Abstract
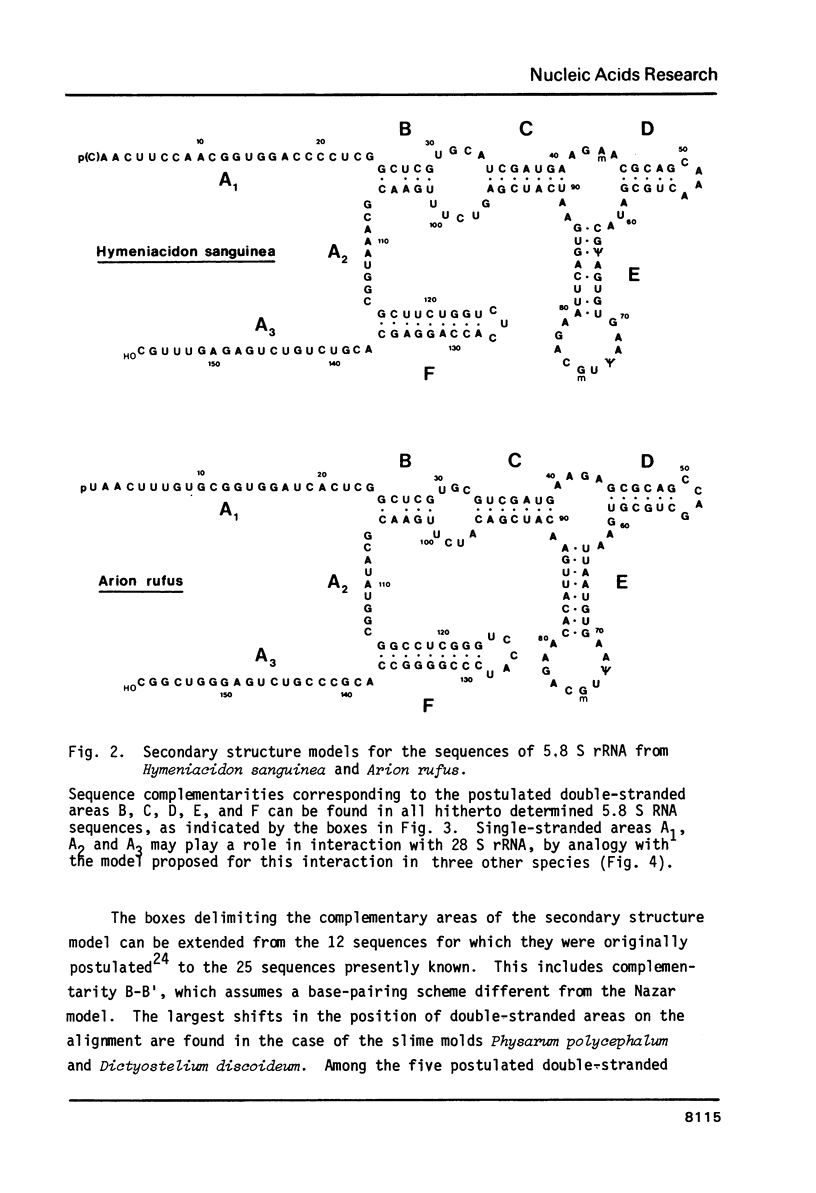
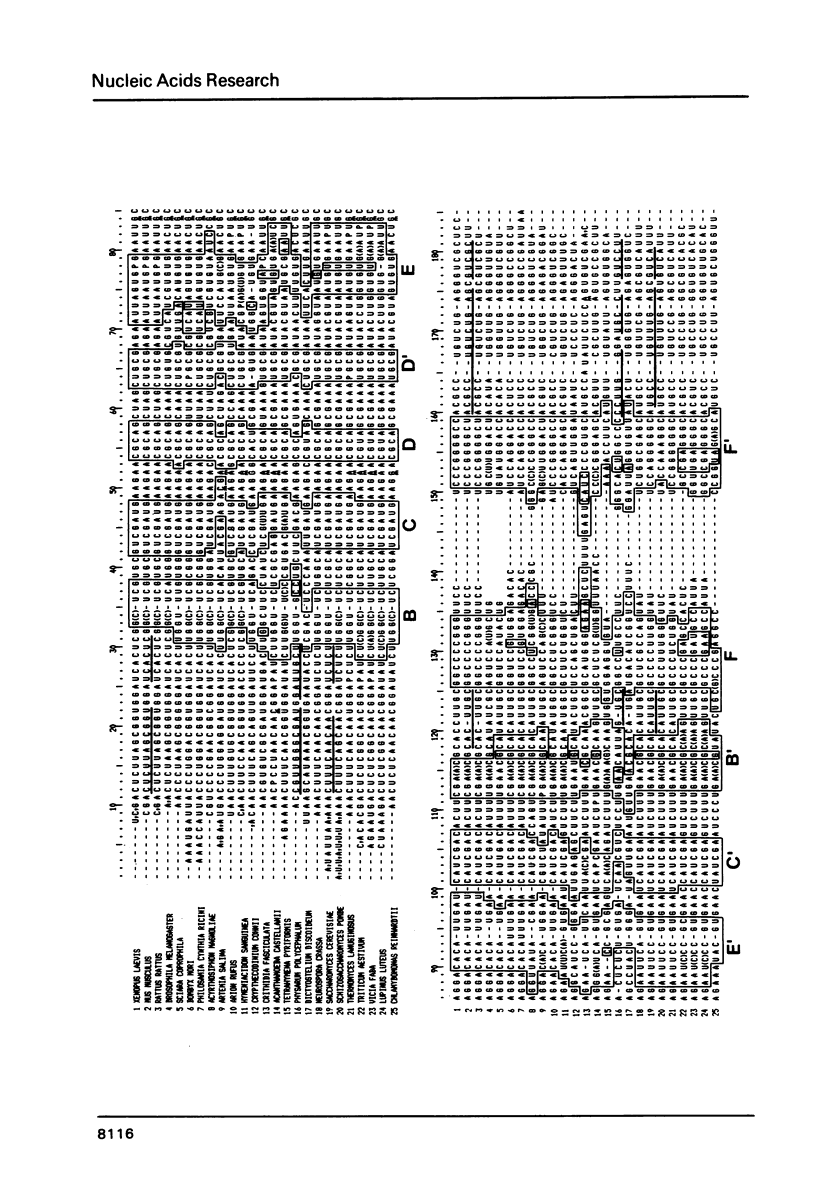
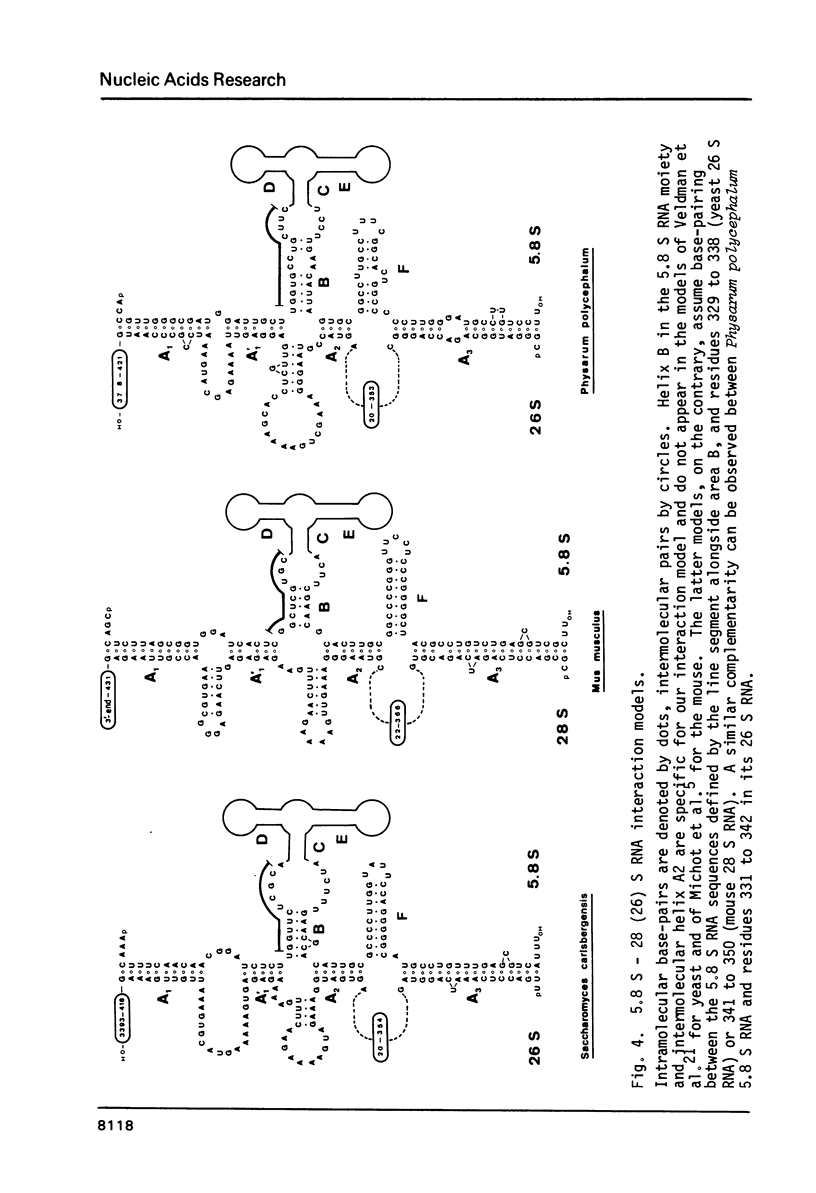
We report the primary structures of the 5.8 S ribosomal RNAs isolated from the sponge Hymeniacidon sanguinea and the snail Arion rufus. We had previously proposed (Ursi et al., Nucl. Acids Res. 10, 3517-3530 (1982)) a secondary structure model on the basis of a comparison of twelve 5.8 S RNA sequences then known, and a matching model for the interaction of 5.8 S RNA with 26 S RNA in yeast. Here we show that the secondary structure model can be extended to the 25 sequences presently available, and that the interaction model can be extended to the binding of 5.8 S RNA to the 5'-terminal domain of 28 S (26 S) RNA in three species.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dams E., Vandenberghe A., De Wachter R. Nucleotide sequences of three poriferan 5 S ribosomal RNAs. Nucleic Acids Res. 1982 Sep 11;10(17):5297–5302. doi: 10.1093/nar/10.17.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dams E., Vandenberghe A., De Wachter R. Sequences of the 5S rRNAs of Azotobacter vinelandii, Pseudomonas aeruginosa and Pseudomonas fluorescens with some notes on 5S RNA secondary structure. Nucleic Acids Res. 1983 Mar 11;11(5):1245–1252. doi: 10.1093/nar/11.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge P., Kastelein R. A., Planta R. J. Non-ribosomal nucleotide sequences in 7-S RNA, the immediate precursor of 5.8-S ribosomal RNA in yeast. Eur J Biochem. 