Background: Metabolic disorders are associated with chronic inflammation.
Results: Energy-sensing factor PGC-1α regulates cytokine expression in hepatocytes. PGC-1α, AMPK, and metformin induce expression of interleukin 1 receptor antagonist.
Conclusion: PGC-1α and AMPK mediate effects of fasting, physical exercise, and antidiabetic drug metformin on hepatic inflammatory gene expression.
Significance: PGC-1α and AMPK are regulatory interlinks between energy homeostasis and inflammation.
Keywords: Cellular Immune Response, Coactivator Transcription, Diabetes, Liver, Nuclear Receptors, Inflammation, IL15Rα, IL1Rn, Tweak, Metformin
Abstract
Obesity and insulin resistance are associated with chronic, low grade inflammation. Moreover, regulation of energy metabolism and immunity are highly integrated. We hypothesized that energy-sensitive coactivator peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α) and AMP-activated protein kinase (AMPK) may modulate inflammatory gene expression in liver. Microarray analysis revealed that PGC-1α up-regulated expression of several cytokines and cytokine receptors, including interleukin 15 receptor α (IL15Rα) and, even more importantly, anti-inflammatory interleukin 1 receptor antagonist (IL1Rn). Overexpression of PGC-1α and induction of PGC-1α by fasting, physical exercise, glucagon, or cAMP was associated with increased IL1Rn mRNA and protein expression in hepatocytes. Knockdown of PGC-1α by siRNA down-regulated cAMP-induced expression of IL1Rn in mouse hepatocytes. Furthermore, knockdown of peroxisome proliferator-activated receptor α (PPARα) attenuated IL1Rn induction by PGC-1α. Overexpression of PGC-1α, at least partially through IL1Rn, suppressed interleukin 1β-induced expression of acute phase proteins, C-reactive protein, and haptoglobin. Fasting and exercise also induced IL15Rα expression, whereas glucagon and cAMP resulted in reduction in IL15Rα mRNA levels. Finally, AMPK activator metformin and adenoviral overexpression of AMPK up-regulated IL1Rn and down-regulated IL15Rα in primary hepatocytes. We conclude that PGC-1α and AMPK alter inflammatory gene expression in liver and thus integrate energy homeostasis and inflammation. Induction of IL1Rn by PGC-1α and AMPK may be involved in the beneficial effects of exercise and caloric restriction and putative anti-inflammatory effects of metformin.
Introduction
The liver is a major regulator of energy and glucose homeostasis. Insulin resistance and dysregulation of hepatic glucose release are key events in the pathogenesis of type 2 diabetes. The exact mechanisms behind the development of insulin resistance and type 2 diabetes remain obscure, but numerous studies, both animal and human, point to the role of chronic inflammation and altered balance of proinflammatory and anti-inflammatory signals regulated by diet and lifestyle factors, such as physical inactivity and obesity (1–4). Low grade inflammation precedes type 2 diabetes by several years and is manifested by increased blood levels of several proinflammatory cytokines, acute phase protein, and other inflammatory mediators (5). Adipose tissue is considered to be the major source of inflammatory factors in metabolic syndrome, whereas liver is both a major source and a target tissue (6). Development of hepatic insulin resistance by inflammation probably involves several molecular pathways, including JNK, NF-κB, and SOCS family members. In addition, endoplasmic reticulum stress and reactive oxygen species activation may contribute to the process (7, 8). Given the important links between metabolism and immunity, it comes as no surprise that the regulatory mechanisms that govern metabolism and immunity intersect at numerous levels.
Hepatic energy metabolism is controlled by the balance between insulin and glucagon. In the fasted state, insulin levels decrease, whereas glucagon secretion is up-regulated, leading to stimulation of glycogenolysis and gluconeogenesis. The transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α)3 is a major regulator of hepatic energy metabolism. During fasting, hepatic PGC-1α expression increases markedly in response to glucagon's second messenger cAMP via cAMP-response element-binding protein, leading to induction of key gluconeogenetic enzymes, such as phosphoenolpyruvate carboxykinase 1 (PCK-1) and glucose-6-phosphatase (9, 10). Type 2 diabetes involves abnormal activation of these energy-releasing processes. PGC-1α has been implicated in the pathogenesis of diabetes, and both hepatic PGC-1α expression and gluconeogenesis are aberrantly induced in mouse models of insulin resistance and type 2 diabetes (10).
PGC-1α does not bind DNA itself but serves as a coactivator for a number of nuclear receptors and other transcription factors, including peroxisome proliferator-activated receptors, hepatocyte nuclear factor 4α, glucocorticoid receptor, and liver X receptor (11).
Nuclear receptors are major mediators linking inflammatory and metabolic gene networks (12, 13). Because PGC-1α is involved in signaling by numerous nuclear receptors, we hypothesized that PGC-1α could be among the key factors integrating regulation of energy homeostasis and inflammatory responses. Indeed, PGC-1α is physiologically induced by caloric restriction and physical exercise that reduce systemic inflammation (14, 15). On the other hand, pathological, extended activation of PGC-1α is observed in obesity and diabetes (10). We therefore aimed to characterize the role of PGC-1α in hepatic regulation of inflammatory process. The results presented here suggest that PGC-1α alters inflammatory gene expression in liver and thus links regulation of energy homeostasis and inflammation. Three novel PGC-1α-regulated genes, belonging to the cytokine or cytokine receptor family, were identified. These included interleukin 1 receptor antagonist (Il1rn; also called Il-1ra), interleukin 15 receptor α (Il15rα), and tumor necrosis factor-like weak inducer of apoptosis (Tweak; systematic name, tumor necrosis factor (ligand) superfamily, member 12 (Tnfsf12)). Cytokines are small, cell-signaling protein molecules that regulate immunity and are mediators of inflammatory reactions. In addition, we show that up-regulation of IL1Rn by PGC-1α restricts induction of hepatic acute phase proteins by interleukin 1β. Furthermore, we provide evidence that IL1Rn and IL15Rα are regulated also by another key energy sensor, AMP-activated protein kinase (AMPK).
EXPERIMENTAL PROCEDURES
Materials
Dibutyryl-cAMP (Bt2cAMP), 8-bromo-cAMP (8-Br-cAMP), glucagon, and metformin were purchased from Sigma-Aldrich, and recombinant human IL1β protein (ab9617) was from Abcam (Cambridge, UK).
Animals and Animal Treatment
The investigations were carried out according to the requirements of the European Union Guiding Principles for Care and Use of Animals and were approved by the local ethics committees for laboratory animal welfare at the University of Oulu and Semmelweis University. Rodents were handled daily for at least 5 days before beginning the experiment. All animals were housed in a temperature- and humidity-controlled room with a 12-h light/dark cycle. Standard rodent chow and water were provided ad libitum.
High Intensity Exercise
Rats were introduced to the motor-driven treadmill the day before the experiment and allowed to get used to the pad for 10 min at a speed of 4 m/min, without incline. Each member of the experimental group ran simultaneously. Stainless steel grids at the end of the lines provided an electrical stimulus of 3.2 mA, 200 V at a frequency of 4 Hz, and brushes prevented the rats from pinching feet between grid and treadmill. The running speed was set to 8 m/min, on a 15% incline pad for 5 min and increased to 14 m/min by gradually changing it every 20 min (16). All experiments were performed during the beginning of the dark cycle. Animals were anesthetized with pentobarbital sodium (60 mg/kg body weight intraperitoneally), and livers were excised rapidly and frozen in liquid nitrogen and stored at −80 °C.
Fasting
Rats were deprived of food for 24 or 48 h and killed as described. Liver samples were collected and stored as above.
Preparation of Primary Culture Hepatocytes and Cell Treatment
Primary hepatocytes were isolated from male DBA/2 (OlaHsd) or C57BL/6 mice (Center for Experimental Animals, University of Oulu) aged 8–10 weeks and cultured as described previously (17). The cultures were maintained for 24 h before adenoviral infections or chemical treatments.
