Abstract
Circadian clocks maintain temporal homeostasis by generating daily output rhythms in molecular, cellular, and physiological functions. Output rhythms, such as sleep/wake cycles and hormonal fluctuations, tend to deteriorate during aging in humans, rodents, and fruit flies. However, it is not clear whether this decay is caused by defects in the core transcriptional clock, or weakening of the clock-output pathways, or both. The authors monitored age-related changes in behavioral and molecular rhythms in Drosophila melanogaster. Aging was associated with disrupted rest/activity patterns and lengthening of the free-running period of the circadian locomotor activity rhythm. The expression of core clock genes was measured in heads and bodies of young, middle-aged, and old flies. Transcriptional oscillations of four clock genes, period, timeless, Par domain protein 1ε, and vrille, were significantly reduced in heads, but not in bodies, of aging flies. It was determined that reduced transcription of these genes was not caused by the deficient expression of their activators, encoded by Clock and cycle genes. Interestingly, transcriptional activation by CLOCK-CYCLE complexes was impaired despite reduced levels of the PERIOD repressor protein in old flies. These data suggest that aging alters the properties of the core transcriptional clock in flies such that both the positive and the negative limbs of the clock are attenuated.
Keywords: Aging, Circadian clock, Clock gene expression, Drosophila
BACKGROUND
Circadian rhythms at the molecular, behavioral, and physiological levels are important for maintaining temporal homeostasis (Reddy & O’Neill, 2010). Whereas robust high-amplitude circadian rhythms are observed in young individuals, these often lose their strength with age. Daily rhythms in hormone levels, body temperature, sleep/wake cycles, and other physiological and behavioral variables are diminished during aging (Touitou & Haus, 2000; Weinert & Waterhouse, 2007; Zhdanova et al., 2011). Loss of temporal coordination in humans is correlated with a variety of diseases, including Alzheimer’s disease and cancer (Van Someren & Riemersma-Van Der Lek, 2007; Wu & Swaab, 2007). Functional studies in mice demonstrated that disruption of circadian rhythms by the knockout of specific clock genes or chronic jet-lag accelerates the onset of age-related pathologies and may reduce life span (Antoch et al., 2008; Davidson et al., 2006; Kondratov et al., 2006; Lee, 2006). Similar to vertebrates, the strength of the sleep/wake rhythm is also reduced in aged Drosophila, indicated by fragmentation of sleep and decreased length of activity bouts in light/dark cycles (Koh et al., 2006). These data suggest that links between the circadian system and aging are evolutionarily conserved. Given that circadian coordination has a pronounced impact on physiological functions, overall health, and disease susceptibility, it is important to determine why circadian rhythms diminish during aging, and whether this process could be reversed. Consequently, there is a need to understand how the core clock mechanism is altered during aging.
At the molecular level, the circadian clock is based on transcription-translation feedback loops that are largely conserved from Drosophila to mammals (Stanewsky, 2003; Yu & Hardin, 2006). In fruit flies, the key activator complex is composed of two transcription factors encoded by the genes Clock (Clk) and cycle (cyc), the latter known as Bmal1 in mammals. The CLK-CYC complex stimulates the expression of genes period (per) and timeless (tim) in the early night. PER and TIM proteins accumulate in cell nuclei late at night, and PER represses CLK-CYC transcriptional activity, resulting in suppression of per and tim transcription (Hardin, 2004). CLK-CYC complexes also induce the expression of transcription factors Par domain protein 1ε (Pdp1ε) and vrille (vri), which contribute to the rhythmic expression of Clk (Cyran et al., 2003; Glossop et al., 2003; Zheng & Sehgal, 2008). Another gene, clockwork orange (cwo), was recently shown to regulate core clock gene expression (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). Posttranslational modifications that affect the phosphorylation status and degradation of clock proteins are also important modulators of period and amplitude of circadian oscillations (Allada & Chung, 2010; Bae & Edery, 2006).
Whereas the core transcriptional clock is well understood in young animals, much less is known about changes in the oscillations of clock genes during aging. Studies performed in aging mammals and zebra fish reported either reduced or normal expression of different clock genes, depending on the organs and species examined (Asai et al., 2001; Kolker et al., 2003; Yamazaki et al., 2002; Zhdanova et al., 2008). Studies of age-related changes in the vertebrate clock mechanism are challenging, because most clock genes have paralogs with partially overlapping functions (Dibner et al., 2010; Ko & Takahashi, 2006). In flies, there is a single ortholog for every clock gene, and each one cycles with its characteristic phase in all the cells due to direct light sensitivity (Giebultowicz, 2000). Here, we investigated the effects of age on the molecular clock mechanism in heads and bodies of Drosophila melanogaster.
We have previously shown that per mRNA oscillations become attenuated in old flies (Krishnan et al., 2009). To obtain further insights into age-related changes in the clock mechanism in Drosophila, we investigated the expression profiles of seven clock-related genes acting in circadian feedback loops. We report that the expression levels of four genes regulated by the CLK-CYC activator complex are reduced in fly heads, but not in bodies. We then investigated whether the expression of Clk or cyc is affected by age. Finally, we measured profiles of PER and TIM proteins in aging flies to elucidate changes in the repressive phase of the transcriptional feedback loop. Our study suggests that aging weakens the positive limb of the circadian feedback loop in a manner that may also affect the negative limb of the clock.
METHODS
All of the experimental protocols conform to international ethical standards (Portaluppi et al., 2010).
