.64 M sodium citrate, 0.3–0.6 M LiCl, 1 mM NaN3 and 0.33% β-OG (n-octyl-β-d-glucopyranoside) at pH 6.5 and equilibrating over reservoir solutions containing 0.64–0.84 M sodium citrate, 0.3–0.6M LiCl and 1 mM NaN3 at 20°C. Crystals were flash-frozen in liquid propane after transfer into 0.9 M sodium citrate, pH 6.5, 1.0 M LiCl, 0.15% β-OG and 24% (w/v) sucrose, and then subsequently transferred into liquid nitrogen for transport to the Argonne National Laboratory (Chicago, IL, U.S.A.). X-ray data collection was performed with the BioCARS Beamline 14-BM-C at the Advanced Photon Source on a ADSC-Q4 detector. X-ray data were scaled and processed using Denzo and Scalepack as described in [19]. The structure of COX-1 in complex with resveratrol was determined by molecular replacement using the co-ordinates of the ovine prostaglandin H synthase-1 complexed with arachidonic acid (Protein Data Bank code 1DIY) as a search model. Structural refinement was performed with the program CNS [20] and manual model building was performed using the program O [21]. X-ray data collection and refinement statistics are provided in Supplementary Table S1 (at http://www.BiochemJ.org/bj/429/bj4290273add.htm).
Inhibition of COX-1 and COX-2 activity by quantification of PGE2 (prostaglandin E2)
The IC50 values for the inhibition of the COX-1 and COX-2 enzymes by resveratrol and its metabolites were measured by an ELISA-based kinetic assay that quantifies the PGE2 produced by enzymatic conversion of arachidonic acid. Detailed protocols for this assay have been described by Cuendet et al. [22].
Molecular docking protocol
We used the X-ray structure of resveratrol in complex with COX-1 to validate the docking procedure. The X-ray structure of resveratrol was also used as a template to build the three-dimensional structure of the resveratrol metabolites using the program SYBYL (Tripos). Gesteiger-Marsili charges were assigned to all the ligands. Dock version 4.0.1 [23] was used to generate and evaluate the orientations of resveratrol and its metabolites inside the COX active sites. Since there were no substantial differences in the crystallization parameters between the COX-2–inhibitor complexes derived from the Brookhaven Protein Data Bank [24], the COX-2–flurbiprofen complex was chosen because of the similarity of flurbiprofen with resveratrol. The same procedure was applied to both complexes, COX-1–resveratrol, whose structure was determined in our laboratory, and COX-2–flurbiprofen (Protein Data Bank code 3PGH). The docking procedure can be described by four major steps: identification of the receptor site and its three-dimensional characterization, calculation of scoring grids for evaluating docked structures, docking, and evaluation of the resulting ligand orientations. A molecular surface representation of the binding site was created with the MS algorithm [25], and then used in the SPHGEN routine [26] to calculate spheres for docking. All molecular surface points were used in the calculation of the sphere set. The maximum and the minimum sphere radius were 4.0 and 1.4 Å (1 Å = 0.1 nm) respectively. Spheres output by SPHGEN that were outside the known COX channel were rejected. The Dock routine GRID generated pre-calculated energy grids, which are used to calculate the energy score for any ligand-receptor atom pair in the docked solutions. Atomic partial charges were assigned to the protein prior to calculating these grids using SYBYL. The intermolecular interaction energies were modelled using a combined Van der Waals and electrostatic interaction potential. Electrostatic interactions were modelled using a distance-dependent dielectric and an initial dielectric constant of 4.
SIRT1 deacetylation assay
The SIRT1 fluorimetric activity assay/drug discovery kit (AK-555; BIOMOL) was used to measure the lysyl-deacetylase activity of recombinant hSIRT1. The assay was performed according to the manufacturer's guidelines using 50 μM Fluor de Lys®-SIRT1 peptide, 500 μM NAD+ and 1 unit SIRT1, in the absence and presence of resveratrol and its sulfate metabolites (100 μM) in assay buffer (25 mM Tris/HCl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2 and 1 mg/ml BSA). Resveratrol and its sulfate metabolites were dissolved at 100 mM stock concentration in DMSO on the day of the assay. For the negative control, 1% (v/v) DMSO was added to the assay. SIRT1 was pre-incubated for 10 min with DMSO (negative control), resveratrol and resveratrol metabolites. Reactions were initiated by the addition of 2× concentrations of the Fluor de Lys® peptide and NAD+. The reactions were quenched at different time points by the addition of 50 μl of 2 mM nicotinamide/Developer II solution to 50 μl of the reaction mixture. The quenched samples were kept at 37°C for 45 min prior to fluorescence reading. The in vitro fluorescence assay was read in a white half-volume 96-well microplate (COSTAR) with a GeniosPro fluorescence spectrophotometer (TECAN) (excitation 360 nm, emission 460 nm).
For the SIRT1 resveratrol dose–response experiments, values were derived from the slopes of fluorescence against time plots with data obtained at 0, 5, 10 and 20 min of deacetylation using 25 μM each of NAD+ and Fluor de Lys® acetylated peptide in the absence or presence of 10, 25, 50, 100, 200 and 500 μM resveratrol and its sulfate metabolites. Calculation of net fluorescence included the subtraction of a blank value by adding an inhibitor (2 mM nicotinamide) to the reaction. The data were fitted to the Michaelis–Menten equation.
ITC (isothermal titration calorimetry) experiments
Calorimetric titrations of QR2 and SIRT1 were performed using a VP-ITC microcalorimeter (MicroCal). Protein samples were extensively dialysed against ITC buffer consisting of 50 mM Hepes, pH 7.5, 100 mM NaCl and 1 mM TCEP [Tris(2-carboxyethyl)phosphine]. All samples were filtered through a 0.22 μM sterile filter membrane (Millipore), and thoroughly degassed by gentle stirring under vacuum. The 1.35 ml sample cell was filled with a 10 μM solution of QR2, or 20 μM SIRT1, and the 250 μl syringe was filled with a 250 μM titrating resveratrol. Each titration consisted of a first 1 μl injection followed by 60 subsequent 2 μl injections all spaced by 5 min intervals. The data were fitted using the one-site model present in Origin 7.0 (MicroCal).
Radioligand-binding assays
[3H]Resveratrol (3 Ci/mmol) was purchased from Moravek Biochemicals. Binding experiments were performed by incubating 10 or 90 μM SIRT1 with various concentrations of [3H]resveratrol from 10 to 500 μM in a final volume of 70 μl. Radioligand binding was allowed to equilibrate at 4°C for 16 h, and then bound radioactivity was isolated by filtration on to a PD SpinTrap G-25 (GE Healthcare). Each eluted fraction was monitored for protein concentration and radioactivity. Concomitantly, triplicates of radioligand dilutions were made in the absence of protein to assay total radioactivity. Experiments were repeated in the presence of 25 μM of each substrate (Fluor de Lys®-SIRT1 or NAD+). Saturation binding experiments were performed in the presence or absence of 500 μM unlabelled resveratrol. Radioactivity was measured using a Perkin Elmer Top Count NXT microplate scintillation and luminescence counter.
QR2 steady-state kinetics
Steady-state activity of QR2 was measured by monitoring the decrease in absorbance of NMeH at 360 nm at 25°C following the procedure described by Kwiek et al. [27] with only slight modifications. Reactions were initiated by the addition of 5 nM QR2 to 200 μl of 50 mM Tris/HCl, pH 8.0, 100 mM NaCl and 0.1% Triton X-100, containing 3–50 μM menadione (Sigma), and 10–150 μM NMeH. For the inhibitory assay, QR2 (5 nM) in assay buffer (50 mM Tris, pH 8.0, 100 mM NaCl and 0.1% Triton X-100) was incubated with 100 μM NMeH and 100 μM inhibitor (resveratrol, resveratol 3-O-sulfate or resveratrol 4′-O-sulfate) for 10 min on ice prior to the addition of the substrate (30 μM menadione). Reaction rates were converted to specific activity using the NMeH absorption coefficient reported previously (ε360 = 7060 M—1 · cm—1) [15]. Each plotted value is the mean of three independent rate measurements calculated from the linear portion of the kinetic plot. Standard equations describing steady-state kinetics for substrate binding and competitive, uncompetitive and non-competitive inhibition were used in order to fit the data sets and derive Km, Ki and Vmax values. The Km, Ki and Vmax parameters were calculated by fitting the curves with the kinetic module embedded in SigmaPlot 9.0. The kinetic mechanism for inhibition was determined by inspection of double-reciprocal plots, and inhibition models were excluded if the substantial error in the derived parameters were large and if a poor fit of the data to the model was indicated. A complete comparison of the final fitted models is given in Supplementary Table S2 (at http://www.BiochemJ.org/bj/429/bj4290273add.htm).