Recombinant adenoviruses expressing mouse PGC-1α (PGC-1α-Ad), green fluorescent protein (GFP-Ad), or human constitutively active AMPKα1 subunit containing amino acids 1–321 (AMPK-Ad) have been described previously (17, 18). LacZ-Ad control virus was kindly provided by Dr. Heikki Ruskoaho (University of Oulu). PGC-1α-L2L3M-Ad was constructed using the same adenoviral vector as PGC-1α-Ad. Plasmid coding L2L3M PGC-1α mutant was a generous gift from Prof. Donald P. McDonnell (19). Adenovirus infections and primary hepatocyte treatments were performed in serum-free Williams E medium.
siRNA Transfection
siIL1Rn (siGENOME SMARTpool) and appropriate control were purchased from Thermo Scientific Dharmacon, all other siRNAs were synthesized by Sigma-Aldrich. Mouse primary hepatocytes were transfected with Lipofectamine 2000 (Invitrogen) using the following siRNAs: siPGC-1α, 5′-AAGACGGAUUGCCCUCAUUUG(dT)(dT) (20); siPPARα, 5′-GAUCGGAGCUGCAAGAUUC(dT)(dT) (21); scramble, 5′-AAGCUUCAUAAGGCGCAUAGC(dT)(dT). After a 5-h incubation in transfection mixture, cells were overlaid with serum-free Williams E medium complemented with 50 μm 8-Br-cAMP and collected 48 and 72 h later (siPGC-1α) or transduced with PGC-1α-Ad for 48 h (siPPARα). Expression of genes of interest was measured with SYBR Green qPCR.
Microarray Experiment
Primary hepatocytes were isolated from a male 9-week-old C57BL/6 mouse as described. Three parallel RNA samples were pooled. RNA was subsequently purified and DNase-treated using the SV Total RNA Isolation System (Promega, Madison, WI). RNA quality was measured on an Agilent 2100 Bioanalyzer (Agilent Technologies, Amsterdam, The Netherlands) using 6000 Nano Chips. 5 μg of RNA were used for one-cycle cRNA synthesis (Affymetrix, Santa Clara, CA). Affymetrix Mouse Genome 430 2.0 arrays were used according to instructions from the manufacturer. Probe sets were redefined according to Dai et al. (22), using remapped CDF version 9, based on the Entrez Gene database (build 36, version 2). Expression estimates were obtained applying the multichip modified γ model for oligonucleotide signal (23). Differentially expressed probe sets were identified using probability of positive log ratio, which calculates the probability of differentially expressed genes in a Bayesian fashion instead of p values. A probability of positive log ratio and of negative log ratio was calculated for up-regulated and down-regulated genes, respectively (24).
RNA Preparation and Quantitative RT-PCR
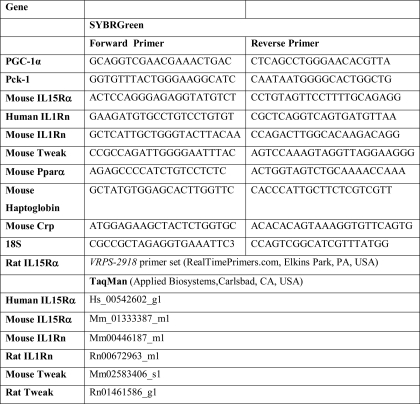
Total RNA was isolated using TRI-Reagent (Sigma) according to the manufacturer's protocol and treated with DNase (Promega, Madison, WI). 1 μg of RNA was reverse transcribed to produce cDNA using p(dN)6 random primers (Roche Applied Science) and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). The quantitative PCRs were performed using either SYBR Green or hydrolysis probes (TaqMan, Applied Bisosystems, Carlsbad, CA). The primers are indicated in Table 1. The fluorescence values of the qPCR products were corrected with the fluorescence signals of the passive reference dye (ROXTM). The RNA levels of target genes were normalized against the 18 S control levels using the comparative CT (ΔΔCT) method.
TABLE 1.
Primer sequences/assay numbers used for real-time PCR
Protein Isolation and Immunoblotting
Whole cell lysate and cytosolic protein fractions were prepared as described previously (25). Soluble proteins were precipitated with ice-cold acetone from culture medium. The protein fractions were separated on SDS-polyacrylamide gel transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA) and incubated with primary goat anti-IL1Rn antibody (sc-8482; 1:500) or goat anti-IL15Rα antibody (sc-1524; 1:500) in 5% skimmed milk, 0.1% Tris-buffered saline followed by HRP anti-goat IgG (1:20,000). Antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). After washing, the immunoreactive bands were visualized with a chemiluminescent peroxidase substrate 1 reaction (Sigma-Aldrich) using Quantity One software (Bio-Rad). β-Actin (anti-β-actin antibody A1978; Sigma-Aldrich) was detected as loading control.
Transient Transfection
1900 bp of mouse Il1rn promoter fragment was inserted in front of the luciferase reporter gene in pGL3-Basic as described (26). Mouse PPARα and PGC-1α WT expression plasmids have been described previously (17, 26). PGC-1α T177A/S538A mutant (AMPK phosphorylation sites mutated) was obtained from Addgene (Cambridge, MA) (Addgene plasmid 180903) (27). Mouse primary hepatocytes were transfected using Fugene HD reagent (Roche Applied Science) according to the manufacturer's protocol. pRL3-TK plasmid with Renilla reniformis luciferase reporter was cotransfected as an internal control. Luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, Madison, WI), and firefly luciferase activities were normalized to Renilla luciferase signals.
RESULTS
Identification of PGC-1α-regulated Hepatic Inflammatory Genes
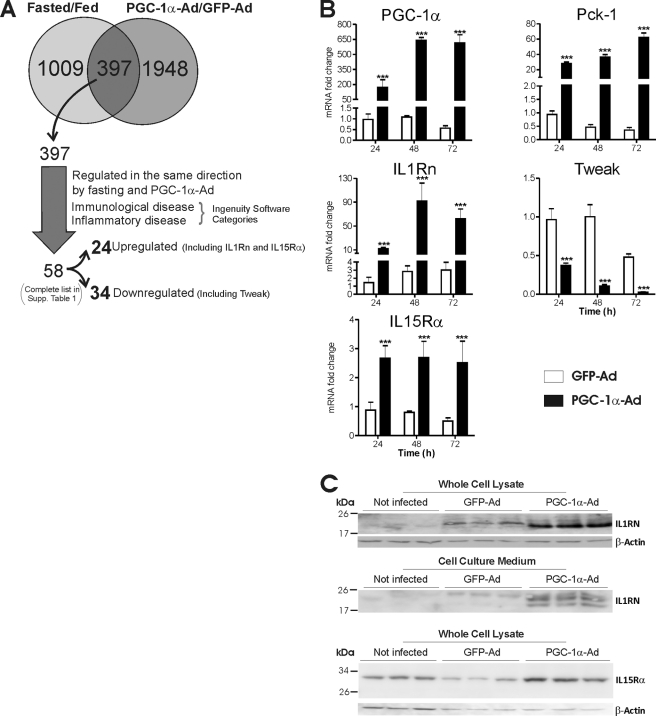
To explore the role of PGC-1α in hepatic regulation of inflammatory genes, mouse primary hepatocytes were transduced with PGC-1α adenovirus (PGC-1α-Ad) or GFP control adenovirus (GFP-Ad), and gene expression was analyzed by microarray. Genes up- or down-regulated at least 1.5-fold by PGC-1α-Ad compared with GFP-Ad were chosen for further analysis. Canonical pathway analysis identified several PGC-1α-regulated pathways from the Ingenuity Pathway Analysis library, and inflammation-related, PGC-1α-regulated pathways were identified, including eicosanoid signaling and communication between innate and adaptive immune cells (supplemental Fig. 1). In order to include only genes with physiological significance, we compared the data with previous microarray data from fasting mice (GSE17865). The data were filtered to contain only genes up- or down-regulated at least 1.5-fold in the same direction by both PGC-1α-Ad and fasting. To identify the genes involved in inflammatory processes, the data were analyzed with the Ingenuity software, and the genes falling into the categories of immunological disease and inflammatory disease were selected. The analysis yielded 58 genes, of which 24 were up-regulated and 34 were down-regulated (Fig. 1A) (supplemental Table 1). The genes regulated by PGC-1α and fasting encode proteins with different basic molecular functions like enzymes, transcription factors, transporters, and signaling molecules, including several cytokines and cytokine receptors. Remarkably, PGC-1α down-regulated several inflammatory cytokines, including Cxcl1 (chemokine (CXC motif) ligand 1), Ccl9 (Chemokine (CC motif) ligand 9), and Tweak. Conversely, PGC-1α up-regulated a key anti-inflammatory cytokine, IL1Rn, as well as IL15Rα, a high affinity-specific receptor subunit for interleukin 15. Altogether, the PGC-1α-regulated gene profile suggests that PGC-1α influences inflammatory gene expression in liver.