Fly Stocks and Rearing
D. melanogaster were reared on 1% agar, 6.25% cornmeal, 6.25% molasses, and 3.5% Red Star yeast at 25°C in 12-h light:12-h dark (LD 12:12) cycles (with an average light intensity of ~2000 lux). By convention, lights-off is denoted as zeitgeber time (ZT) 12. For experiments on aging flies, cohorts of 100 Canton Special (CS) mated males, were housed in 8-oz round bottom poly-propylene bottles (Genesee Scientific, San Diego, CA) inverted over 60-mm Falcon Primaria Tissue culture dishes (Becton Dickinson Labware, Franklin Lakes, NJ) containing 15 mL of diet. Diet dishes were replaced daily without CO2, after tapping flies to the bottom of the bottle.
Locomotor Activity Analysis
Flies were entrained in LD 12:12 at 25°C. Locomotor activity of 5-, 35-, and 50-day-old males was recorded for 3 d in LD 12:12, followed by 7 d in constant darkness (DD) using the Trikinetics locomotor activity monitor (Waltham, MA). For a quantitative measure of circadian rhythmicity, fast Fourier transform (FFT) analysis was conducted using ClockLab software (Actimetrics; Coulbourn Instruments, Whitehall, PA). Flies with FFT values <.04 were classified as arrhythmic, ones with values of .04–.08 were classified as weakly rhythmic, whereas flies with FFT values >.08 were considered strongly rhythmic. Flies with both weak and strong rhythms, which showed a single peak in the periodogram, were included in the calculation of the free-running period using the ClockLab software (Actimetrics, Wilmette, IL).
Quantitative Real-Time Polymerase Chain Reaction
Three independent bioreplicates of flies were collected at 4-h intervals around the clock on days 5, 35, and 50. Total RNA was extracted from fly heads and bodies separately using TriReagent (Sigma, St. Louis, MO). The samples were purified using the RNeasy mini kit (Qiagen, Valencia, CA) with on-column DNase digestion (Qiagen). Synthesis of cDNA was achieved with the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative real-time PCR (qRT-PCR) was performed on the StepOnePlus Real-Time machine (Applied Biosystems, Carlsbad, CA) under default thermal cycling conditions with a dissociation curve step. Every reaction contained iTaq SYBR Green Supermix with ROX (Bio-Rad), .6–1ng cDNA, and 80 nM primers. Primer sequences are available upon request. Data were analyzed using the 2−ΔΔCT method with mRNA levels normalized to the gene rp49. Relative mRNA amplitude was calculated with respect to the trough levels set as 1 for each age.
Western Blotting
Three independent bioreplicates of 5- and 50-day-old males were collected at ZT 16, 20, 0, and 4. About 5–10 fly heads/time point were homogenized on ice in Laemmli buffer, sonicated, boiled at 100°C for 5 min, and centrifuged at 12000 × g at 4°C. A constant ratio of the buffer (7 μL/head) was used to ensure equal protein loading and separation on 5.7% acrylamide gel. Proteins were transferred to the .45-μm polyvinylidene fluoride (PVDF) Immobilon-FL membrane (Millipore, Billerica, MA) and incubated in 1X TBST (10mM Tris, .15M NaCL, .1% Tween-20, pH 7.5) + 5% milk (for PER) or Odyssey Blocking buffer (LI-COR Biosciences, Lincoln, NE) (for TIM) for 2 h, then overnight at 4°C with 1:15 000 anti-PER (Muskus et al., 2007) or 1:2500 anti-TIM (Giebultowicz & Emery, unpublished) in their respective buffers. Membranes were treated for 2 h with 1:20 000 goat anti-rabbit IRDye680 (LI-COR Biosciences) and 1:5000 goat anti-guinea pig IRDye700 (LI-COR Biosciences), respectively. Proteins were quantified using the LI-COR Odyssey Infrared Imaging System software (v. 3.0).
Immunofluorescence
PER immunofluorescence in the retinal photoreceptors was examined in 5- and 50-day-old males collected at ZT 8, 20, and 24. Heads were fixed in 4% paraformaldehyde in phosphate buffer saline (PBS) (.1 M, pH 7) for 2 h, cryoprotected in 12.5% sucrose for 10 min followed by 25% sucrose overnight at 4°C, embedded in Tissue Tek, frozen in liquid nitrogen, and cut into 20-μm cryo-sections. Sections were washed in PBS, then PBST (PBS with .2% of Triton X 100), and incubated in 5% normal goat serum in PBST and .5% bovine serum albumin (BSA) for 1 h at room temperature. Sections were incubated with 1:1000 polyclonal rabbit anti-PER (Muskus et al., 2007) for 48 h at 4°C, followed by overnight incubation at 4°C with the 1:800 secondary goat anti-rabbit conjugated with Cy3 (Jackson Immuno-Research Laboratories, Westgrove, PA). For negative control, per01 mutants were used. Sections were mounted in Vectashield with 4′-6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) and examined on the Zeiss Meta 510 LSM confocal microscope. PER levels were evaluated by measuring the fluorescence intensity in photoreceptor cell nuclei after converting the mean level of fluorescence to the mean gray value that was quantified using ImageJ (v.1.4, NIH, http://rsb.info.nih.gov/ij/) software. The same brightness parameters and other image settings were used for comparison of young and old tissues. For whole mounts, dissected abdominal organs were fixed and processed as described (Kotwica et al., 2009) and stained with anti-PER followed by Alexa 488 (Invitrogen, xx) secondary antibodies.