RESULTS
Resveratrol-4′-O-sulfate inhibits COX-1 and COX-2 activity
One of the molecular mechanisms through which resveratrol is proposed to exert its cardioprotective and anticancer effects is by inhibiting the COX-1 and COX-2 enzymes [4,28,29]. However, whether resveratrol and/or its main metabolites are responsible for this effect in vivo is not known. Therefore, we determined the IC50 values of resveratrol and its metabolites towards the COX-1 and COX-2 enzymes (Table 1). Strikingly, it was observed that both resveratrol and the 4′-O-sulfate metabolite are potent inhibitors of both COX-1 and COX-2, with resveratrol-4′-O-sulfate being only slightly less potent than the parent compound. In contrast, the resveratrol-3-O-sulfate and -3-O-glucuronide were found to be only weak inhibitors of both enzymes. Since human plasma concentrations as high as 0.5 μM for resveratrol and 2 to 10 μM for its 4′-O-sulfate conjugate have been reported after an oral dose of pure resveratrol [8], effective in vivo inhibition of both COX-1 and COX-2 could be achieved.
Table 1. Summary of kinetic constants and computationally derived binding energies for the interactions of resveratrol and its metabolites with various molecular targets.
Experimental procedures for measurement of the inhibition constants (IC50) and computational binding energies of resveratrol and its metabolites for COX-1 and COX-2, as well as for determination of the Ka and Ki values of resveratrol and its metabolites for SIRT1 and QR2 are described in the Supplementary Methods. NA, not assayed; NI, no inhibition.
| Compound | COX-1 IC50 (μM) | Computational binding energy (kcal/mol) | COX-2 IC50 (μM) | Computational binding energy (kcal/mol) | SIRT1 Ka(μM) | QR2 Ki (nM) |
|---|---|---|---|---|---|---|
| Resveratrol | 1.1±0.44 | –26 to –25 | 1.3±0.40 | –26 to –25 | 32.2±3.4 | 88±25 (Kd = 54±0.6*) |
| Resveratrol-3-O-sulfate | 68±2.8 | –10 to +13 | >300 | –12 to +145 | 52.6±6.6 | NI |
| Resveratrol-4′-O-sulfate | 5.1±0.55 | –20 to –3 | 2.5±0.35 | –19 to –15 | 36.4±6.7 | NI |
| Resveratrol-3-O-glucuronide | 150±5.8 | +2.7 | >300 | –17 to +7.8 | NA | NI |
Equilibrium dissociation constant (Kd) of resveratrol from the QR2–resveratrol complex. This value was determined via ITC by titrating resveratrol into free QR2. The resulting thermodynamic parameters were ΔH = –14 kcal/mol, –TΔS = 3.9 kcal/mol (1 kcal = 4.184 kJ) and N = 0.9.
X-ray structural basis for inhibition of COX-1 by resveratrol
To directly determine the precise location of the resveratrol-binding site and to gain a better understanding of its mechanism of inhibition, we determined the X-ray crystal structure of the COX-1–resveratrol complex to a resolution of 3.0 Å (Figure 2A and Supplementary Table S1). Analysis of electron density difference maps within the COX and peroxidase active sites of COX-1 revealed that resveratrol binds exclusively to the COX active site (Figure 2A). Residual electron density near the haem cofactor of the peroxidase site was not observed (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/429/bj4290273add.htm). A detailed analysis of the resveratrol-binding conformation shows that one of the 3-O-hydroxy groups of resveratrol makes a hydrogen bond with Ser530 (2.7 Å), the residue selectively acetylated by aspirin [30]. The other 3-O-hydroxy group is capable of forming a hydrogen bond with the backbone carbonyl oxygen of Met522 (Figure 2B). These and other contacts participate in stabilizing resveratrol within the COX active site, allowing for the exclusion of arachidonic acid binding and therefore inhibition of catalysis. A superposition of the COX-1–resveratrol structure with other COX structures reveals that resveratrol binds in a similar manner to that observed for the popular NSAIDs (non-steroidal anti-inflammatory drugs) indomethacin, ibuprofen, diclofenac, celebrex and aspirin (Figure 2C).
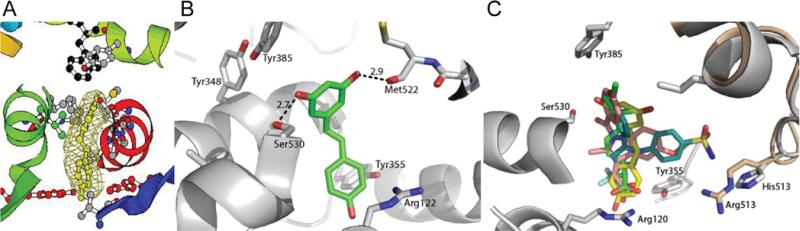
Figure 2. Crystal structure of resveratrol bound to COX-1.
(A) Fo–Fc difference electron density maps surrounding resveratrol contoured at 3σ. The orientation of COX-1–resveratrol and the electron density map are the same as for the COX-1–arachidonic acid structure reported by Malkowski et al. [55] (B) Resveratrol forms hydrogen bonds (broken lines) with the backbone carbonyl of Met522 and the side chain of Ser530, the residue that is selectively acetylated by aspirin. (C) Resveratrol and NSAIDs bind to the same site of COX. COX-1 is shown as a cartoon representation and is coloured in grey. COX-2 is superimposed on to COX-1, but only the portion of COX-2 that is not conserved in COX-1 is shown (wheat). Resveratrol (yellow), indomethacin (pink), ibuprofen (green), and the selective COX-2 inhibitor SC-558 (turquoise) are shown in stick representation. The phenylsulfonamide group of SC-588 extends into the COX-2 side pocket made accessible by Val523 and interacts with Arg513.
In silico prediction of the binding modes of resveratrol and metabolites to COX-1 and COX-2
To elucidate the binding mode of resveratrol in COX-2, we performed molecular docking studies on resveratrol binding to the active sites of both COX-1 and COX-2. We first used the X-ray structure of the COX-1–resveratrol complex as a structural guide to parameterize and validate our computational strategy. The similarities between the X-ray structure and computationally derived poses of resveratrol are shown in Figure 3. The energies of binding of resveratrol to COX-1 and COX-2 were calculated and were found to be remarkably similar, consistent with the nearly identical IC50 values of resveratrol for each enzyme (Table 1). Moreover, the binding orientations and geometries for resveratrol in both the COX-1 and COX-2 channels were superimposeable and consistent with the X-ray structure (Figure 3). Together, the IC50 data, the X-ray structure of the COX-1–resveratrol complex and the computational modelling studies support a model whereby resveratrol binds within the COX active site of both COX-1 and COX-2.
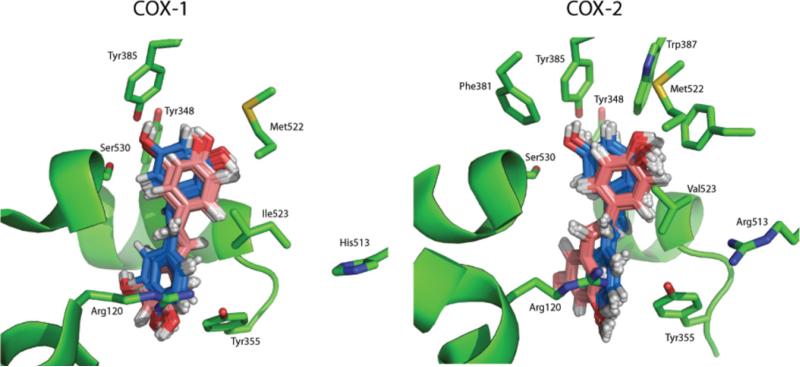
Figure 3. Computational docking orientations for resveratrol binding to the active site of COX-1 and COX-2.
The top 50 energetically favourable solutions for each enzyme are illustrated. The two major orientations resulting from the docking calculation are shown in blue and pink. In blue are shown the solutions with an orientation analogous to that of resveratrol in the X-ray structure. The orientations coloured in pink are instead rotated ~180° compared with resveratrol in the X-ray structure.
Since our computational results with resveratrol were validated with the crystal structure of COX-1–resveratrol complex, we next performed a series of docking studies on the binding of the three resveratrol metabolites to the COX-1 and COX-2 active sites. We found that the binding of the 3- and 4′-O-resveratrol sulfates within both COX active sites were energetically favourable (Table 1). Moreover, their binding geometries resembled those of resveratrol bound to the COX-1 enzyme, since all of the computational solutions were superimposeable with resveratrol within the COX active site (Figure 4). In contrast, the binding free-energies for resveratrol-3-O-glucuronide were found to be mostly unfavourable, which is most likely to be due to the fact that the structure of the glucuronide moiety is too bulky to fit within the COX channel (results not shown). Although our docking studies did not predict the exact binding energies expected based on the IC50 values of the compounds, the results do correctly discriminate between the tight- and poor-binding analogues qualitatively, since the rank order of the binding energies were well correlated with the rank order of the IC50 values: resveratrol > resveratrol-4′-O-sulfate > resveratrol-3-O-sulfate>resveratrol-3-O-glucuronide (Table 1).