1978 Feb;83(2):537–546. doi: 10.1111/j.1432-1033.1978.tb12121.x. [DOI] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r105–r133. [PMC free article] [PubMed] [Google Scholar]
- Fang B. L., De Baere R., Vandenberghe A., De Wachter R. Sequences of three molluscan 5 S ribosomal RNAs confirm the validity of a dynamic secondary structure model. Nucleic Acids Res. 1982 Aug 11;10(15):4679–4685. doi: 10.1093/nar/10.15.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. X., Krupp G., Gross H. J. The nucleotide sequence of 5.8S rRNA from the posterior silk gland of the silkworm Philosamia cynthia ricini. Nucleic Acids Res. 1982 Oct 25;10(20):6383–6387. doi: 10.1093/nar/10.20.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. The nucleotide sequences of 5.8-S ribosomal RNA from Xenopus laevis and Xenopus borealis. Eur J Biochem. 1978 Jun 1;87(1):199–214. doi: 10.1111/j.1432-1033.1978.tb12367.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Ishikawa H. Primary and secondary structures of Tetrahymena and aphid 5.8S rRNAs: structural features of 5.8S rRNA which interacts with the 28S rRNA containing the hidden break. Nucleic Acids Res. 1982 Sep 11;10(17):5173–5182. doi: 10.1093/nar/10.17.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Kawata Y., Ishikawa H. Primary and secondary structure of 5.8S rRNA from the silkgland of Bombyx mori. Nucleic Acids Res. 1982 Apr 10;10(7):2415–2418. doi: 10.1093/nar/10.7.