Statistical Analysis
Data were statistically analyzed with GraphPad Prism (v.5.0) and GraphPad Instat (v.3.0; San Diego, CA). For analysis of locomotor activity data in 5-, 35-, and 50-day-old flies, average FFT values and period were subjected to one-way analysis of variance (ANOVA) with Tukey’s post hoc test. For qRT-PCR data, statistical significance was evaluated by two-way ANOVA with Bonferroni’s post hoc test. For Western data, the relative strength of the signals was quantified using LI-COR Image analysis software (v.3.0) and subjected to two-way ANOVA with Bonferroni’s post hoc test. For immunofluorescence (IF) data, mean fluorescence levels were converted to the mean gray value, which was quantified using ImageJ software (v.1.4). The numerical values obtained were subjected to Wilcoxon matched pairs test.
RESULTS
Aging Lengthens the Free-Running Period of Locomotor Activity Rhythms
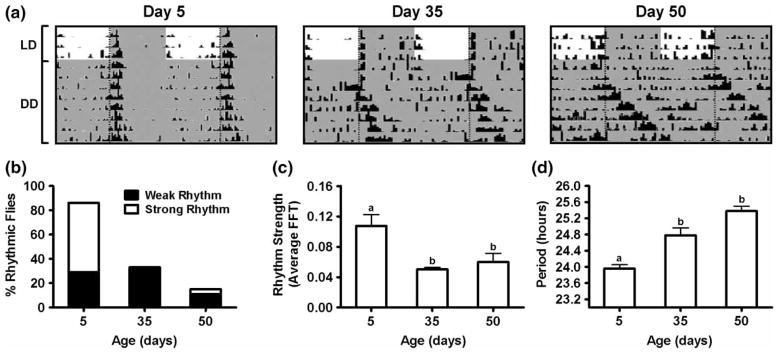
The free-running period of locomotor activity provides a sensitive measure of the circadian clock mechanism. The median life span of CS males is approximately 62 d (Krishnan et al., 2009), and we recorded locomotor activity rhythms in 5–15-day-old (young), 35–45-day-old (middle-aged), and 50–60-day-old (old) flies to assess age-related changes in period and other circadian parameters. Males were monitored for 3 d in LD 12:12 followed by 7 d in DD. We observed that the activity of aging flies was fragmented and extended into the night in LD (Figure 1A), similar to previous reports (Koh et al., 2006; Rezaval et al., 2008). Analysis of fly activity in DD revealed decreased proportion of rhythmic flies in 35- and 50-day-old flies (Figure 1B). We observed a significant decrease in rhythm strength (F2,22 = 6.35, p < .01) in middle-aged and old flies (Figure 1C). Flies that remained rhythmic on days 35 and 50 showed significant age-dependent lengthening of the free-running period (F2,22 = 20.13, p < .001), compared to day 5 (Figure 1D).
FIGURE 1.
(A) Locomotor activity profiles of representative 5-, 35-, and 50-day-old CS males. Flies of each age were monitored in LD (12:12) for 3 d, followed by 7 d in DD at 25°C. Shaded areas represent periods of darkness. Vertical dotted lines indicate time of lights-off (ZT/CT 12). (B) Percentage of rhythmic flies on days 5, 35, and 50. Flies with FFT values >.08 were considered strongly rhythmic, whereas flies with FFT values .04–.08 were classified as weakly rhythmic. (C) Average rhythm strength on days 5, 35, and 50. Values are mean ± SEM (n = 14 for day 5, n = 27 for day 35, and n = 28 for day 50). Statistical significance was determined using one-way ANOVA with Tukey’s post hoc test, and bars with different letters are significantly different (days 5 vs. 35, p < .01; 5 vs. 50, p < .01; 35 vs. 50, p > .05). (D) Average free-running period of locomotor activity on days 5, 35, and 50. Values are mean ± SEM (n = 14 for day 5, n = 27 for day 35, and n = 28 for day 50). Statistical significance was determined using one-way ANOVA with Tukey’s post hoc test, and bars with different letters are significantly different (days 5 vs. 35, p < .001; 5 vs. 50, p < .001; 35 vs. 50, p > .05).
Expression of CLK-CYC Transcriptional Targets Dampen With Age in Fly Heads
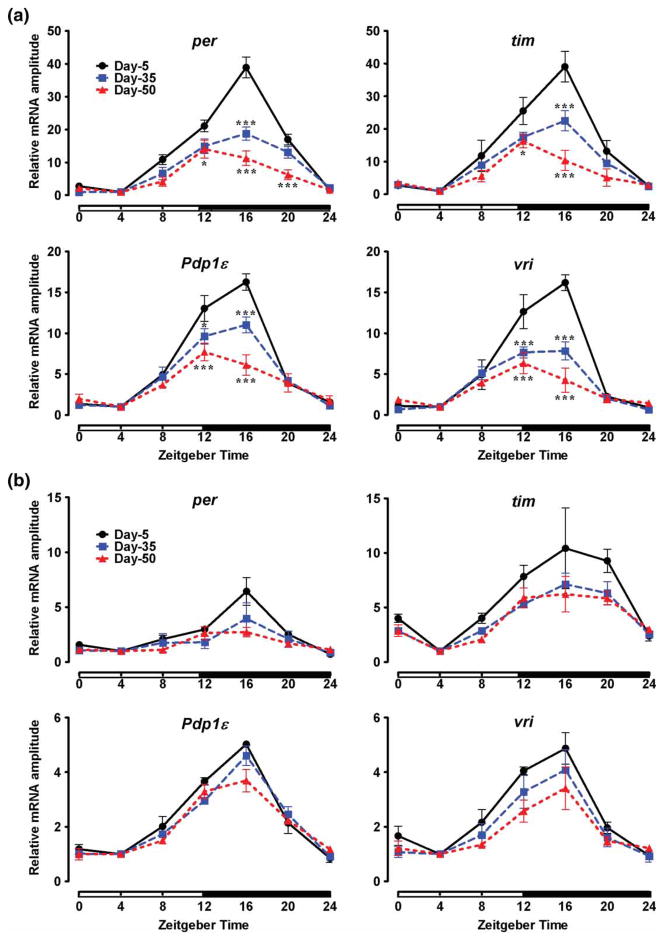
Decrease in the strength of behavioral rhythms prompted us to examine whether the molecular cycling of clock genes might be altered during aging. Hence, we obtained daily expression profiles of two core clock genes, per and tim, in the heads and bodies of 5-, 35-, and 50-day-old CS males. In heads of young flies, per mRNA showed expected daily oscillations, with trough at ZT 4 and peak at ZT 16. The trough-to-peak amplitude of per mRNA oscillations was significantly dampened in 35-day-old flies and further reduced on day 50 (Figure 2A, Table 1). Similar changes were detected in the expression profile of tim, which encodes TIM protein that forms heterodimers with PER. High-amplitude oscillations of tim mRNA observed in young flies were significantly dampened on day 35, and further significant reduction was observed on day 50 (Figure 2A, Table 1).