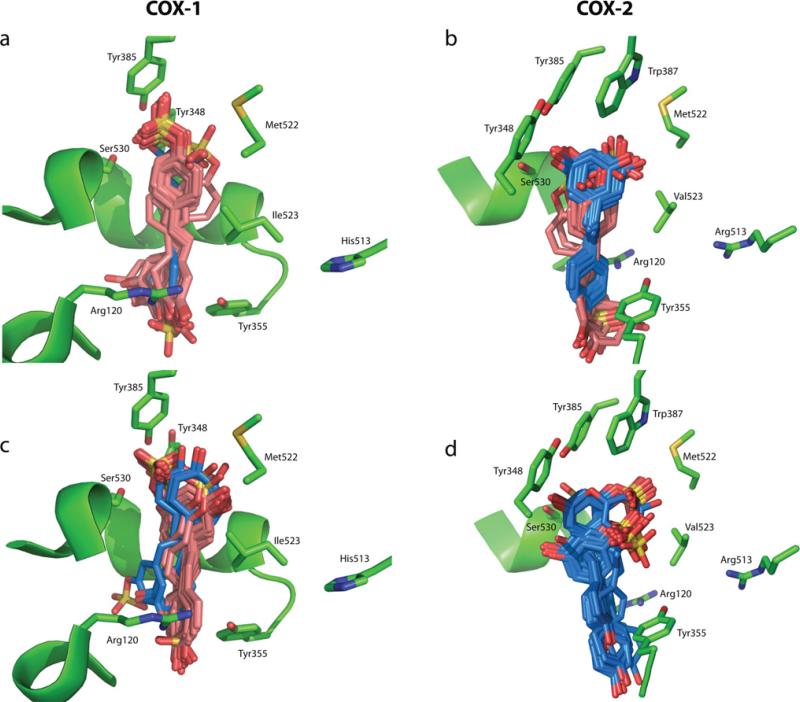
Figure 4. Docking orientations for the resveratrol sulfate metabolites binding to the active sites of COX-1 and COX-2.
Computational docking orientations for the resveratrol sulfate metabolites binding to the active sites of COX-1 (a and c) and COX-2 (b and d). The docking results for each of the sulfate metabolites are as follows: resveratrol-4′-O-sulfate (a and b) and resveratrol-3-O-sulfate (c and d). The two major orientations resulting from the docking calculation are shown in blue and pink for both resveratrol-4′-O-sulfate (a and b) and resveratrol-3-O-sulfate (c and d). Resveratrol-3-O-glucuronide-binding geometries are not shown.
Resveratrol and its sulfate metabolites activate SIRT1, but do not bind directly to the SIRT1 apoenzyme
Resveratrol is proposed to promote organismal longevity and resistance to age-related diseases by targeting and activating the human NAD+-dependent protein deacetylase SIRT1, although results have been inconsistent [5,31–36]. In particular, the activation of SIRT1 by resveratrol in vitro has been shown to be dependent on the presence of a fluorescently-labelled peptide (Fluor de Lys®-SIRT1) that is analogous to one of the SIRT1 natural substrates, the p53 tumor suppressor peptide [31,33]. Nonetheless, we decided to first evaluate whether resveratrol metabolites are capable of activating SIRT1 with the Fluor de Lys®-SIRT1 substrate as does resveratrol. We focused only on the two sulfate metabolites, since they are the most prevalent metabolites in human plasma and since they are the only metabolites that inhibit COX-1 and COX-2. When the two sulfate metabolites were incubated with SIRT1, the enzyme activity was stimulated to the same extent as observed for resveratrol (results not shown) [32]. We then performed a dose–response experiment in which the increase in SIRT1 activity as a function of increasing concentrations of resveratrol or the two sulfate metabolites was measured (Figure 5). From a fit of the data in Figure 5 to a simple binding isotherm, apparent activator constants (Ka values) were determined (Table 1). The nearly equivalent Ka values indicate that the two sulfate analogues are able to activate hSIRT1 to the same extent as resveratrol. These data suggest that resveratrol and its metabolites have an analogous mechanism of action when the Fluor de Lys®–SIRT1 substrate is used.
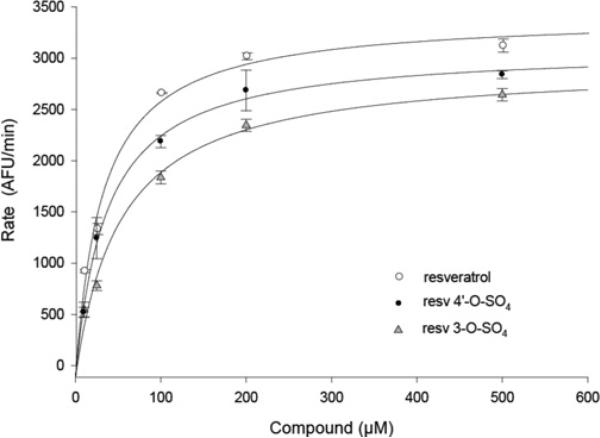
Figure 5. Kinetic response of SIRT1 to the binding of resveratrol and its metabolites.
Initial deacetylation rates of SIRT1 were determined at 37°C using 25 μM of Fluor de Lys®-SIRT1 and 25 μM NAD+ in the presence of 10, 25, 100, 200 and 500 μM of the indicated compound. Lines represent nonlinear least-square fits to the Michaelis–Menten equation. Ka resveratrol (○) = 32.2 ± 3.4 μM, Ka resveratrol-3-O-sulfate (resv 3-O-SO4; ▲) = 52.6 ± 6.6 μM; Ka resveratrol-4′-O-sulfate (resv 4′-O-SO4; ●) = 36.4 ± 6.7 μM. AFU, arbitrary fluorescence units.
It has been hypothesized that resveratrol binds to SIRT1 and induces a conformational change in the enzyme that allows the enzyme to better accommodate the substrate's fluorescent moiety and thereby activate enzyme catalysis. We therefore tested if resveratrol could bind directly to SIRT1. We first investigated the binding properties of resveratrol to SIRT1 using ITC. The addition of resveratrol to the enzyme resulted in no detectable change in heat upon binding, indicating that either resveratrol interacts weakly with SIRT1 or that little enthalpy is associated with binding (results not shown). Therefore, to help distinguish between these two possibilities, we determined whether SIRT1 is capable of binding [3H]resveratrol using a radioligand-binding assay where free SIRT1 is incubated with increasing concentrations of [3H]resveratrol. Under the experimental conditions tested, and in the presence or absence of either the substrate Fluor de Lys®-SIRT1 or NAD+, we could not detect significant binding of [3H]resveratrol to SIRT1 to a level above control reactions without enzyme (results not shown). The radioligand-binding data support that the absence of any observable binding isotherm in ITC experiments is likely the result of a weak interaction of resveratrol with SIRT1 in the absence of its substrates. Our radioligand-binding data do not rule out the possibility that one or both substrates are required for the binding of resveratrol, only that we could not detect any significant binding under the experimental conditions used. Unfortunately, we were unable to perform ITC or radioligand-binding experiments on SIRT1 at concentrations higher than 90 μM due to protein precipitation. As this manuscript was under review, Pacholec et al. [35] showed that another small molecule activator of SIRT1, structurally unrelated to resveratrol, could not bind to the SIRT1–native p53 peptide substrate complex, but was able to bind to the Fluor de Lys®–SIRT1 substrate complex, albeit only weakly. Therefore we conclude that activation of SIRT1 by resveratrol and its sulfate metabolites is dependent on the presence of the fluorogenic moiety on substrates, similar to other reported SIRT1 activators [35].
Resveratrol metabolites display no inhibitory effect on QR2, but resveratrol itself is a potent QR2 inhibitor
One of the highest affinity targets of resveratrol that has been identified to date is the enzyme QR2 [37]. This enzyme has been shown to be important for the prodrug activation of mitomycin C [38,39], an anticancer antibiotic used for treatment of bladder cancer and other tumours, and for product activation of the synthetic anticancer agent CB1954 that is currently in clinical trials [40]. QR2 is capable of metabolically activating various quinones and other compounds, via either 2- or 4-electron reductions, which can ultimately lead to cytotoxicity and cell death [38,41]. In contrast, the NADP-dependent QR1, which is upregulated by cancer chemopreventive agents via the ARE (antioxidant-responsive element), has a completely different in vivo biological function. The induction of QR1 is considered a biomarker for cancer prevention, since the function of QR1 is to inactivate harmful xenobiotic compounds [42]. Therefore inhibition of QR2 by resveratrol or other compounds might protect cells from harmful metabolically activated compounds that can promote DNA damage and therefore cancer.