FIGURE 1.
Effect of PGC-1α and fasting on expression of immune system-related genes in liver. Mouse primary hepatocytes were transduced with PGC-1α-Ad or GFP-Ad (MOI = 1) or left untreated. A, microarray experiments were performed as described. Genes regulated by PGC-1α-Ad in primary hepatocytes (±1.5-fold) were compared with the set of genes from livers of fasting mice (±1.5-fold). The common set of 397 genes was filtered to contain only genes regulated in the same direction both by fasting and PGC-1α-Ad and categorized in Ingenuity Software, resulting in total of 58 genes belonging to the immunological disease or inflammatory disease categories (complete list in supplemental Table 1), of which 24 were up-regulated (including IL1Rn and IL15Rα) and 34 were down-regulated (including Tweak). B, cells were collected at the indicated time points, and isolated mRNA was used for cDNA synthesis and analysis with qPCR using TaqMan probes (IL1Rn, Tweak, and IL15Rα) and SYBR Green chemistry (PGC-1α and Pck-1). Data are presented as -fold change compared with uninfected controls at a given time point. Bars representing uninfected samples (means equal 1) are not shown. Values represent means ± S.D. (error bars), n = 3. ***, p < 0.001 statistical significance compared with GFP-Ad samples (Student's two-tailed t test). C, cells and culture medium were collected 72 h after transduction, and protein fractions were prepared as described under “Experimental Procedures” and used for immunoblotting. Top, IL1RN in whole cell lysate; middle, IL1RN in cell culture medium protein precipitate; bottom, IL15Rα in whole cell lysate. β-Actin was detected as a loading control.
PGC-1α Modulates Expression of IL1Rn, IL15Rα, and Tweak in Hepatocytes
We selected for further analysis three genes, Il1rn, Il15rα, and Tweak, that all appear to be connected to the regulation of insulin resistance and metabolic disorders (28–31). Mouse primary hepatocytes were transduced with PGC-1α-Ad or control virus GFP-Ad, and temporal changes of mRNA levels were measured up to 72 h by qPCR. The established PGC-1α target gene Pck-1 was induced as expected. Consistent with the microarray data, IL1Rn and IL15Rα were induced by PGC-1α, whereas Tweak was down-regulated (Fig. 1B). IL1Rn was most powerfully regulated by PGC-1α, showing a 26-fold increase at the 48 h time point compared with GFP-Ad. Similar treatment was performed for human hepatoma HepG2 cells, and IL1Rn and IL15Rα mRNA expression was induced about 2-fold after 24 h (data not shown).
To confirm the functional significance of IL1Rn and IL15Rα regulation by PGC-1α, we studied the effect of PGC-1α on IL1RN and IL15Rα protein levels. Adenoviral overexpression of PGC-1α induced one major IL1RN protein band of about 20 kDa in the whole cell lysate fraction (Fig. 1C). Previous reports have indicated that hepatocytes express a soluble, secreted form of IL1RN, whereas expression of an intracellular form is negligible (26, 32). Therefore, the detected band appears to represent soluble IL1RN. To confirm induction of soluble IL1RN in primary hepatocytes, the culture medium was analyzed for IL1RN protein by immunoblotting. Two proteins corresponding in size with glycosylated and unglycosylated forms of IL1RN were detected and strongly up-regulated by PGC-1α (Fig. 1C). Furthermore, overexpression of PGC-1α induced a 3-fold increase of a 30-kDa protein corresponding in size to the predicted molecular weight of mouse IL15Rα (Fig. 1C).
Effect of PGC-1α-up-regulating Hormonal Stimuli on IL1Rn, IL15Rα, and Tweak Expression
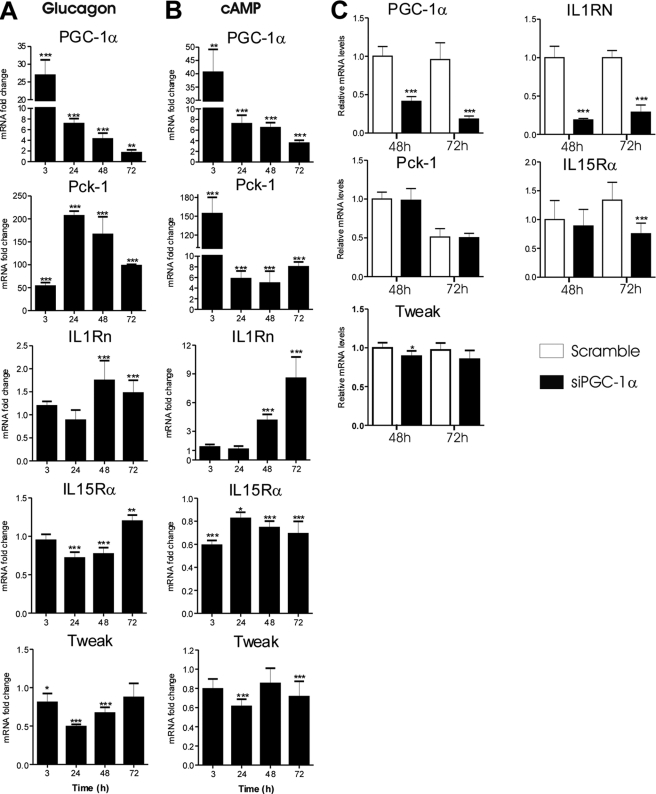
Fasting and exercise increase excretion of the pancreatic hormone glucagon, which is a major regulator of hepatic PGC-1α expression and mediates its effects through the second messenger cAMP. We therefore studied whether glucagon and cAMP have similar effects as PGC-1α on Il1rn, Il15rα, and Tweak expression in cultured cells. Glucagon and cAMP induced PGC-1α mRNA by 27- and 40-fold, respectively, already after 3 h of treatment. Furthermore, although the PGC-1α induction declined after 3 h, PGC-1α mRNA levels remained elevated until at least 72 h. As expected, Pck-1 was also induced by cAMP and glucagon. IL1Rn was induced after 48 h by both cAMP and glucagon, and similar to PGC-1α, IL1Rn was induced more efficiently by cAMP than glucagon (Fig. 2, A and B). Tweak expression was down-regulated in cells exposed to glucagon or cAMP in concordance with the down-regulation by PGC-1α (Fig. 2, A and B). Unexpectedly, expression of IL15Rα mRNA decreased after treatment with cAMP at all time points. In addition, glucagon down-regulated IL15Rα expression at the 24 and 48 h time points (Fig. 2, A and B). Induction of IL1Rn and suppression of Tweak by glucagon and cAMP are consistent with a potential role of PGC-1α.
FIGURE 2.
A and B, effect of glucagon (5 μg/ml porcine glucagon) (A) and cAMP (50 μm 8-Br-cAMP) (B) on expression of selected genes in mouse primary hepatocytes. Cells were collected at the indicated time points, and relative mRNA levels were measured with qPCR using TaqMan probes or SYBR Green chemistry (PGC-1α and Pck-1). Data are presented as -fold change compared with untreated controls. Values represent means ± S.D. (error bars), n = 3–6/point. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with controls at a given time point (Student's two-tailed t test). C, effect of PGC-1α siRNA on cAMP-induced expression of IL1Rn, IL15Rα, and Tweak in mouse primary hepatocytes. Cells were transfected with 150 pmol/μl control (Scramble) or PGC-1α siRNA before treatment with 50 μm 8-Br-cAMP and collected at the indicated time points. mRNAs were measured with SYBR Green qPCR. Values are presented relative to the control (Scramble) sample at the 48 h time point and represent means ± S.D. (n = 3). ***, p < 0.001 compared with the control (Student's two tailed t test).