FIGURE 2.
Daily mRNA profiles of four CLK-CYC–controlled transcripts on days 5, 35, and 50 in (A) heads and (B) bodies of CS males, normalized to the trough (ZT 4) values set at 1 for each age. White and black horizontal bars mark periods of light and dark, respectively. Each data point represents mean ± SEM for three independent RNA samples. Statistical significance between day 5 vs. 35 and day 5 vs. 50 values was determined using two-way ANOVA with Bonferonni’s post hoc test, and is denoted by ***p < .001 and *p < .05.
TABLE 1.
Statistical analysis of gene expression (qPCR) data by two-way ANOVA with Bonferroni’s post hoc test
| Gene | Effects of ZT | Effects of age | ||
|---|---|---|---|---|
| Heads | ||||
| per | F6,42 = 253.29 | p < .0001 | F2,42 = 134.60 | p < .0001 |
| tim | F6,42 = 45.56 | p < .0001 | F2,42 = 18.63 | p < .0001 |
| Pdp1ε | F6,42 = 93.04 | p < .0001 | F2,42 = 15.97 | p < .0001 |
| vri | F6,42 = 58.06 | p < .0001 | F2,42 = 17.62 | p < .0001 |
| cwo | F6,42 = 21.81 | p < .0001 | F2,42 = 1.71 | p = .1926 |
| Clk | F6,42 = 20.27 | p < .0001 | F2,42 = .41 | p = .6685 |
| cyc | F6,42 = .17 | p = .9843 | F2,42 = 1.32 | p = .1970 |
| Bodies | ||||
| per | F6,42 = 14.76 | p < .0001 | F2,42 = 1.74 | p = .0926 |
| tim | F6,42 = 19.04 | p < .0001 | F2,42 = .81 | p = .6495 |
| Pdp1ε | F6,42 = 122.83 | p < .0001 | F2,42 = 2.25 | p = .0663 |
| vri | F6,42 = 31.43 | p < .0001 | F2,42 = .85 | p = .6022 |
| Clk | F6,42 = 23.26 | p < .0001 | F2,42 = .55 | p = .5817 |
| cyc | F6,42 = 1.68 | p = .1486 | F2,42 = .61 | p = .8176 |
Subscripted values indicate the degrees of freedom in numerator (DFn) and denominator (DFd), respectively.
Reduced oscillations of both per and tim suggest that CLK-CYC–driven transcription may be weakened during aging. To test this, we examined the expression of Pdp1ε and vri, which are also activated by the CLK-CYC complex and oscillate with a phase similar to per and tim. We observed that the amplitude of mRNA oscillations for both genes was considerably dampened at the peak (ZT 12–16) in heads of middle-aged and old flies (Figure 2A, Table 1). Interestingly, the mRNA for per, tim, and Pdp1ε initially increased with a similar slope in heads of both young and old flies until ZT 12 (or until ZT 8 in the case of vri), suggesting that the initiation of cyclic transcription is not affected by age. However, the peak expression of all four genes was prematurely truncated in old flies (Figure 2A).
In parallel with heads, we measured the expression profiles of the same clock genes in male bodies. As previously reported, the amplitude of per oscillations was lower in bodies compared to the heads of young flies (Hardin, 1994). We determined that mRNA for other clock genes tim, vri, and Pdp1ε also cycled with a lower amplitude in bodies than in heads of young males (Figure 2B). Importantly, in contrast to heads, there was no statistically significant effect of age on the oscillatory amplitude of these genes (p > .05), although there was a declining trend in the peak levels of all four genes (Figure 2B, Table 1).
Reduced expression of the CLK-CYC target genes was reported in young flies with disrupted clockwork orange (cwo) (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). It is not known whether the expression of this gene changes during aging; therefore, we examined the cwo mRNA profile as a function of age. As reported previously (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008), cwo mRNA cycled in the heads of young flies with a trough at ZT 4 and a peak at ZT 12 (Figure 3). There was no significant effect of age on cwo cycling, albeit individual comparison of 5- vs. 50-day-old samples revealed that levels of cwo were significantly lower (p < .05) at ZT 16 (Figure 3, Table 1).
FIGURE 3.