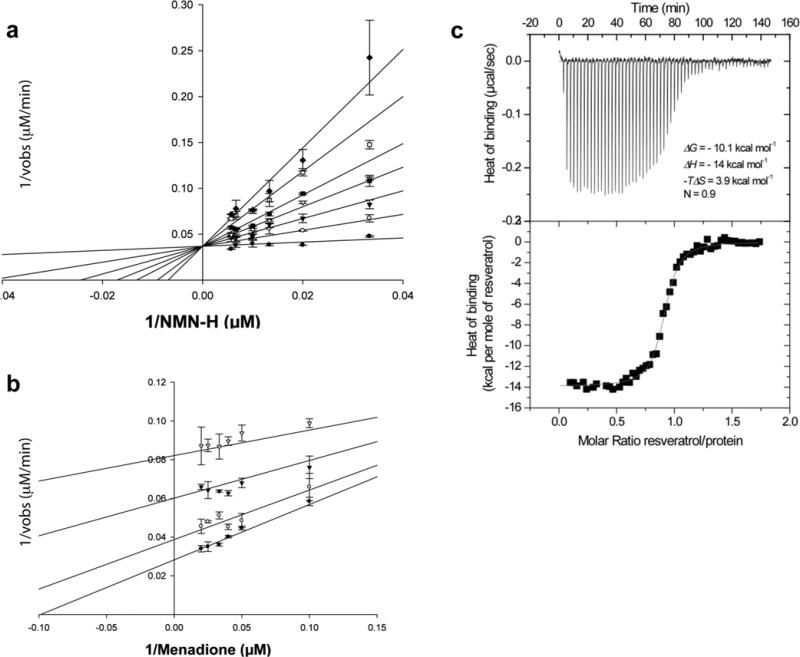
Since the mechanism of QR2 inhibition by resveratrol has not been rigorously established, we measured the kinetics of inhibition of QR2 by resveratrol by varying the substrate concentrations of menadione and NMeH (Figure 6). Resveratrol acts as a competitive inhibitor against NMeH with a Ki value of 80 ± 20 nM (Figure 6A), and as an uncompetitive inhibitor against menadione with a Ki value of 0.9 ± 0.1 μM (Figure 6B). These results suggest that resveratrol inhibits the oxidized state of the FAD cofactor of QR2. The potent inhibition of QR2 by resveratrol prompted us to confirm the tight-binding affinity of the enzyme for resveratrol by ITC (Figure 6C). From three independent measurements, we determined that resveratrol binds tightly to the oxidized, free-enzyme form of QR2 with a dissociation constant (Kd) of 54.0 ± 0.6 nM, which is consistent with the Ki value of 80 ± 20 nM derived from kinetic data. In contrast, the two sulfate metabolites of resveratrol do not inhibit QR2 up to concentrations as high as 100 μM (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/429/bj4290273add.htm). The QR2 inhibition data suggests that since plasma concentrations of resveratrol are able to reach levels as high as 500 nM, QR2 could be almost completely inhibited by resveratrol in vivo. In contrast with its inhibitory properties on QR2, resveratrol has no effect on QR1 activity, being only a weak activator of the enzyme (see Supplementary Table S3 at http://www.BiochemJ.org/bj/429/bj4290273add.htm), as previously reported [43,44].
Figure 6. Potent binding and inhibition of QR2 by resveratrol.
(a) A double-reciprocal plot is shown of the kinetic rate data against various concentrations of NMeH at various concentrations of resveratrol; 0 (●), 0.25 (○), 0.5 (▼), 0.75 (▽), 1 (■), 1.5 (□) and 2 μM (◆). Menadione concentration was 30 μM. (b) A double-reciprocal plot of the kinetic rate data against various concentrations of menadione at various concentrations of resveratrol; 0 (●), 0.5 (○), 2 (▼) and 5 μM (▽). NMeH concentration was 100 μM. Continuous lines represent the best-fit of the data to the linearized form of the Michaelis–Menten equation using the kinetics module in SigmaPlot 9.0. (c) ITC traces (top panel) and the binding isotherm (bottom panel) are shown for the calorimetric titration of resveratrol into QR2. The continuous line in the bottom panel represents the fit of the data to a single binding site model with Kd = 54 ± 0.6 nM. The resulting thermodynamic parameters were ΔH = –14 kcal/mol (1 kcal = 4.184 kJ), TΔS = 3.9 kcal/mol and N = 0.9.
DISCUSSION
The activity of a series of human metabolites of resveratrol on different biological targets of resveratrol was investigated. Here, we show that both resveratrol and resveratrol-4′-O-sulfate are able to inhibit the activity of COX-1 and -2, and that resveratrol alone is able to inhibit QR2, at concentrations that have been shown to be present in human plasma. These results indicate that resveratrol and its 4′-O-sulfate metabolite may mediate or contribute to the health benefits attributed previously only to resveratrol. Our inhibition results indicate that resveratrol and its 4′-O-sulfate metabolite inhibit COX-1 and COX-2 with similar efficacy, and our X-ray structural and computational studies indicate that these compounds bind to the COX sites of the enzymes. In a previous study [45], however, resveratrol was identified as a potent, mechanistic-based peroxidase inhibitor of COX-1 that acts via a ‘hit and run’ mechanism. In the same study, resveratrol was also found to be an ineffective inhibitor against COX-2, a result that we also observed previously using the same assay [4]. These discrepancies could be attributed to the fact that these results, and those of others [4,45,46], are based on different kinetic assays, including those that measure the inhibition of the hydroperoxidase activity of the enzymes using high concentrations of H2O2 for enzymatic detection. These assays are less direct and could obscure interpretation. Additionally, substantially different IC50 values have been observed for different NSAIDs depending on the assay utilized to measure COX inhibition [47]. Therefore, we utilized an assay that is directly coupled to the formation of the product PGH2 (prostaglandin H2), the precursor to important prostanoids involved in disease such as PGE2 and thromboxane [22]. Several studies [48–50], which measured the PGE2 production using this immunoassay system, have shown that resveratrol is an inhibitor of both COX-1 and -2 enzymatic activities, in agreement with our results. In addition, Zykova et al. [49] clearly demonstrated that resveratrol binds directly to COX-2 and that this binding is absolutely required for the inhibition of cancer cell growth by resveratrol, thereby supporting our findings. Therefore the observation that resveratrol and its 4′-O-sulfate metabolite inhibit both COX-1 and COX-2 enzymes with nearly the same efficacy is of importance, since selective inhibition of either one of the enzymes has shown to lead to serious side effects such as gastric ulcer, heart attack and stroke. Compounds that target both enzymes equipotently instead might provide beneficial effects without the complications due to single enzyme inhibition.
We also show by X-ray crystallography that resveratrol binds directly within the arachidonic-acid-binding site of COX-1 and predict via in silico methods that resveratrol is also capable of binding to the arachidonic acid site of COX-2. Murias et al. [48] also predicted via computational approaches that resveratrol and synthetic analogues bind within the arachidonic acid site of COX-1 and -2. They also showed via kinetic studies using a similar assay to ours that both COX-1 and -2 are inhibited by resveratrol. The aforementioned results contrast with the interpretation of kinetics results of Szewczuk et al. [45], who suggest that resveratrol binds only to the peroxidase site of the enzyme. A possible explanation for this inconsistency may be attributed to the use of different assays to measure the activity of the enzyme as discussed above. It is also possible that COX-1 is capable of binding resveratrol on both the COX and peroxidase sites, but binding to the peroxidase site occurs only in the presence of certain substrates, e.g. via an uncompetitive mechanism, as suggested by Szewczuk et al. [45]. Although we cannot rule out the possibility that resveratrol binds within the peroxidase site of COX-1 under different experimental conditions, we believe that kinetic data alone are insufficient to define a precise location of a binding site, since such data are indirect. However, our kinetic, crystallographic and computational modeling results together suggest that resveratrol and its 4′-O-sulfate metabolite are capable of binding within the arachadonic acid sites of COX-1 and COX-2 and, as such, this information could be exploited for the rational design of resveratrol analogues with higher inhibitory potencies against the two COX enzymes. Interestingly, modifying the m-hydroquinone moiety (3,5-di-OH group) of resveratrol, as in the 3-O-sulfate metabolite, results in loss of inhibitory activity towards COX-1 and -2, an observation that supports previous results [45].