We next studied the involvement of PGC-1α in regulation of IL1Rn and IL15Rα by cAMP. Primary mouse hepatocytes were transfected with PGC-1α siRNA or scramble control siRNA and treated with cAMP. cAMP-induced expression of PGC-1α and IL1Rn was significantly attenuated by PGC-1α siRNA (Fig. 2C). Also, IL15Rα was slightly down-regulated by PGC-1α siRNA at the 72 h time point, consistent with the observed up-regulation by PGC-1α overexpression. In contrast, induction of Pck-1 by cAMP was not affected by PGC-1α knockdown, and the effect on Tweak was minor.
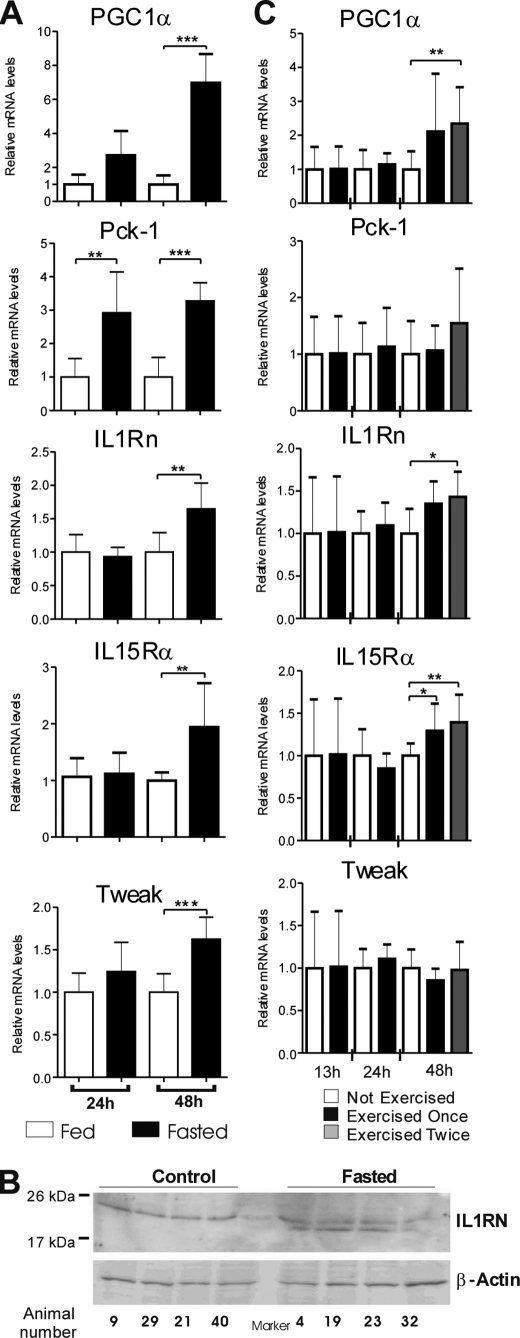
Fasting and Physical Exercise Induce IL1Rn and IL15Rα in Rat Livers
IL1Rn, IL15Rα, and Tweak were among the genes regulated by fasting in mouse liver (microarray data, GSE17865). We investigated whether a similar response occurs also in another species and performed a fasting experiment on rats. Fasting has previously been shown to potently stimulate PGC-1α in liver (9). We found that Pck-1 was significantly induced after 24 h of fasting, whereas PGC-1α induction was not statistically significant. However, mRNA levels of PGC-1α, Pck-1, IL1Rn, IL15Rα, and Tweak were induced after 48 h of fasting (Fig. 3A). Although the fasting data of PGC-1α, Pck-1, IL1Rn, and IL15Rα are consistent with the previous mouse fasting array results, the Tweak results were opposite in mice and rats, indicating a species-specific fasting response. IL1RN protein expression was also measured in livers of fasting rats. The cytosolic protein fraction of whole liver tissue lysates revealed two anti-IL1RN immunoreactive proteins instead of one visualized in a control group. The fasting-induced band corresponded in size with the soluble form of IL1RN (Fig. 3B).
FIGURE 3.
Effect of fasting and exercise on IL1Rn, IL15Rα, and Tweak mRNA and IL1Rn protein expression in rat livers. A, animals were fasted for 24 or 48 h and sacrificed. Livers were isolated immediately and snap frozen in liquid nitrogen. Random liver sections of a minimum of 80 mg were used for mRNA isolation. mRNA expression was measured with SYBR Green qPCR or TaqMan probes (IL1Rn and Tweak). Values are presented relative to the mean of control sample (Fed) for each time point (Student's two-tailed t test; **, p < 0.01; ***, p < 0.001). B, IL1RN was detected in cytosolic protein fraction of random rat liver sections of a minimum of 80 mg. Animals in the experimental group were fasted for 48 h. Proteins were separated in 15% SDS-PAGE gel, and β-actin was used as a loading control. Migration of molecular weight markers is indicated. C, animals were forced to run once or twice for 1 h each time and sacrificed as specified. mRNA was isolated and measured as stated above. Hours represent time after the first exercise period. Values are presented relative to the mean of corresponding control samples (Not Exercised) (Student's two-tailed t test; *, p < 0.05; **, p < 0.01). Error bars, S.D.
Besides fasting, exercise is another physiological condition requiring energy output by liver and is known to induce hepatic PGC-1α expression. We therefore investigated if IL1Rn, IL15Rα, and Tweak are regulated by exercise. Untrained animals were forced to run one or two times for 1 h (with a 24-h break), and livers were collected at different time intervals after the exercise period. PGC-1α and IL1Rn were induced at 48 h after the first exercise in the animals that exercised twice, whereas there was a tendency toward up-regulation in the rats that exercised once (Fig. 3C). Il15rα gene expression was modestly but significantly induced in both exercise groups. Exercise had no effect on Tweak expression, and the exercise-induced increase in Pck-1 mRNA levels was not statistically significant.
Regulation of IL1Rn and IL15Rα Expression by PGC-1α Involves PPARα
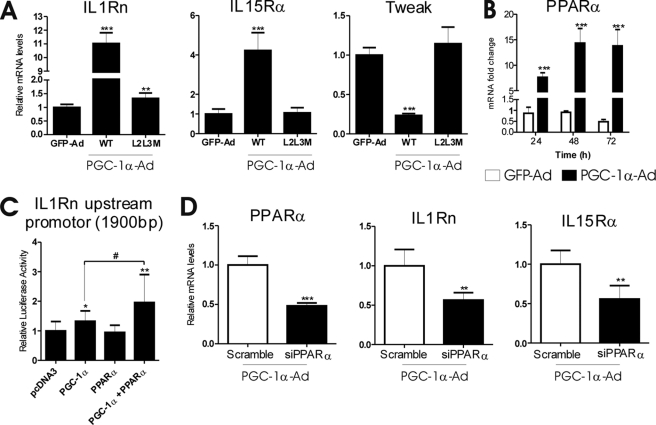
PGC-1α coactivates several nuclear receptors but also other transcription factors. We transduced mouse primary hepatocytes with PGC-1α-Ad wild type or L2L3M mutant that lacks a functional nuclear receptor interaction domain (19) or control virus GFP-Ad. As before, wild type PGC-1α-Ad induced IL1Rn and IL15Rα but down-regulated Tweak. In contrast, L2L3M mutant had a negligible effect or no effect at all compared with GFP-Ad, suggesting that nuclear receptor interaction is necessary for a PGC-1α-Ad effect on the studied inflammatory factors (Fig. 4A). PPARα has been described to directly regulate Il1rn (26). Furthermore, PPARα is both induced and coactivated by PGC-1α. We analyzed previous microarray data from wild type and PPARα null fasting mice (GEO series GSE17865). Compared with fed animals, IL1Rn and IL15Rα were induced in wild type but not in PPARα null animals, indicating that PPARα is necessary for the fasting response of IL1Rn and IL15Rα. We next measured the effect of PGC-1α-Ad on PPARα expression in our experimental conditions. PPARα mRNA was induced about 15-fold by PGC-1α-Ad in mouse primary hepatocytes (Fig. 4B).
FIGURE 4.