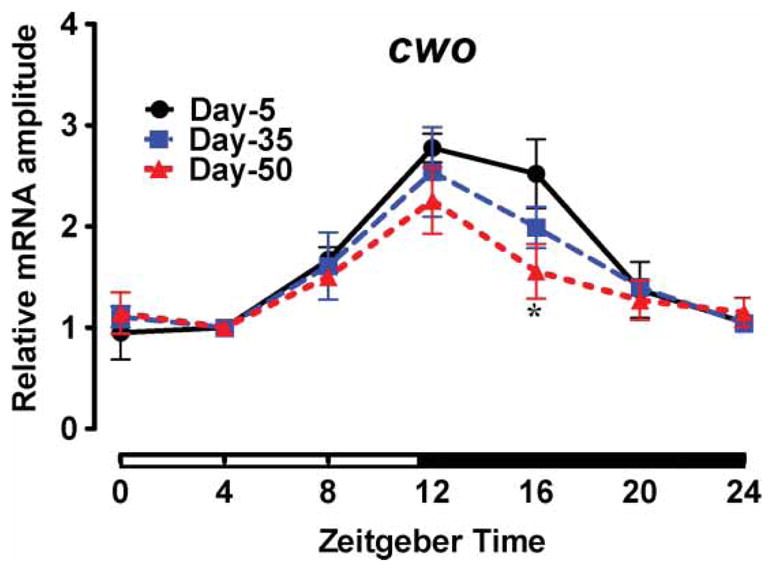
Daily mRNA profiles of cwo in the heads of 5-, 35-, and 50-day-old CS males, normalized to the trough (ZT 4) values set at 1 for each age. Each data point represents mean ± SEM for three independent RNA samples. Statistical significance between day 5 vs. 35 and day 5 vs. 50 values was determined using two-way ANOVA with Bonferonni’s post hoc test, and is denoted by *p < .05.
Aging Does Not Alter the Expression of Clk and cyc mRNAs
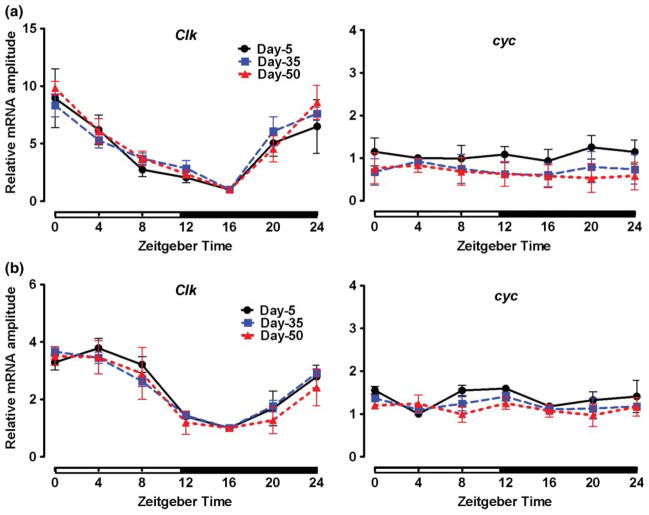
Since our data suggest that aging is associated with reduced activity of the CLK-CYC complexes, we next measured the expression levels of Clk and cyc mRNAs in young, middle-aged, and old flies. Young flies showed expected Clk mRNA oscillations, with peak levels at ZT 4 and trough at ZT 16. Similar oscillations were detected in the heads of middle-aged and old flies, and there was no significant reduction in Clk mRNA amplitude with age (Figure 4A, Table 1).
FIGURE 4.
Daily mRNA profiles of Clk and cyc on days 5, 35, and 50 in (A) heads and (B) bodies of CS males, normalized to ZT 16 (Clk) and ZT 4 (cyc) values set at 1 for each age. Each data point represents mean ± SEM for three independent RNA samples. Statistical significance between day 5 vs. 35 and day 5 vs. 50 values was determined using two-way ANOVA with Bonferonni’s post hoc test (p > .05).
The expression of cyc is nonrhythmic in young flies (Bae et al., 2000), and we determined that the same was true in heads of old flies (Figure 4A, Table 1). There was a declining trend in cyc mRNA in heads of old flies; however, the reduction was not statistically significant. Similar as in heads, Clk and cyc expression did not change significantly (p > .05) in bodies of old flies (Figure 4B, Table 1).
PER and TIM Protein Levels Are Substantially Reduced in Old Flies
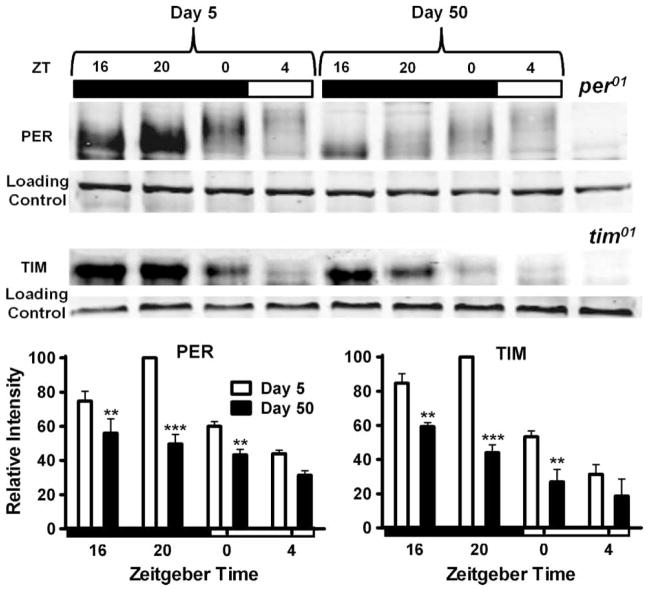
Given the age-related decline in per and tim transcription (Figure 2), we compared PER and TIM protein profiles in head extracts of 5- and 50-day-old flies. In young flies, PER showed expected cycling, with peak at ZT 20 and decline at ZT 0–4, with retarded band migration indicative of progressive phosphorylation (Figure 5). By comparison, PER levels were lower in old flies across all time points, with an especially significant reduction observed at ZT 20 (effect of age: F1,16 = 59.77, p < .0001; effect of ZT: F3,16 = 26.02, p < .0001). Interestingly, the retarded migration of residual PER was observed at ZT 0–4 in old flies, suggesting that PER phosphorylation was similar as in young flies. In addition to PER, we analyzed TIM by Western blotting and found that levels of this protein also declined in old flies, with the most significant difference observed at ZT 20 (effect of age: F1,16 = 59.96, p < .0001; effect of ZT: F3,16 = 35.01, p < .0001) (Figure 5). Consequently, the peak of both PER and TIM was prematurely truncated in old flies, consistent with age-related changes in the mRNA profiles for both genes (Figure 2A).