Buryanovskyy et al. [37] identified QR2 as a high-affinity target for resveratrol using a resveratrol-affinity column, and they showed resveratrol binds in the active site cleft via X-ray crystallography. Although we also find that resveratrol binds tightly to QR2, with a Kd of 54 nM and a Ki value of 80 nM, we cannot detect any inhibitory activity of resveratrol-4′-O-sulfate and resveratrol-3-O-sulfate against QR2. Analysis of the binding mode of resveratrol within the active site cleft of QR2 indicates it requires a planar binding interaction with the isoalloxazine ring of the FAD cofactor. The addition of a large negatively charged moiety, such as a sulfate moiety, to resveratrol would require a substantial geometric alteration within the active site that would break a series of favourable resveratrol–QR2 interactions. Since resveratrol does inhibit QR2 enzyme at nanomolar concentrations, it is quite possible that in vivo, the plasma concentrations of unmetabolized resveratrol are high enough to completely suppress QR2 activity.
Finally, we show that although resveratrol and its sulfate metabolites are able to activate the SIRT1 enzyme to the same extent when the Fluor de Lys®-SIRT1 peptide substrate is used, resveratrol itself binds only weakly, or not at all, to SIRT1 in the absence of fluorescently-labelled peptide substrates. On the basis of our data, and on the basis of previous findings which show that: (i) resveratrol and other SIRT1 activators increase SIRT1 activity only in the presence of the fluorescently labelled substrate [31,35,51]; (ii) resveratrol and other SIRT1 activators increase the affinity of SIRT1 for the Fluor de Lys®-SIRT1, but not for the native peptide [33]; and (iii) resveratrol and other SIRT1 activators are not able to bind to the enzyme in the absence of the Fluor de Lys®-SIRT1 substrate (the present study and [35]), we propose a model whereby resveratrol, NAD+ and the Fluor de Lys®-SIRT1 substrate mutually co-operate to increase their binding to the enzyme, which eventually results in an enhanced enzyme catalytic activity. Although collectively these results strongly indicate that in vitro SIRT1 is not a direct target of resveratrol, nonetheless it is still possible that in vivo the beneficial effects attributed to resveratrol are in part mediated by SIRT1 and perhaps are mediated by a yet unknown substrate. In fact, several studies in yeast, worms and flies have shown that resveratrol effects are dependent on the Sir2 gene [32,52]. In light of the results from the present study and previous findings, which indicated that SIRT1 lacks specificity in substrate recognition [53], we cannot discount the hypothesis that resveratrol in vivo may either increase the binding affinity of an unidentified endogenous substrate to SIRT1 or allow the enzyme to distinguish between different endogenous targets.
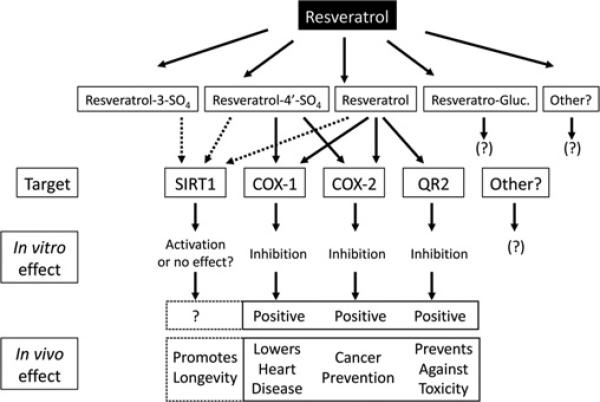
In summary, the results from the present study provide significant new insight into the molecular basis for resveratrol biological activity and, collectively, our results add to those of others and strongly support a mechanistic model whereby resveratrol and its metabolites act pleiotropically through different biological targets to alter their function and therefore the biochemical processes these targets mediate (Figure 7). Consequently, the activity of resveratrol cannot be summarized by a unique mechanism of action. Rather, the biological activity of resveratrol and its metabolites depends on concurrent interactions with various molecular targets of diverse function. We compared the action of resveratrol and its metabolites on three of these targets, COX, SIRT1 and QR2, which are involved in diverse biochemical processes important in cancer chemoprevention, cardiovascular maintenance, inflammation and aging. We have shown that by inhibiting the COX and QR2 enzymes, resveratrol and its metabolites may engender ‘positive’ cardioprotective, cancer preventive and anti-aging properties attributed to red wine and other foods (Figure 7). Regarding the SIRT1 enzyme, whether resveratrol and its metabolites act directly to activate SIRT1 catalytic activity in vivo, or whether the results obtained using a synthetic fluorescent peptide in vitro are experimental artefacts and do not have any validity in vivo, is still matter of considerable debate [54]. However, the micromolar concentrations of resveratrol and its metabolites required to activate SIRT1 are probably beyond the concentrations achievable through diet alone.
Figure 7.
Summary of the pleiotropic effects of resveratrol and its metabolites, and resulting biological responses in cells
Supplementary Material
SUPPLEMENTARY ONLINE DATA
Pleiotropic mechanisms facilitated by resveratrol and its metabolites
Barbara CALAMINI*1,2, Kiira RATIA*, Michael G. MALKOWSKI†, Muriel CUENDET‡, John M. PEZZUTO§, Bernard D. SANTARSIERO* and Andrew D. MESECAR*2
*Department of Medicinal Chemistry and Pharmacognosy and Center for Pharmaceutical Biotechnology, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60607, U.S.A., †Hauptman-Woodward Medical Research Institute and Department of Structural Biology, State University of New York at Buffalo, Buffalo, NY 14203, U.S.A., ‡School of Pharmaceutical Sciences, University of Geneva, 1211 Geneva, Switzerland, and §College of Pharmacy, University of Hawai'i at Hilo, Hilo, HI 96720, U.S.A.
1 Current address: Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, IL 60208, U.S.A.
2 Correspondence should be addressed to either of these authors (b-calamini@northwestern.edu or mesecar@uic.edu).
The atomic co-ordinates for the crystal structure of the cyclo-oxygenase-1–resveratrol complex will appear in the Protein Data Bank under accession code 3BGN.
METHODS
Expression and purification of hSIRT1
Wild-type hSIRT1 from the SIRT1-pHex vector was purified from 4 litres of Rosetta pLysS Escherichia coli cells. Cells were grown at 37°C in LB (Luria–Bertani) broth containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Once the D600 reached 0.7–0.8, protein expression was induced with 1 mM IPTG (isopropyl β-d-thiogalactopyranoside) for 16–20 h at 16°C. Cells were collected by centrifugation at 4300 rev./min for 15 min in a Sorvall Super T21 centrifuge (ST-H750 rotor) and resuspended in 60 ml of Buffer A [25 mM Tris/HCl, pH 8.0, 250 mM NaCl and 1 mM DTT (dithiothreitol)] containing EDTA-free Complete™ mini protease inhibitor cocktail tablets (Roche Applied Science), a pinch of solid DNase I, lysozyme (1 mg/ml) and 0.5 mM PMSF. The cell suspension was sonicated at 600 W for a total of 9 min using a high intensity ultrasonic processor Sonic processor VCX600, with a pulse of 6.6 s on and 9.9 s off. The lysed cells were pelleted by centrifugation at 18000 rev./min for 45 min in a Sorvall Super T21 centrifuge (SL-50T rotor) at 4°C. The cleared lysate was then loaded on to a Hitrap chelating HP affinity column (GE Healthcare) charged with cobalt and equilibrated with 25 mM Tris/HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole and 1 mM DTT. The protein was eluted using a linear gradient from 5 to 50% buffer B (25 mM Tris/HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole and 1 mM DTT). Fractions containing the enzyme were pooled and dialysed overnight at 4°C in 25 mM Tris/HCl, pH 8.0, 10 mM NaCl and 1 mM DTT. The resulting enzyme pool was then applied to a MonoQ (GE Healthcare) anion-exchange column. The protein was eluted using a linear gradient ranging from 5% to 50% of 25 mM Tris/HCl, pH 8.0, 1M NaCl and 1 mM DTT. Fractions containing the enzyme of interest were pooled and finally applied to a Superdex 200 gel filtration column (GE Healthcare), and eluted with 50 mM Tris/HCl, pH 7.5, 300 mM NaCl and 1 mM DTT. The purest fractions were pooled, buffer-exchanged to 50 mM Tris/HCl, pH 8.0, 150 mM NaCl and 10% (v/v) glycerol, and concentrated up to 5 mg/ml using the Centricon Plus-20 device (Millipore). SIRT1 was then stored at –80°C until needed. During the purification, the protein concentration was measured using the determined absorption coefficient (ε = 43810 M–1 · cm–1).