Effect of PGC-1α-Ad on IL1Rn, IL15Rα, and Tweak is mediated by a nuclear receptor. A, mouse primary hepatocytes were transduced with wild type (WT) or L2L3M mutant PGC-1α-Ad or GFP-Ad at MOI = 1. Cells were collected after 48 h, and mRNA expression was analyzed with SYBR Green qPCR. Data are presented as -fold change compared with GFP-Ad controls. Values represent means ± S.D. (error bars) (n = 4). ***, p < 0.001 statistical significance compared with GFP-Ad samples. (Student's two-tailed t test). B, cells were treated as described (Fig. 1B), and expression of PPARα was measured with qPCR. Data are presented as -fold change compared with uninfected controls at a given time point. Values represent means ± S.D. (n = 3). ***, p < 0.001, statistical significance compared with GFP-Ad samples, Student's two-tailed t test. C, mouse primary hepatocytes were transfected with the Il1rn upstream promoter luciferase construct and expression vectors as indicated. Cells were collected 48 h later, and luciferase activity was measured. The values are presented relative to pcDNA3 control and represent means ± S.D. *, statistical significance compared with pcDNA3 control samples (Student's two-tailed t test; *, p < 0.05; **, p < 0.01; #, p < 0.05; n = 10–12/point). D, PGC-1α-Ad effect involves PPARα. Cells were transfected with 150 pmol/μl of control (Scramble) or PPARα siRNA before transduction with PGC-1α-Ad and collected after 48 h. mRNAs were measured with SYBR Green qPCR. Values are presented relative to the control (Scramble) sample and represent means ± S.D. (n = 3). **, p < 0.01; ***, p < 0.001 compared with the control (Student's two-tailed t test).
A functional PPARα element has been previously identified in the Ilrn promoter at about 700 bp upstream from the transcription start site (26). To evaluate the significance of the Il1rn gene proximal promoter for PGC-1α response, we transfected mouse primary hepatocytes with a reporter construct under the control of 1900 bp of the Il1rn promoter. PGC-1α cotransfection activated luciferase activity only 1.3-fold. However, cotransfection of PPARα and PGC-1α together activated transcription 2-fold, suggesting PPARα coactivation by PGC-1α (Fig. 4C). The modest response of Il1rn proximal promoter to PGC-1α, compared with mRNA induction, suggests that other regulatory elements located outside the studied region are required for full response.
To confirm the role of PPARα in PGC-1α-mediated induction of IL1Rn, we performed a knockdown experiment. Primary mouse hepatocytes were transfected with PPARα siRNA or scramble control siRNA and transduced with PGC-1α-Ad. Induction of IL1Rn and also IL15Rα by PGC-1α-Ad was reduced by about 50% (Fig. 4D).
PGC-1α Antagonizes Effects of IL1β in Hepatocytes
IL1RN binds to IL1 receptors and functions as a naturally occurring antagonist for IL1. We therefore investigated if PGC-1α-mediated induction of IL1Rn affects induction of haptoglobin and C-reactive protein (CRP) by IL1β. Haptoglobin and CRP are acute phase proteins, and their plasma concentrations rise during inflammation. Mouse primary hepatocytes were transduced with PGC-1α-Ad or GFP-Ad and treated with two different doses of IL1β. As expected, IL1β induced IL1Rn; nevertheless, the PGC-1α-Ad effect was stronger. IL1β did not significantly affect IL15Rα or Tweak mRNA. Haptoglobin and CRP were induced by IL1β; however, this effect was abolished by PGC-1α-Ad (supplemental Fig. 2).
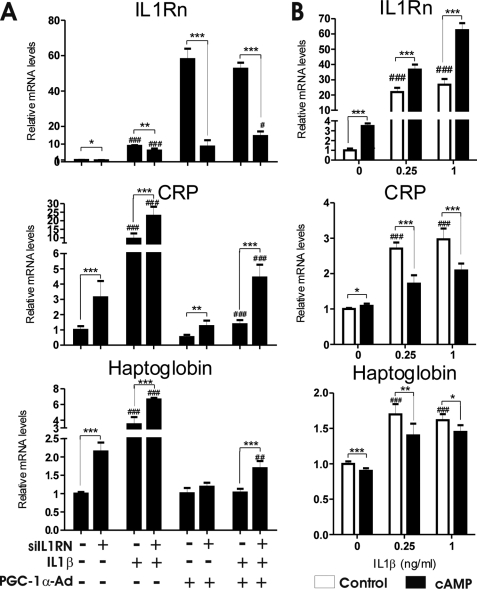
To further elucidate role of IL1Rn in anti-inflammatory response by PGC-1α, we knocked down expression of IL1Rn in hepatocytes treated with IL1β and transduced with PGC-1α-Ad. As expected, IL1Rn mRNA was induced by PGC-1α-Ad and IL1β but reduced by IL1Rn siRNA (Fig. 5A). IL1β increased CRP mRNA 10-fold, whereas transduction with PGC-1α-Ad reduced this effect by 85%. In IL1Rn siRNA-treated cells, PGC-1α-Ad inhibited CRP expression by only 53%, indicating a contribution of IL1Rn to the PGC-1α-Ad-mediated anti-inflammatory effect. Analogous results were observed for haptoglobin, although all of the -fold changes were smaller (Fig. 5A). IL1Rn siRNA was able to increase CRP and haptoglobin expression even in the absence of IL1β treatment. This is probably due to inflammatory response and up-regulation of IL1Rn induced by control adenovirus, as seen also in Fig. 1. IL1Rn siRNA did not have any effect on another PGC-1α target gene, Pck-1, measured as a control (data not shown).
FIGURE 5.
PGC-1α-Ad and cAMP attenuate expression of CRP and haptoglobin in response to IL1β in hepatocytes. A, mouse primary hepatocytes were transfected with 100 pmol/μl control (scramble) or IL1Rn siRNA before transduction with PGC-1α-Ad or LacZ-Ad (MOI = 1). After 48 h, cells were treated with 0.5 ng/ml IL1β for an additional 24 h. Hepatocytes were collected, and mRNA was isolated and analyzed with SYBR Green qPCR. Data are presented as -fold change compared with LacZ-Ad + scramble siRNA sample without IL1β (normalized as 1). *, statistical significance between scramble and siIL1Rn; #, statistical significance between cells with added IL1β and corresponding untreated samples. Values represent means ± S.D. (n = 3). **, p < 0.01; ***, p < 0.001; ##, p < 0.01; ###, p < 0.001 (Student's two-tailed t test). B, mouse primary hepatocytes were treated with 75 μm Bt2cAMP, and controls were left untreated for 24 h before IL1β was added to the cells as indicated. After an additional 24 h, samples were collected, and mRNA was analyzed with SYBR Green qPCR. Data are presented as -fold change compared with untreated controls without IL1β. Values represent means ± S.D. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001, statistical significance compared with control samples. ###, p < 0.001, controls without cAMP compared with 0 ng/ml IL1β sample (Student's two-tailed t test).
In addition, we studied whether cAMP affects expression of acute phase proteins induced by IL1β. Primary hepatocytes were first treated with cAMP for 24 h, after which IL1β was added, and the cells were incubated for an additional 24 h. IL1Rn was induced by both cAMP and IL1β, and the two compounds had an additive effect. Haptoglobin and CRP were induced by IL1β; however, treatment with cAMP attenuated this response (Fig. 5B).