FIGURE 5.
Western blots showing PER and TIM protein profiles and phosphorylation status on days 5 and 50. Bar graphs (below) indicate the relative band intensity for different ages and time points, with signal intensity at peak (ZT 20) in 5-day-old flies set as 100. Values are mean ± SEM of three independent bioreplicates. Statistical significance between day 5 vs. 50 values was determined using two-way ANOVA with Bonferonni’s post hoc test, and is denoted by ***p < .001 and **p < .01.
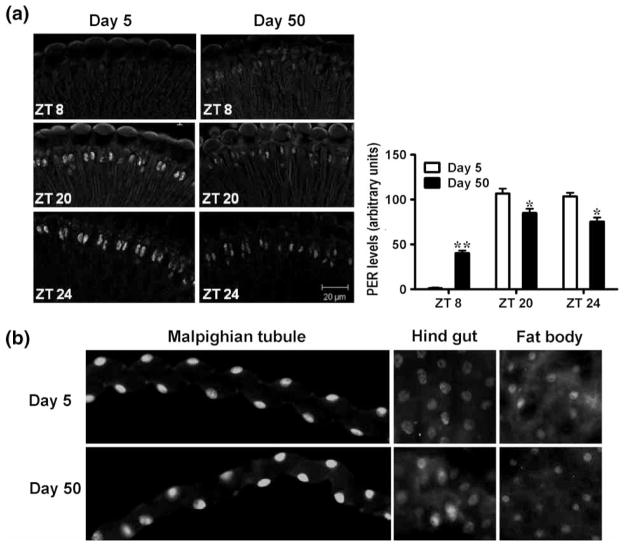
PER and TIM proteins detected by Western blotting in whole-head homogenates are derived from both central and peripheral oscillators, but the bulk of both proteins are from retinal photoreceptors (Cheng & Hardin, 1998). Therefore, we examined the pattern of PER in photoreceptor cells of the retina by immunofluorescence on fly-head sections. In both young and old flies, PER was localized in cell nuclei; however, PER levels were significantly reduced in 50-day-old flies at ZT 20 and 24 (Figure 6A). The highest signal from individual photoreceptor nuclei in old flies did not reach the levels observed in the young flies, suggesting loss of PER amplitude in individual cells. In addition to photoreceptors, we also compared PER in several abdominal organs of young and old flies and determined that the levels of PER protein were not altered significantly with age. Similar levels of nuclear PER were detected in Malpighian tubules, gut, and abdominal fat of young and old flies at ZT 24 (Figure 6B), whereas PER staining was absent at ZT 8 (not shown). These data agree with per mRNA profiles, which did not show significant decline with age in fly bodies (Figure 2B).
FIGURE 6.
(A) PER protein levels in the nuclei of retinal photoreceptors of 5- and 50-day-old males at ZT 8, 20, and 24. Bar graph (right) shows quantification of the signal averaged from >20 flies for each time point and age. Statistical significance was determined using Wilcox-on matched pairs test, and is denoted by **p < .01 and *p < .05. (B) PER protein levels in the nuclei of Malpighian tubule, hind gut, and abdominal fat body cells of 5- and 50-day-old males at ZT 24. Images represent typical staining based on 6–8 flies analyzed for each age.
DISCUSSION
It has been widely observed that circadian output rhythms decay during aging in different animals, including humans. This decay has been suggested to have detrimental effects on health, and promote aging; therefore, it is important to determine how aging alters the clock mechanism at the molecular level. We addressed this question using Drosophila, which are short-lived and have a well-understood circadian system. First, we confirmed previous reports of deterioration of the circadian locomotor activity rhythm in aging flies (Koh et al., 2006; Rezaval et al., 2008). In our study, flies that remained rhythmic in DD showed significant lengthening of the free-running period of locomotor activity. Although the previous study on aging flies reported a similar tendency, period length did not show a statistically significant difference (Rezaval et al., 2008). Interestingly, one of the studies examining the period of wheel-running activity as a function of age in mammals reported significant lengthening of the free-running period in 2-year-old mice (Valentinuzzi et al., 1997).