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Coding and spacer sequences in the 5.8S-2S region of Sciara coprophila ribosomal DNA. Nucleic Acids Res. 1980 Aug 25;8(16):3565–3573. doi: 10.1093/nar/8.16.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Sequence and secondary structure of mouse 28S rRNA 5'terminal domain. Organisation of the 5.8S-28S rRNA complex. Nucleic Acids Res. 1982 Sep 11;10(17):5273–5283. doi: 10.1093/nar/10.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Nazar R. N., Wildeman A. G. Altered features in the secondary structure of Vicia faba 5.8s rRNA. Nucleic Acids Res. 1981 Oct 24;9(20):5345–5358. doi: 10.1093/nar/9.20.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Sogin M. L. Nucleotide sequence of Dictyostelium discoideum 5.8S ribosomal ribonucleic acid: evolutionary and secondary structural implications. Biochemistry. 1982 May 11;21(10):2335–2343. doi: 10.1021/bi00539a010. [DOI] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Sakaki Y., Takagi Y. Nucleotide sequence of Physarum polycephalum 5.8S rRNA gene and its flanking regions. Nucleic Acids Res. 1982 Apr 10;10(7):2379–2385. doi: 10.1093/nar/10.7.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. A., Walker T. A., Pace N. R. Independent binding sites in mouse 5.8S ribosomal ribonucleic acid for 28S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2329–2335. doi: 10.1021/bi00539a009. [DOI] [PubMed] [Google Scholar]