Cloning, expression and purification of human QR2 (quinone reductase 2)
Human QR2 cDNA (MGC-12729) was purchased from ATCC. The primers used in cloning were: 5′-AACCATGGCAGGTAAGAAAGTACTC (the added NcoI site is underlined) and 5′-AACTCGAGTTATTGCCCGAAGTGCCAGTG (the added XhoI site is underlined). The PCR product was digested with NcoI and XhoI restriction enzymes, purified and then ligated into a pET-23d expression vector (Novagen). The parental vector was digested with the methylation-sensitive restriction enzyme DpnI, whereas the unmethylated PCR-amplified vector remains intact. The ligation product was transformed into E. coli XL1 Blue and several colonies were screened for positive gene insertion in the vector by restriction digestion. The entire insert containing the human QR2 gene was sequenced via the Research Resources Center DNA core facility at University of Illinois at Chicago. Human QR2 was purified from 3 litres of E. coli BL21(DE3) in LB broth containing 100 μg/ml ampicillin grown at 37°C. The expression of human QR2 was induced by the addition of 1 mM IPTG when the culture attenuance at 600 nm reached 0.7–0.8. Cells were then transferred at 16°C for 12 h. The enzyme expression was evaluated using SDS/PAGE and enzyme activity
Table S1 X-ray data collection and refinement statistics for the COX-1–resveratrol complex assay. The cells were collected by centrifugation at 4300 rev./min for 15 min in a Sorvall Super T21 centrifuge (ST-H750 rotor) and resuspended in 60 ml of Buffer A1 (25 mM Tris/HCl, pH 8.0, 10 mM NaCl and 1 mM DTT) containing 2 tablets of EDTA-free Complete™ mini protease inhibitor cocktail tablets and 0.5 mM PMSF. The cell suspension was sonicated at 600 W for a total of 6 min using a high intensity ultrasonic processor Sonic processor VCX600, with a pulse of 6.6 s on and 9.9 s off. The lysed cells were pelleted by centrifugation at 18000 rev./min for 45 min in a Sorvall Super T21 centrifuge (SL-50T rotor) at 4°C. The supernatant was checked for human QR2 activity using the enzyme activity assay described in the Experimental section of the main text, and then was applied to a DEAE-Sepharose column (Amersham Pharmacia Biotech) equilibrated with buffer A1. The enzyme was eluted with a linear gradient of increasing buffer B1 (25 mM Tris/HCl, pH 8.0, 1 M NaCl and 1 mM DTT). Fractions containing the enzyme (detected by SDS/PAGE) were pooled and concentrated using an 80 ml Centricon Plus-20 concentration device from Millipore (molecular mass cut-off 10000 Da). The enzyme was then applied to a Superdex 75 (Amersham Pharmacia Biotech) column equilibrated with buffer A. The enzyme was eluted with buffer A and the fractions containing pure enzyme were pooled and finally applied to a MonoQ (GE Healthcare) anion-exchange column equilibrated with buffer A. The protein was eluted using a gradient ranging from 5% to 50% buffer B. Protein was collected in 3 ml fractions. The purest fractions were pooled, the buffer was exchanged to 50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 10% (v/v) glycerol, and concentrated using the Centricon Plus-20 device. QR2 was then stored at –80°C. During the purification, the protein concentration was measured using the Bradford assay [1].
Effect of resveratrol on QR1 activity
This assay was modified from a previously described method [2]. Cultured Hepa 1c1c7 mouse hepatoma cells were plated at a density of 2×104 cells/ml in 96-well plates and incubated for 24 h. The medium was then changed and resveratrol, dissolved in 10% (v/v) DMSO, was added to each well. The cells were incubated for an additional 48 h. QR1 activity was measured by the NADPH-dependent menadiol-mediated reduction of MTT [3-(4,5-dimethylthiazo-2-yl)-2,5-diphenyltetrazolium bromide] to a blue formazan. Enzyme activity was expressed as a ‘CD’ value, the concentration of test material needed to double the specific activity of QR1.
Figure S1 Resveratrol does not bind to the peroxidase site of COX-1
Electron density map (2Fo–Fc contoured at 1σ surrounding the haem cofactor in the peroxidase site of COX-1. Residual electron density for resveratrol is not observed. The haem cofactor (light blue) is shown as stick representation. COX-1 is represented by the surface, and the metal and water molecules are represented as pink and red spheres respectively.
Table S2 Comparison of the steady-state kinetic parameters for various inhibition models for the inhibition of QR2 by resveratrol
Table S3 Effect of resveratrol on the activity of hQR1
Figure S2 Residual activity of QR2 in the absence and presence of resveratrol and resveratrol-sulfate metabolites
The assays were measured using 5 nM of QR2, 100 μM NMeH (N-methylnicotinamide) and 30 μM menadione. The compounds were tested at final concentrations of 100 μM. 3SO4, resveratrol-3-O-sulfate; 4′SO4, resveratrol-4′-O-sulfate.
REFERENCES
1 Bradford, M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254
2 Zhang, Y., Talalay, P., Cho, C. G. and Posner, G. H. (1992) A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc. Natl. Acad. Sci. U.S.A. 89, 2399–2403
ACKNOWLEDGEMENTS
We would like to thank Dr David Sinclair (Department of Pathology, Harvard University, Boston, MA, U.S.A.) for the gift of the hSIRT1 plasmid. We are grateful to the synchrotron beamline personnel at the Advanced Photon Source (BioCars, Argonne National Laboratory) 14-BM-C beamline.
FUNDING This work is supported by grants from the National Cancer Institute of the National Institutes of Health [grant numbers CA92744 and PO1 CA48112]. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (under Contract W-31-109-Eng-38).
Abbreviations used
- β-OG
n-octyl-β-d-glucopyranoside
- COX
cyclo-oxygenase
- hSIRT
human SIRT1
- ITC
isothermal titration calorimetry
- QR2
quinone reductase 2
- NSAID
non-steroidal anti-inflammatory drug
- NMeH
N-methyldihydronicotinamide
- PGE2
prostaglandin E2
Footnotes
The atomic co-ordinates for the crystal structure of the cyclo-oxygenase-1–resveratrol complex will appear in the Protein Data Bank under accession code 3BGN.
REFERENCES
- 1.Das DK, Sato M, Ray PS, Maulik G, Engelman RM, Bertelli AA, Bertelli A. Cardioprotection of red wine: role of polyphonic antioxidants. Drugs Exp. Clin. Res. 1999;25:115–120. [PubMed] [Google Scholar]
- 2.Holmes-McNary M, Baldwin AS., Jr Chemopreventive properties of trans-resveratrol are associated with inhibition of activation of the IκB kinase. Cancer Res. 2000;60:3477–3483. [PubMed] [Google Scholar]
- 3.Siemann EH, Creasy LL. Concentration of the phytoalexin resveratrol in wine. Am. J. Enol. Vitic. 1992;43:49–52. [Google Scholar]
- 4.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 5.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pezzuto JM. Resveratrol as an inhibitor of carcinogenesis. In: Aggarwal BB, Shishodia S, editors. Resveratrol in Health and Disease. CRC Press; Boca Raton: 2006. pp. 233–383. [Google Scholar]
- 7.Corder R, Crozier A, Kroon PA. Drinking your health? It's too early to say. Nature. 2003;426:119. doi: 10.1038/426119d. [DOI] [PubMed] [Google Scholar]
- 8.Boocock DJ, Faust GE, Patel KR, Schinas AM, Brown VA, Ducharme MP, Booth TD, Crowell JA, Perloff M, Gescher AJ, et al. Phase I dose escalation pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive agent. Cancer Epidemiol. Biomarkers Prev. 2007;16:1246–1252. doi: 10.1158/1055-9965.EPI-07-0022. [DOI] [PubMed] [Google Scholar]
- 9.Boocock DJ, Patel KR, Faust GE, Normolle DP, Marczylo TH, Crowell JA, Brenner DE, Booth TD, Gescher A, Steward WP. Quantitation of trans-resveratrol and detection of its metabolites in human plasma and urine by high performance liquid chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;848:182–187. doi: 10.1016/j.jchromb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walle T, Hsieh F, DeLegge MH, Oatis JE, Jr, Walle UK. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab. Dispos. 2004;32:1377–1382. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- 11.Yu C, Shin YG, Chow A, Li Y, Kosmeder JW, Lee YS, Hirschelman WH, Pezzuto JM, Mehta RG, van Breemen RB. Human, rat, and mouse metabolism of resveratrol. Pharm. Res. 2002;19:1907–1914. doi: 10.1023/a:1021414129280. [DOI] [PubMed] [Google Scholar]
- 12.Yu C, Shin YG, Kosmeder JW, Pezzuto JM, van Breemen RB. Liquid chromatography/tandem mass spectrometric determination of inhibition of human cytochrome P450 isozymes by resveratrol and resveratrol-3-sulfate. Rapid Commun. Mass Spectrom. 2003;17:307–313. doi: 10.1002/rcm.918. [DOI] [PubMed] [Google Scholar]