In addition to the behavioral changes in old flies, we observed dampening of the molecular clock oscillations, manifested as significantly reduced peak levels of per, tim, Pdp1ε, and vri mRNAs in heads of aging flies. Given that we analyzed gene expression in whole-head extracts (or retinal photoreceptors), whereas the circadian locomotor activity rhythms are regulated by ~150 central brain pacemaker cells, a direct link between the observed behavioral and molecular changes cannot be made. Nevertheless, previous studies have shown that flies with lower amplitude of per and tim oscillations in head extracts show a tendency to lengthen the period of behavioral rhythms (Allada et al., 1998; Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). On the other hand, flies expressing CYC-VP16 have strongly enhanced transcriptional activity relative to that of wild-type CLK-CYC and display shortened period, implicating the strength of circadian transcription in period determination (Kadener et al., 2008).
In young flies, the transcription of per, tim, Pdp1ε, and vri is activated by the CLK-CYC complex that forms the positive limb of the clock. Synchronous dampening of CLK-CYC target genes in aging flies suggests weakened transcriptional activity of the CLK-CYC complexes. We determined that this is not caused by Clk and cyc deficiency, as the expression of Clk mRNA did not differ between young and old flies. Although we show that the expression of both the activator (Pdp1ε) and the repressor (vri) of Clk transcription is dampened during aging, the net effect may be negligible due to their opposing action. Expression of cyc showed a declining trend in old flies, but these changes were not statistically significant. In addition to Clk and cyc, we analyzed the expression levels of cwo during aging, because the transcription of CLK-CYC target genes is diminished in cwo mutants, whereas the cwo gene is, itself, a target of CLK-CYC complexes (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). We detected subtle, albeit statistically significant, reduction of cwo levels in 50-day-old flies at ZT 16.
Our data do not reveal strong age-related changes in Clk, cyc, and cwo expression that could account for >50% reduction of CLK-CYC transcriptional targets observed in old flies. Further studies would be needed to identify factors contributing to the dampening of the positive limb of the clock with age. These could include changes in CLK or CYC protein levels, posttranslational modifications (particularly the phosphorylation status of CLK), binding of CLK-CYC complexes to DNA, as well as age-related alterations in chromatin modifications that result from CLK-CYC binding to DNA (Bae et al., 2000; Menet et al., 2010; Taylor & Hardin, 2008; Yu & Chung, 2001; Yu et al., 2006). Interestingly, dampened amplitude of per, tim, Pdp1ε, and vri was consistently observed in the heads, but not bodies, of aging flies despite similar profiles of Clk and cyc mRNAs in both tissues. This could be connected to our previous findings that levels of oxidative damage are substantially higher in the heads than bodies of aging flies (Krishnan et al., 2009). Consistent with this idea, a strong oxidative stressor reduced the amplitude of per-reporter oscillations in young flies and prevented CLK from activating transcription from per E-box in transfected cells (Zheng et al., 2007). The relative insensitivity of clocks to aging in abdominal tissues could be related to feeding rhythms identified in Drosophila (Xu et al., 2008). It has been shown recently that feeding affects the phase of rhythmic gene expression in the fat body, without affecting clocks in the brain (Xu et al., 2011). If the feeding rhythm remains strong in aging flies, it could support clock gene oscillations in abdominal tissues.
Robust molecular circadian oscillations in the heads of young flies are generated by transcription-translation negative feedback loops (Zeng et al., 1994). PER/TIM proteins accumulate in cell nuclei late at night and PER represses the CLK-CYC activating complexes, resulting in the inhibition of per and tim transcription until PER is removed (Hardin, 2005; Zheng & Sehgal, 2008). Based on this model, a plausible cause of weakened CLK-CYC activity in old flies could have been the persistence of PER, perhaps due to age-related defects in phosphorylation, or degradation of this protein (Bae & Edery, 2006; Grima et al., 2002; Ko et al., 2002). However, a quantitative measure of protein levels by Western blotting showed significant reduction of PER as well as TIM in the heads of old flies, in agreement with their mRNA profiles. Additionally PER, albeit significantly reduced, appeared to have normal phosphorylation profile in heads and displayed normal nuclear localization in the retinal photoreceptor cells of old flies.
Taken together, the above data demonstrate that aging weakens both the positive and negative limbs of the clock feedback loop, such that reduced per mRNA expression results in reduced PER protein levels. It remains to be determined whether inadequate repression due to PER deficiency may contribute to the dampening of CLK-CYC activity during aging. This line of reasoning is supported by studies highlighting the complex involvement of PER in sequestering DNA-bound CLK, which leads to the nighttime decrease of CLK-CYC activity. Later, the complex is released from DNA, leading to the re-activation of CLK-CYC activity in the next circadian cycle (Menet et al., 2010; Sun et al., 2010; Yu et al., 2009).
Our analysis of the expression of clock genes and proteins across the life span determined that aging affects the molecular oscillations in heads more strongly than in the body tissues of Drosophila. Thus, the extent of age-related changes in clock gene expression in flies depends on the specific clock component and tissue examined, similar to what was previously reported in vertebrates (Asai et al., 2001; Jud et al., 2009; Kolker et al., 2003; Yamazaki et al., 2002; Zhdanova et al., 2008). Importantly, weakened transcriptional activity of the CLK-CYC complexes during aging in Drosophila provides a foundation to investigate how this dampening affects clock-controlled pathways. Previous microarray studies identified several oscillating genes in fly heads, whose expression became arrhythmic in ClkJrk mutants (McDonald & Rosbash, 2001). Further, several studies have shown widespread gene activation by CLOCK-BMAL1 complexes in mammals (Panda et al., 2002; Rey et al., 2011). Dampening of the positive limb of the clock during aging is likely to impair the rhythmic expression of clock-controlled effector genes that are involved in many pathways maintaining temporal homeostasis.