AMPK Modulates Expression of IL1Rn and IL15Rα
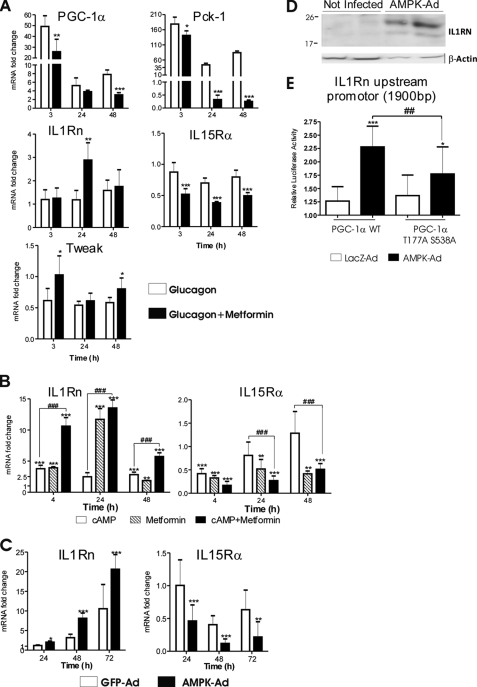
Besides PGC-1α, AMPK is another key cellular sensor of energy status. PGC-1α and AMPK pathways are known to cross-talk, and AMPK may affect both PGC-1α expression and function (27, 33). We therefore studied whether AMPK modulates the PGC-1α effect on inflammatory signaling in hepatocytes. Metformin is a widely used antidiabetic drug that activates AMPK and attenuates PGC-1α induction by cAMP (34). First, we investigated the effect of metformin on glucagon-induced changes in PGC-1α, Pck-1, IL1Rn, IL15Rα, and Tweak expression in mouse primary hepatocytes. As expected, metformin attenuated the PGC-1α induction by glucagon. Consistent with these data, metformin attenuated Pck-1 up-regulation and Tweak down-regulation by PGC-1α (Fig. 6A). In contrast, metformin increased expression of IL1Rn compared with glucagon alone, suggesting that metformin has PGC-1α-independent effects on IL1Rn. On the other hand, IL15Rα was down-regulated by metformin (Fig. 6A). Because IL15Rα is paradoxically down-regulated by glucagon (although induced by PGC-1α), it is difficult to determine how much of the observed change is related to metformin-mediated reduction of PGC-1α.
FIGURE 6.
Effect of AMPK on expression of IL1Rn and IL15Rα in mouse primary hepatocytes. A, mouse primary hepatocytes were treated with 5 μg/ml porcine glucagon alone or together with 1 mm metformin and compared with untreated control. Cells were collected at the indicated time points, and mRNA expression was measured with TaqMan or SYBR Green (PGC-1α and Pck-1) qPCR. Data are presented as -fold change compared with untreated controls at a given time point. Values represent means ± S.D. (error bars) (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001, statistical significance compared with glucagon alone (Student's two-tailed t test). B, mouse primary hepatocytes were treated with 25 μm Bt2cAMP, 1 mm metformin, or a combination of the two. Cells were collected at specified time points. mRNA expression was measured with SYBR Green qPCR. Data are represented as -fold change compared with untreated controls at a given time point. Values represent means ± S.D. (n = 4). *, statistical significance compared with untreated controls; #, statistically significant difference between Bt2cAMP alone and Bt2cAMP plus metformin (Student's two-tailed t test). **, p < 0.01; ***, p < 0.001; ###, p < 0.001. C, mouse primary hepatocytes were infected with AMPK-Ad or GFP-Ad (MOI = 2). Cells were collected at the indicated time points, and mRNA expression was measured with SYBR Green qPCR. Values represent means ± S.D. (n = 4). Data represent -fold change compared with untreated controls at a given time point. *, p < 0.05; **, p < 0.01; ***, p < 0.001, statistical significance compared with GFP-Ad (Student's two-tailed t test). D, detection of IL1RN protein by immunoblotting in whole cell lysates of mouse primary hepatocytes 72 h after infection with AMPK-Ad (MOI = 2). Control samples were uninfected. β-Actin was detected as a loading control. E, mouse primary hepatocytes were transfected with Il1rn promoter luciferase construct and cotransfected with PGC-1α or PGC-1α T177A/S538A mutant expression vectors or empty pCDNA3 plasmid. After 24 h, cells were transduced with LacZ-Ad or AMPK-Ad at MOI = 1 and collected after an additional 24 h, and luciferase activity was measured. Values are presented as -fold change compared with pcDNA3 control samples and represent means ± S.D. *, statistical significance compared with LacZ-Ad samples. #, statistical significance compared with PGC-1α WT (Student's two-tailed t test). *, p < 0.05; ***, p < 0.001; ##, p < 0.01 (n = 14–16/point). Results are representative of two independent experiments.
Consequently, we investigated the effect of metformin alone or in combination with cAMP on expression of IL1Rn. IL1Rn was up-regulated by both cAMP and metformin, and the combination of the two had a synergistic effect (Fig. 6B).
Subsequently, we studied whether AMPK can directly regulate IL1Rn. Mouse primary hepatocytes were transduced with a constitutively active form of AMPK-Ad or GFP-Ad, and IL1Rn mRNA levels were measured up to 72 h. Similar to metformin, AMPK up-regulated IL1Rn (Fig. 6C). Consequently, we measured the effect of AMPK-Ad on IL1RN protein in mouse primary hepatocytes. AMPK-Ad treatment for 72 h strongly up-regulated IL1RN protein expression compared with untreated control cells (Fig. 6D).
In muscle cells, AMPK-mediated phosphorylation enhances PGC-1α transcriptional activity (27). We therefore investigated if AMPK affects PGC-1α function in the context of the Ilrn promoter. The Luciferase reporter construct regulated by the Il1rn promoter (1900 bp) was cotransfected with PGC-1α expression plasmid to primary hepatocytes. As described above, PGC-1α activated luciferase activity 1.3-fold. When the cells were concomitantly transduced with AMPK-Ad, the luciferase activity was activated more than 2-fold (Fig. 6E). AMPK alone did not induce IL1Rn promoter activity (data not shown). PGC-1α T177A/S538A mutant (sites directly phosphorylated by AMPK (27)) activated Il1rn promoter similarly to wild type PGC-1α. However, mutation of the phosphorylation sites significantly attenuated the AMPK effect compared with the wild type PGC-1α (Fig. 6E). These results suggest that AMPK modulates PGC-1α function also in the context of the IL1Rn promoter in liver cells. Nevertheless, the fact that AMPK activation promotes IL1Rn expression while simultaneously attenuating PGC-1α induction by glucagon (or cAMP) suggests that PGC-1α does not play a major role in induction of IL1Rn by AMPK.
IL15Rα was down-regulated by the combination of metformin and cAMP but also by metformin alone (Fig. 6B), indicating an AMPK-related mechanism involved in regulation of the gene. Consequently, AMPK-Ad reduced expression of IL15Rα mRNA (Fig. 6C).
In the previous experiments, we showed that PPARα is involved in induction of IL1Rn and IL15Rα by PGC-1α. Therefore, we investigated if AMPK and metformin affects PPARα expression. Metformin down-regulated PPARα mRNA 2–5-fold (depending on the time point), whereas AMPK-Ad had no effect (data not shown), suggesting that PPARα is not involved in regulation of IL1Rn and IL15Rα by AMPK.
DISCUSSION
Growing evidence supports the importance of inflammation in the metabolic syndrome, a combination of risk factors for cardiovascular diseases and diabetes. Factors that integrate nutrient- and pathogen-sensing pathways are of great interest in understanding the mechanisms of chronic metabolic pathologies. PGC-1α is a powerful regulator of energy metabolism in key tissues, including liver, adipose tissue, and skeletal muscle. In skeletal muscle, PGC-1α suppresses the production of inflammatory cytokines, such as TNFα and IL6 (35), suggesting that PGC-1α is one of the interlinks between regulation of energy homeostasis and inflammation and that activation of PGC-1α in muscle may have beneficial anti-inflammatory effects.
In the current study, we investigated the role of PGC-1α in cross-talk of energy metabolism and inflammation in liver. PGC-1α overexpression down-regulated several inflammatory cytokines, including Tweak as well as CxCl1 and Ccl9 chemokines. Simultaneously, PGC-1α up-regulated anti-inflammatory signals, especially IL1Rn, and also the more pleiotropic IL15Rα. These results suggest that, similar to muscle, hepatic PGC-1α restrains inflammatory response.