- 13.Kaldas MI, Walle UK, Walle T. Resveratrol transport and metabolism by human intestinal Caco-2 cells. J. Pharm. Pharmacol. 2003;55:307–312. doi: 10.1211/002235702612. [DOI] [PubMed] [Google Scholar]
- 14.Yu C, Shin YG, Chow A, Li Y, Kosmeder JW, Lee YS, Hirschelman WH, Pezzuto JM, Mehta RG, van Breemen RB. Human, rat, and mouse metabolism of resveratrol. Pharm. Res. 2002;19:1907–1914. doi: 10.1023/a:1021414129280. [DOI] [PubMed] [Google Scholar]
- 15.Ortiz-Maldonado M, Gatti D, Ballou DP, Massey V. Structure-function correlations of the reaction of reduced nicotinamide analogues with p-hydroxybenzoate hydroxylase substituted with a series of 8-substituted flavins. Biochemistry. 1999;38:16636–16647. doi: 10.1021/bi991603u. [DOI] [PubMed] [Google Scholar]
- 16.Malkowski MG, Theisen MJ, Scharmen A, Garavito RM. The formation of stable fatty acid substrate complexes in prostaglandin H2 synthase-1. Arch. Biochem. Biophys. 2000;380:39–45. doi: 10.1006/abbi.2000.1906. [DOI] [PubMed] [Google Scholar]
- 17.Smith T, Leipprandt J, DeWitt D. Purification and characterization of the human recombinant histidine-tagged prostaglandin endoperoxide H synthases-1 and -2. Arch. Biochem. Biophys. 2000;375:195–200. doi: 10.1006/abbi.1999.1659. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Hsieh TC, Zhang Z, Ma Y, Wu JM. Identification and purification of resveratrol targeting proteins using immobilized resveratrol affinity chromatography. Biochem. Biophys. Res. Commun. 2004;323:743–749. doi: 10.1016/j.bbrc.2004.08.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology: Volume 276: Macromolecular Crystallography, Part A. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Cuendet M, Mesecar AD, Dewitt DL, Pezzuto JM. An ELISA method to measure inhibition of the Cox enzymes. Nature Protocols. 2006;1:1–7. doi: 10.1038/nprot.2006.308. [DOI] [PubMed] [Google Scholar]
- 23.Kuntz ID, Ewing JA. Critical evaluation of search algorithms used in automated molecular docking. J. Comput. Chem. 1997;18:1175–1189. [Google Scholar]
- 24.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 26.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 27.Kwiek JJ, Haystead TA, Rudolph J. Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry. 2004;43:4538–4547. doi: 10.1021/bi035923w. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg DM, Hahn SE, Parkes JG. Beyond alcohol: beverage consumption and cardiovascular mortality. Clin. Chim. Acta. 1995;237:155–187. doi: 10.1016/0009-8981(95)06069-p. [DOI] [PubMed] [Google Scholar]
- 29.Pace-Asciak CR, Hahn S, Diamandis EP, Soleas G, Goldberg DM. The red wine phenolics trans-resveratrol and quercetin block human platelet aggregation and eicosanoid synthesis: implications for protection against coronary heart disease. Clin. Chim. Acta. 1995;235:207–219. doi: 10.1016/0009-8981(95)06045-1. [DOI] [PubMed] [Google Scholar]
- 30.Roth GJ, Stanford N, Majerus PW. Acetylation of prostaglandin synthase by aspirin. Proc. Natl. Acad. Sci. U.S.A. 1975;72:3073–3076. doi: 10.1073/pnas.72.8.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 32.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 33.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 34.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pacholec M, Chrunyk BA, Cunningham D, Flynn D, Griffith DA, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buryanovskyy L, Fu Y, Boyd M, Ma Y, Hsieh TC, Wu JM, Zhang Z. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry. 2004;43:11417–11426. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celli CM, Tran N, Knox R, Jaiswal AK. NRH:quinone oxidoreductase 2 (NQO2) catalyzes metabolic activation of quinones and anti-tumor drugs. Biochem. Pharmacol. 2006;72:366–376. doi: 10.1016/j.bcp.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 39.Jamieson D, Tung AT, Knox RJ, Boddy AV. Reduction of mitomycin C is catalysed by human recombinant NRH:quinone oxidoreductase 2 using reduced nicotinamide adenine dinucleotide as an electron donating co-factor. Br. J. Cancer. 2006;95:1229–1233. doi: 10.1038/sj.bjc.6603414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knox RJ, Jenkins TC, Hobbs SM, Chen S, Melton RG, Burke PJ. Bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by human NAD(P)H quinone oxidoreductase 2: a novel co-substrate-mediated antitumor prodrug therapy. Cancer Res. 2000;60:4179–4186. [PubMed] [Google Scholar]
- 41.Long DJ, 2nd, Iskander K, Gaikwad A, Arin M, Roop DR, Knox R, Barrios R, Jaiswal AK. Disruption of dihydronicotinamide riboside:quinone oxidoreductase 2 (NQO2) leads to myeloid hyperplasia of bone marrow and decreased sensitivity to menadione toxicity. J. Biol. Chem. 2002;277:46131–46139. doi: 10.1074/jbc.M208675200. [DOI] [PubMed] [Google Scholar]
- 42.Kang Y-H, Pezzuto J. Quinones and Quinone Enzymes. In: Sies H, Packer L, editors. Methods in Enzymology. Elsevier Science; San Diego: 2004. pp. 380–415. [Google Scholar]
- 43.Zahid M, Gaikwad NW, Ali MF, Lu F, Saeed M, Yang L, Rogan EG, Cavalieri EL. Prevention of estrogen-DNA adduct formation in MCF-10F cells by resveratrol. Free Radical Biol. Med. 2008;45:136–145. doi: 10.1016/j.freeradbiomed.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu F, Zahid M, Wang C, Saeed M, Cavalieri EL, Rogan EG. Resveratrol prevents estrogen-DNA adduct formation and neoplastic transformation in MCF-10F cells. Cancer Prev. Res. 2008;1:135–145. doi: 10.1158/1940-6207.CAPR-08-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szewczuk LM, Forti L, Stivala LA, Penning TM. Resveratrol is a peroxidase-mediated inactivator of COX-1 but not COX-2: a mechanistic approach to the design of COX-1 selective agents. J. Biol. Chem. 2004;279:22727–22737. doi: 10.1074/jbc.M314302200. [DOI] [PubMed] [Google Scholar]
- 46.Johnson JL, Maddipati KR. Paradoxical effects of resveratrol on the two prostaglandin H synthases. Prostaglandins Other Lipid Mediat. 1998;56:131–143. doi: 10.1016/s0090-6980(98)00052-5. [DOI] [PubMed] [Google Scholar]
- 47.Gierse JK, Koboldt CM, Walker MC, Seibert K, Isakson PC. Kinetic basis for selective inhibition of cyclo-oxygenases. Biochem. J. 1999;339:607–614. [PMC free article] [PubMed] [Google Scholar]
- 48.Murias M, Handler N, Erker T, Pleban K, Ecker G, Saiko P, Szekeres T, Jager W. Resveratrol analogues as selective cyclooxygenase-2 inhibitors: synthesis and structure-activity relationship. Bioorg. Med. Chem. 2004;12:5571–5578. doi: 10.1016/j.bmc.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Zykova TA, Zhu F, Zhai X, Ma WY, Ermakova SP, Lee KW, Bode AM, Dong Z. Resveratrol directly targets COX-2 to inhibit carcinogenesis. Mol. Carcinog. 2008;47:797–805. doi: 10.1002/mc.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subbaramaiah K, Chung WJ, Michaluart P, Telang N, Tanabe T, Inoue H, Jang M, Pezzuto JM, Dannenberg AJ. Resveratrol inhibits cyclooxygenase-2 transcription and activity in phorbol ester-treated human mammary epithelial cells. J. Biol. Chem. 1998;273:21875–21882. doi: 10.1074/jbc.273.34.21875. [DOI] [PubMed] [Google Scholar]
- 51.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 52.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 53.Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J. Biol. Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt C. GSK/Sirtris compounds dogged by assay artifacts. Nat. Biotechnol. 2010;28:185–186. doi: 10.1038/nbt0310-185. [DOI] [PubMed] [Google Scholar]
- 55.Malkowski MG, Ginell SL, Smith WL, Garavito RM. The productive conformation of arachidonic acid bound to prostaglandin synthase. Science. 2000;289:1933–1937. doi: 10.1126/science.289.5486.1933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY ONLINE DATA
Pleiotropic mechanisms facilitated by resveratrol and its metabolites
Barbara CALAMINI*1,2, Kiira RATIA*, Michael G. MALKOWSKI†, Muriel CUENDET‡, John M. PEZZUTO§, Bernard D. SANTARSIERO* and Andrew D. MESECAR*2
*Department of Medicinal Chemistry and Pharmacognosy and Center for Pharmaceutical Biotechnology, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60607, U.S.A., †Hauptman-Woodward Medical Research Institute and Department of Structural Biology, State University of New York at Buffalo, Buffalo, NY 14203, U.S.A., ‡School of Pharmaceutical Sciences, University of Geneva, 1211 Geneva, Switzerland, and §College of Pharmacy, University of Hawai'i at Hilo, Hilo, HI 96720, U.S.A.