Acknowledgments
We are grateful to Eileen Chow for help with qRT-PCR in fly bodies and locomotor activity analysis. We thank Jeff Price for gift of anti-PER, and Amita Sehgal for sharing unpublished data. This research was supported by NIH 1R21AG038989 and R21-NS-075500 grants to J.M.G, and Jagiellonian University K/ZDS/001964 grant to E.P. K.R. is supported by NSF IGERT in Aging Sciences Fellowship at Oregon State University (DGE 0965820).
Footnotes
Declaration of Interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, Lee C, Nikitin AY. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai M, Yoshinobu Y, Kaneko S, Mori A, Nikaido T, Moriya T, Akiyama M, Shibata S. Circadian profile of Per gene mRNA expression in the suprachiasmatic nucleus, paraventricular nucleus, and pineal body of aged rats. J Neurosci Res. 2001;66:1133–1139. doi: 10.1002/jnr.10010. [DOI] [PubMed] [Google Scholar]
- Bae K, Edery I. Regulating a circadian clock’s period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem. 2006;140:609–617. doi: 10.1093/jb/mvj198. [DOI] [PubMed] [Google Scholar]
- Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER- TIM complex. J Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Hardin PE. Drosophila photoreceptors contain an autonomous circadian pacemaker that can function without period mRNA cycling. J Neurosci. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Giebultowicz JM. Molecular mechanism and cellular distribution of insect circadian clocks. Annu Rev Entomol. 2000;45:767–791. doi: 10.1146/annurev.ento.45.1.769. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Jud C, Chappuis S, Revell VL, Sletten TL, Saaltink DJ, Cajochen C, Skene DJ, Albrecht U. Age-dependent alterations in human PER2 levels after early morning blue light exposure. Chronobiol Int. 2009;26:1462–1469. doi: 10.3109/07420520903385564. [DOI] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:pe119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker DE, Fukuyama H, Huang DS, Takahashi JS, Horton TH, Turek FW. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J Biol Rhythms. 2003;18:159–169. doi: 10.1177/0748730403251802. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwica J, Larson MK, Bebas P, Giebultowicz JM. Developmental profiles of PERIOD and DOUBLETIME in Drosophila melanogaster ovary. J Insect Physiol. 2009;55:419–425. doi: 10.1016/j.jinsphys.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Kretzschmar D, Rakshit K, Chow E, Giebultowicz J. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging. 2009;1:937–948. doi: 10.18632/aging.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, Hardin PE, Tanimura T, Ueda HR. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Portaluppi F, Smolensky MH, Touitou Y. Ethics and methods for biological rhythm research on animals and human beings. Chronobiol Int. 2010;27:1911–1929. doi: 10.3109/07420528.2010.516381. [DOI] [PubMed] [Google Scholar]
- Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:pe1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaval C, Berni J, Gorostiza EA, Werbajh S, Fagilde MM, Fernandez MP, Beckwith EJ, Aranovich EJ, Sabio y Garcia CA, Ceriani MF. A functional misexpression screen uncovers a role for enabled in progressive neurodegeneration. PLoS One. 2008;3:pe3332. doi: 10.1371/journal.pone.0003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richier B, Michard-Vanhee C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms. 2008;23:103–116. doi: 10.1177/0748730407313817. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Sun WC, Jeong EH, Jeong HJ, Ko HW, Edery I, Kim EY. Two distinct modes of PERIOD recruitment onto dCLOCK reveal a novel role for TIMELESS in circadian transcription. J Neurosci. 2010;30:14458–14469. doi: 10.1523/JNEUROSCI.2366-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Hardin PE. Rhythmic E-box binding by CLK-CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol. 2008;28:4642–4652. doi: 10.1128/MCB.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou Y, Haus E. Alterations with aging of the endocrine and neuroendocrine circadian system in humans. Chronobiol Int. 2000;17:369–390. doi: 10.1081/cbi-100101052. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Scarbrough K, Takahashi JS, Turek FW. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am J Physiol. 1997;273:R1957–R1964. doi: 10.1152/ajpregu.1997.273.6.R1957. [DOI] [PubMed] [Google Scholar]
- Van Someren EJ, Riemersma-Van Der Lek RF. Live to the rhythm, slave to the rhythm. Sleep Med Rev. 2007;11:465–484. doi: 10.1016/j.smrv.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Weinert D, Waterhouse J. The circadian rhythm of core temperature: effects of physical activity and aging. Physiol Behav. 2007;90:246–256. doi: 10.1016/j.physbeh.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Wu Y-H, Swaab DF. Disturbance and strategies for reactivation of the circadian rhythm system in aging and Alzheimer’s disease. Sleep Med. 2007;8:623–636. doi: 10.1016/j.sleep.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13:639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci U S A. 2002;99:10801–10806. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BP, Chung HY. Stress resistance by caloric restriction for longevity. Ann N Y Acad Sci. 2001;928:39–47. doi: 10.1111/j.1749-6632.2001.tb05633.x. [DOI] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova IV, Yu L, Lopez-Patino M, Shang E, Kishi S, Guelin E. Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res Bull. 2008;75:433–441. doi: 10.1016/j.brainresbull.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova IV, Masuda K, Quasarano-Kourkoulis C, Rosene DL, Killiany RJ, Wang S. Aging of intrinsic circadian rhythms and sleep in a diurnal nonhuman primate, Macaca mulatta. J Biol Rhythms. 2011;26:149–159. doi: 10.1177/0748730410395849. [DOI] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]