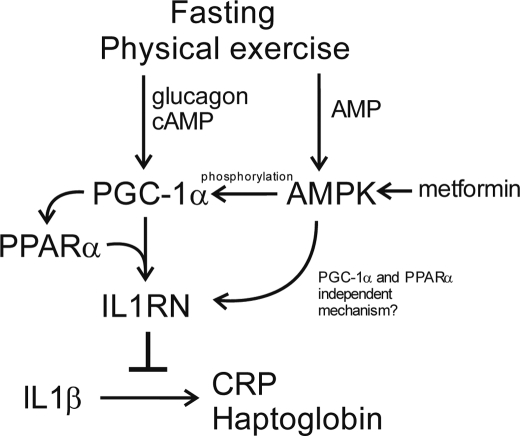
IL1Rn was found to be consistently up-regulated by PGC-1α overexpression, fasting, and physical exercise (the physiological conditions up-regulating PGC-1α) as well as by PGC-1α-regulating hormonal stimulus (glucagon and cAMP). Furthermore, PGC-1α siRNA down-regulated cAMP-stimulated expression of both PGC-1α and IL1Rn. These findings strongly suggest that Il1Rn gene expression is under strict control of PGC-1α. Analysis of public microarray data revealed that IL1Rn induction by fasting is absent in PPARα-null animals. PGC-1α induction by fasting is not affected by PPARα knock-out (36). Therefore, PGC-1α appears to be upstream of PPARα in the signal cascade, resulting in IL1Rn induction. Indeed, PPARα has been described to directly regulate IL1Rn (26). PPARα is both induced and coactivated by PGC-1α, making this pathway very robust. Finally, we showed that knockdown of PPARα in hepatocytes suppressed PGC-1α-mediated up-regulation of IL1Rn. These data indicate that IL1Rn induction by PGC-1α involves PPARα. Our model of IL1Rn regulation by PGC-1α is summarized in Fig. 7.
FIGURE 7.
Model of PGC-1α- and AMPK-mediated regulation of IL1Rn expression in hepatocytes. Fasting and physical exercise increase blood concentration of glucagon, leading to increased cAMP level in hepatocytes. cAMP induces PGC-1α, which in turn triggers expression of PPARα and IL1RN in a PPARα-dependent manner. Consequently, IL1RN interferes with IL1β-mediated inflammation and inhibits expression of acute phase proteins (CRP and haptoglobin) in hepatocytes. Fasting, physical exercise, and antidiabetic drug metformin also activate AMPK. AMPK phosphorylates PGC-1α and increases its transcriptional activity in the context of the Il1rn promoter. In addition, AMPK and metformin affect IL1RN expression in a mechanism independent of the PGC-1α and PPARα pathway.
IL1RN belongs to the interleukin 1 family and binds to IL1 receptors without signal transduction, serving as a naturally occurring antagonist. Accordingly, the balance between IL1 and IL1RN controls the inflammatory effect of IL1 (37). Indeed, we could demonstrate that PGC-1α suppresses the IL1β-mediated response of acute phase proteins in hepatocytes, and at least in part, this is due to increased expression of IL1RN. A number of studies have previously shown a close connection of IL1RN and metabolic disorders. IL1RN serum levels are increased in obese and type II prediabetic patients (28, 29). However, treatment with recombinant IL1RN (anakinra) has been shown to be antidiabetic, improving both insulin sensitivity and beta cell function (38, 39), suggesting that the increase in IL1RN may represent a compensatory change to proinflammatory signals. The liver is a major source of circulating ILRN (40). PGC-1α is induced in liver by caloric restriction and exercise (both conditions improving insulin sensitivity and glucose tolerance (41)). Thus, PGC-1α-induced IL1RN expression may contribute to the beneficial effects of these physiological stimuli.
Fasting and physical exercise, as well as PGC-1α overexpression, induced IL15Rα in liver. However, cAMP and glucagon had a mild opposite effect. These conflicting results emphasize the complexity of IL15Rα regulation and suggest the existence of inhibitory signals activated by identical hormonal stimuli. One of the possible explanations is activation of the AMPK pathway because AMPK-Ad reduced expression of the IL15Rα, and AMPK is activated in glucagon-treated hepatocytes and livers of fasted mice (42, 43). However, the in vivo experiments indicate that in physiological context, the net effect is induction.
IL15 is a potent proinflammatory cytokine. Its aberrant expression is characteristic for several autoimmune disorders, including type 1 diabetes mellitus (44). The complete IL15 receptor is a α/β/γ heterotrimer (45). Although all three elements are expressed by hepatocytes, fasting and PGC1α-Ad induced only the α subunit. IL15Rα is fully capable of perpetuating signal alone, although the composition of the receptor might affect the type of activated pathways (45). IL15Rα is primarily a membrane-bound receptor, yet it exists also in a soluble form, due to proteolytic shedding from the surface (50). Soluble IL15Rα (sIL15Rα) retains the ability to bind IL15 and functions as its antagonist (46–48). In line with these studies, treatment with sIL15Rα reduced hyperglycemia in autoimmune diabetes in mice (30). However, the fusion IL15-IL15Rα protein and certain forms of sIL15Rα have been shown to have potent agonist properties, emphasizing the pleiotropic nature of IL15Rα (49). Interestingly, IL15 is secreted by muscle, and its circulating levels increase after exercise (50, 51). According to our results, physical activity induces expression of IL15Rα in liver. It is tempting to speculate that hepatic IL15Rα removes muscle-derived IL15 from systemic circulation or, in the case of sIL15Rα, renders it inactive. Clearly, this subject requires more study.
PGC-1α down-regulated several inflammatory cytokines, including Tweak. In mouse hepatocytes, Tweak was down-regulated by fasting, by PGC-1α overexpression, and consistently also by PGC-1α-regulating hormonal stimuli, glucagon and cAMP. TWEAK is a member of the TNF ligand family with proinflammatory, proliferative, apoptotic, and angiogenic effects, depending upon cell type. In hepatocytes, TWEAK induces insulin resistance (31). Therefore, down-regulation of Tweak by PGC-1α can be assumed to have a beneficial effect on glucose tolerance. However, in rat livers, fasting actually induced Tweak expression. This species difference suggests that Tweak does not play a fundamental role in cross-talk of metabolism and inflammation. Mechanisms of PGC-1α-mediated down-regulation are still poorly understood. Recently, heme-regulated transcription repressor Rev-Erbα was found to mediate negative regulation of FGF21 by PGC-1α (52). Most likely, there are several other pathways similarly mediating repressive effects of PGC-1α. Furthermore, inflammation could be regulated through alteration of cellular reactive oxygen species by PGC-1α (35). It will be an important future task to identify the molecular mechanisms involved in PGC-1α-mediated down-regulation of inflammatory signaling pathways.
AMPK is a cellular energy sensor regulated by the ATP/AMP ratio. In muscle, AMPK increases PGC-1α expression and also directly phosphorylates PGC-1α, increasing its transcriptional activity (33). However, in liver, AMPK activator metformin prevents TORC2-mediated up-regulation of PGC-1α (34). Thus, AMPK and PGC-1α signals appear to be closely integrated. To elucidate possible interaction of AMPK and PGC-1α pathways in the regulation of IL1Rn, IL15Rα, and Tweak, we analyzed the effect of the AMPK-activating drug metformin on the response of these factors to glucagon. As expected, metformin attenuated PGC-1α induction by glucagon. In analogy, the effect of glucagon on Tweak expression was decreased by metformin. In contrast, metformin alone or in combination with glucacon or cAMP induced IL1Rn expression. Furthermore, AMPK-Ad up-regulated IL1Rn in hepatocytes. AMPK phosphorylates and regulates the function of numerous transcription factors and coactivators (53). We showed that in hepatocytes, AMPK potentiated PGC-1α-mediated transactivation of the Il1rn promoter. Nevertheless, AMPK promoted IL1Rn expression but at the same time attenuated PGC-1α induction by glucagon. This suggests that AMPK regulation of IL1Rn is predominantly mediated by a transcription factor other than PGC-1α (Fig. 7). Metformin has been for many years suggested to have an anti-inflammatory effect, but the mechanisms have remained elusive (54). Our findings indicate that induction of IL1RN expression by AMPK is a novel mechanism mediating anti-inflammatory effects of metformin.
Expression of IL15Rα is attenuated by AMPK, which at least in part could be explained by reduced PGC-1α levels. However, we cannot exclude contributions of other factors. Nevertheless, the AMPK-related mechanism appears to be independent of PPARα. Taken together, our data indicate that AMPK is another, energy-sensing pathway regulating hepatic expression of IL1Rn and IL15Rα.