1 Current address: Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, IL 60208, U.S.A.
2 Correspondence should be addressed to either of these authors (b-calamini@northwestern.edu or mesecar@uic.edu).
The atomic co-ordinates for the crystal structure of the cyclo-oxygenase-1–resveratrol complex will appear in the Protein Data Bank under accession code 3BGN.
METHODS
Expression and purification of hSIRT1
Wild-type hSIRT1 from the SIRT1-pHex vector was purified from 4 litres of Rosetta pLysS Escherichia coli cells. Cells were grown at 37°C in LB (Luria–Bertani) broth containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Once the D600 reached 0.7–0.8, protein expression was induced with 1 mM IPTG (isopropyl β-d-thiogalactopyranoside) for 16–20 h at 16°C. Cells were collected by centrifugation at 4300 rev./min for 15 min in a Sorvall Super T21 centrifuge (ST-H750 rotor) and resuspended in 60 ml of Buffer A [25 mM Tris/HCl, pH 8.0, 250 mM NaCl and 1 mM DTT (dithiothreitol)] containing EDTA-free Complete™ mini protease inhibitor cocktail tablets (Roche Applied Science), a pinch of solid DNase I, lysozyme (1 mg/ml) and 0.5 mM PMSF. The cell suspension was sonicated at 600 W for a total of 9 min using a high intensity ultrasonic processor Sonic processor VCX600, with a pulse of 6.6 s on and 9.9 s off. The lysed cells were pelleted by centrifugation at 18000 rev./min for 45 min in a Sorvall Super T21 centrifuge (SL-50T rotor) at 4°C. The cleared lysate was then loaded on to a Hitrap chelating HP affinity column (GE Healthcare) charged with cobalt and equilibrated with 25 mM Tris/HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole and 1 mM DTT. The protein was eluted using a linear gradient from 5 to 50% buffer B (25 mM Tris/HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole and 1 mM DTT). Fractions containing the enzyme were pooled and dialysed overnight at 4°C in 25 mM Tris/HCl, pH 8.0, 10 mM NaCl and 1 mM DTT. The resulting enzyme pool was then applied to a MonoQ (GE Healthcare) anion-exchange column. The protein was eluted using a linear gradient ranging from 5% to 50% of 25 mM Tris/HCl, pH 8.0, 1M NaCl and 1 mM DTT. Fractions containing the enzyme of interest were pooled and finally applied to a Superdex 200 gel filtration column (GE Healthcare), and eluted with 50 mM Tris/HCl, pH 7.5, 300 mM NaCl and 1 mM DTT. The purest fractions were pooled, buffer-exchanged to 50 mM Tris/HCl, pH 8.0, 150 mM NaCl and 10% (v/v) glycerol, and concentrated up to 5 mg/ml using the Centricon Plus-20 device (Millipore). SIRT1 was then stored at –80°C until needed. During the purification, the protein concentration was measured using the determined absorption coefficient (ε = 43810 M–1 · cm–1).
Cloning, expression and purification of human QR2 (quinone reductase 2)
Human QR2 cDNA (MGC-12729) was purchased from ATCC. The primers used in cloning were: 5′-AACCATGGCAGGTAAGAAAGTACTC (the added NcoI site is underlined) and 5′-AACTCGAGTTATTGCCCGAAGTGCCAGTG (the added XhoI site is underlined). The PCR product was digested with NcoI and XhoI restriction enzymes, purified and then ligated into a pET-23d expression vector (Novagen). The parental vector was digested with the methylation-sensitive restriction enzyme DpnI, whereas the unmethylated PCR-amplified vector remains intact. The ligation product was transformed into E. coli XL1 Blue and several colonies were screened for positive gene insertion in the vector by restriction digestion. The entire insert containing the human QR2 gene was sequenced via the Research Resources Center DNA core facility at University of Illinois at Chicago. Human QR2 was purified from 3 litres of E. coli BL21(DE3) in LB broth containing 100 μg/ml ampicillin grown at 37°C. The expression of human QR2 was induced by the addition of 1 mM IPTG when the culture attenuance at 600 nm reached 0.7–0.8. Cells were then transferred at 16°C for 12 h. The enzyme expression was evaluated using SDS/PAGE and enzyme activity
Table S1 X-ray data collection and refinement statistics for the COX-1–resveratrol complex assay. The cells were collected by centrifugation at 4300 rev./min for 15 min in a Sorvall Super T21 centrifuge (ST-H750 rotor) and resuspended in 60 ml of Buffer A1 (25 mM Tris/HCl, pH 8.0, 10 mM NaCl and 1 mM DTT) containing 2 tablets of EDTA-free Complete™ mini protease inhibitor cocktail tablets and 0.5 mM PMSF. The cell suspension was sonicated at 600 W for a total of 6 min using a high intensity ultrasonic processor Sonic processor VCX600, with a pulse of 6.6 s on and 9.9 s off. The lysed cells were pelleted by centrifugation at 18000 rev./min for 45 min in a Sorvall Super T21 centrifuge (SL-50T rotor) at 4°C. The supernatant was checked for human QR2 activity using the enzyme activity assay described in the Experimental section of the main text, and then was applied to a DEAE-Sepharose column (Amersham Pharmacia Biotech) equilibrated with buffer A1. The enzyme was eluted with a linear gradient of increasing buffer B1 (25 mM Tris/HCl, pH 8.0, 1 M NaCl and 1 mM DTT). Fractions containing the enzyme (detected by SDS/PAGE) were pooled and concentrated using an 80 ml Centricon Plus-20 concentration device from Millipore (molecular mass cut-off 10000 Da). The enzyme was then applied to a Superdex 75 (Amersham Pharmacia Biotech) column equilibrated with buffer A. The enzyme was eluted with buffer A and the fractions containing pure enzyme were pooled and finally applied to a MonoQ (GE Healthcare) anion-exchange column equilibrated with buffer A. The protein was eluted using a gradient ranging from 5% to 50% buffer B. Protein was collected in 3 ml fractions. The purest fractions were pooled, the buffer was exchanged to 50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 10% (v/v) glycerol, and concentrated using the Centricon Plus-20 device. QR2 was then stored at –80°C. During the purification, the protein concentration was measured using the Bradford assay [1].
Effect of resveratrol on QR1 activity
This assay was modified from a previously described method [2]. Cultured Hepa 1c1c7 mouse hepatoma cells were plated at a density of 2×104 cells/ml in 96-well plates and incubated for 24 h. The medium was then changed and resveratrol, dissolved in 10% (v/v) DMSO, was added to each well. The cells were incubated for an additional 48 h. QR1 activity was measured by the NADPH-dependent menadiol-mediated reduction of MTT [3-(4,5-dimethylthiazo-2-yl)-2,5-diphenyltetrazolium bromide] to a blue formazan. Enzyme activity was expressed as a ‘CD’ value, the concentration of test material needed to double the specific activity of QR1.
Figure S1 Resveratrol does not bind to the peroxidase site of COX-1
Electron density map (2Fo–Fc contoured at 1σ surrounding the haem cofactor in the peroxidase site of COX-1. Residual electron density for resveratrol is not observed. The haem cofactor (light blue) is shown as stick representation. COX-1 is represented by the surface, and the metal and water molecules are represented as pink and red spheres respectively.
Table S2 Comparison of the steady-state kinetic parameters for various inhibition models for the inhibition of QR2 by resveratrol
Table S3 Effect of resveratrol on the activity of hQR1
Figure S2 Residual activity of QR2 in the absence and presence of resveratrol and resveratrol-sulfate metabolites
The assays were measured using 5 nM of QR2, 100 μM NMeH (N-methylnicotinamide) and 30 μM menadione. The compounds were tested at final concentrations of 100 μM. 3SO4, resveratrol-3-O-sulfate; 4′SO4, resveratrol-4′-O-sulfate.
REFERENCES
1 Bradford, M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254
2 Zhang, Y., Talalay, P., Cho, C. G. and Posner, G. H. (1992) A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc. Natl. Acad. Sci. U.S.A. 89, 2399–2403