- Schaak J., Mao J., Söll D. The 5.8S RNA gene sequence and the ribosomal repeat of Schizosaccharomyces pombe. Nucleic Acids Res. 1982 May 11;10(9):2851–2864. doi: 10.1093/nar/10.9.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. Nucleotide sequence of an exceptionally long 5.8S ribosomal RNA from Crithidia fasciculata. Nucleic Acids Res. 1982 Mar 25;10(6):2085–2092. doi: 10.1093/nar/10.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam C. S., Cassidy B., Busch H., Rothblum L. I. Nucleotide sequence of the region between the 18S rRNA sequence and the 28S rRNA sequence of rat ribosomal DNA. Nucleic Acids Res. 1982 Jun 25;10(12):3667–3680. doi: 10.1093/nar/10.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursi D., Vandenberghe A., De Wachter R. The sequence of the 5.8 S ribosomal RNA of the crustacean Artemia salina. With a proposal for a general secondary structure model for 5.8 S ribosomal RNA. Nucleic Acids Res. 1982 Jun 11;10(11):3517–3530. doi: 10.1093/nar/10.11.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. A., Johnson K. D., Olsen G. J., Peters M. A., Pace N. R. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2320–2329. doi: 10.1021/bi00539a008. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Studies on the secondary structure of 5.8 S rRNA from a thermophile, Thermomyces lanuginosus. J Biol Chem. 1981 Jun 10;256(11):5675–5682. [PubMed] [Google Scholar]