In summary, our study presents two major findings. (i) We describe a novel function of PGC-1α as an immunomodulating factor and a regulatory interlink between energy homeostasis and the hepatic immune system. More specifically, we show that PGC-1α induces IL1Rn and represses expression of acute phase proteins in response to IL1β. This suggests that PGC-1α is involved in beneficial, anti-inflammatory effects of physical exercise and caloric restriction. (ii) In addition we show that another energy-sensing factor, AMPK, regulates IL1Rn and IL15Rα. Induction of IL1RN by AMPK is a novel pathway mediating anti-inflammatory effects of metformin.
Supplementary Material
Acknowledgments
The skillful technical assistance of Ritva Tauriainen and Päivi Tyni is gratefully acknowledged.
This work was supported by grants from the European Union (Marie Curie RTN NucSys), the Academy of Finland (Contract 110591), the Sigrid Juselius Foundation, and the National Development Agency of Hungary (Grant TÁMOP 4.2.2-08/01/KMR-2008-004).

This article contains supplemental Table 1 and Figs. 1 and 2.
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1 α
- Pck-1
- phosphoenolpyruvate carboxykinase 1
- IL1Rn
- interleukin 1 receptor antagonist
- IL15Rα
- interleukin 15 receptor α
- Tweak
- tumor necrosis factor-like weak inducer of apoptosis
- AMPK
- AMP-activated protein kinase
- PPARα
- peroxisome proliferator-activated receptor α
- CRP
- C-reactive protein
- Bt2cAMP
- dibutyryl-cAMP
- 8-Br-cAMP
- 8-bromo-cyclic AMP
- qPCR
- quantitative PCR
- Ad
- adenovirus
- MOI
- multiplicity of infection.
REFERENCES
- 1. Hotamisligil G. S., Shargill N. S., Spiegelman B. M. (1993) Adipose expression of tumor necrosis factor-α. Direct role in obesity-linked insulin resistance. Science 259, 87–91 [DOI] [PubMed] [Google Scholar]
- 2. Hotamisligil G. S., Arner P., Caro J. F., Atkinson R. L., Spiegelman B. M. (1995) Increased adipose tissue expression of tumor necrosis factor-α in human obesity and insulin resistance. J. Clin. Invest. 95, 2409–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pickup J. C., Mattock M. B., Chusney G. D., Burt D. (1997) NIDDM as a disease of the innate immune system. Association of acute-phase reactants and interleukin-6 with metabolic syndrome X. Diabetologia 40, 1286–1292 [DOI] [PubMed] [Google Scholar]
- 4. Spranger J., Kroke A., Möhlig M., Hoffmann K., Bergmann M. M., Ristow M., Boeing H., Pfeiffer A. F. (2003) Inflammatory cytokines and the risk to develop type 2 diabetes. Results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes 52, 812–817 [DOI] [PubMed] [Google Scholar]
- 5. Donath M. Y., Shoelson S. E. (2011) Type 2 diabetes as an inflammatory disease. Nat. Rev. Immunol. 11, 98–107 [DOI] [PubMed] [Google Scholar]
- 6. Baker R. G., Hayden M. S., Ghosh S. (2011) NF-κB, inflammation, and metabolic disease. Cell. Metab. 13, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hummasti S., Hotamisligil G. S. (2010) Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ. Res. 107, 579–591 [DOI] [PubMed] [Google Scholar]
- 8. Hotamisligil G. S. (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140, 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., Montminy M. (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183 [DOI] [PubMed] [Google Scholar]
- 10. Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., Spiegelman B. M. (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 [DOI] [PubMed] [Google Scholar]
- 11. Soyal S., Krempler F., Oberkofler H., Patsch W. (2006) PGC-1α. A potent transcriptional cofactor involved in the pathogenesis of type 2 diabetes. Diabetologia 49, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 12. Bensinger S. J., Tontonoz P. (2008) Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454, 470–477 [DOI] [PubMed] [Google Scholar]
- 13. Huang W., Glass C. K. (2010) Nuclear receptors and inflammation control. Molecular mechanisms and pathophysiological relevance. Arterioscler. Thromb. Vasc. Biol. 30, 1542–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoene M., Lehmann R., Hennige A. M., Pohl A. K., Häring H. U., Schleicher E. D., Weigert C. (2009) Acute regulation of metabolic genes and insulin receptor substrates in the liver of mice by one single bout of treadmill exercise. J. Physiol. 587, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ranhotra H. S. (2010) Long-term caloric restriction up-regulates PPAR γ co-activator 1 α (PGC-1α) expression in mice. Indian J. Biochem. Biophys. 47, 272–277 [PubMed] [Google Scholar]
- 16. Suwa M., Nakano H., Radak Z., Kumagai S. (2008) Endurance exercise increases the SIRT1 and peroxisome proliferator-activated receptor γ coactivator-1α protein expressions in rat skeletal muscle. Metabolism 57, 986–998 [DOI] [PubMed] [Google Scholar]
- 17. Arpiainen S., Järvenpää S. M., Manninen A., Viitala P., Lang M. A., Pelkonen O., Hakkola J. (2008) Coactivator PGC-1α regulates the fasting inducible xenobiotic-metabolizing enzyme CYP2A5 in mouse primary hepatocytes. Toxicol. Appl. Pharmacol. 232, 135–141 [DOI] [PubMed] [Google Scholar]
- 18. Kerkela R., Woulfe K. C., Durand J. B., Vagnozzi R., Kramer D., Chu T. F., Beahm C., Chen M. H., Force T. (2009) Sunitinib-induced cardiotoxicity is mediated by off-target inhibition of AMP-activated protein kinase. Clin. Transl. Sci. 2, 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaillard S., Grasfeder L. L., Haeffele C. L., Lobenhofer E. K., Chu T. M., Wolfinger R., Kazmin D., Koves T. R., Muoio D. M., Chang C. Y., McDonnell D. P. (2006) Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol. Cell 24, 797–803 [DOI] [PubMed] [Google Scholar]
- 20. Zhang P., Liu C., Zhang C., Zhang Y., Shen P., Zhang J., Zhang C. Y. (2005) Free fatty acids increase PGC-1α expression in isolated rat islets. FEBS Lett. 579, 1446–1452 [DOI] [PubMed] [Google Scholar]
- 21. De Souza A. T., Dai X., Spencer A. G., Reppen T., Menzie A., Roesch P. L., He Y., Caguyong M. J., Bloomer S., Herweijer H., Wolff J. A., Hagstrom J. E., Lewis D. L., Linsley P. S., Ulrich R. G. (2006) Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. 34, 4486–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., Watson S. J., Meng F. (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu X., Milo M., Lawrence N. D., Rattray M. (2005) A tractable probabilistic model for Affymetrix probe-level analysis across multiple chips. Bioinformatics 21, 3637–3644 [DOI] [PubMed] [Google Scholar]
- 24. Liu X., Milo M., Lawrence N. D., Rattray M. (2006) Probe-level measurement error improves accuracy in detecting differential gene expression. Bioinformatics 22, 2107–2113 [DOI] [PubMed] [Google Scholar]
- 25. Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stienstra R., Mandard S., Tan N. S., Wahli W., Trautwein C., Richardson T. A., Lichtenauer-Kaligis E., Kersten S., Müller M. (2007) The interleukin-1 receptor antagonist is a direct target gene of PPARα in liver. J. Hepatol. 46, 869–877 [DOI] [PubMed] [Google Scholar]
- 27. Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meier C. A., Bobbioni E., Gabay C., Assimacopoulos-Jeannet F., Golay A., Dayer J. M. (2002) IL-1 receptor antagonist serum levels are increased in human obesity. A possible link to the resistance to leptin? J. Clin. Endocrinol. Metab. 87, 1184–1188 [DOI] [PubMed] [Google Scholar]
- 29. Carstensen M., Herder C., Kivimäki M., Jokela M., Roden M., Shipley M. J., Witte D. R., Brunner E. J., Tabák A. G. (2010) Accelerated increase in serum interleukin-1 receptor antagonist starts 6 years before diagnosis of type 2 diabetes. Whitehall II prospective cohort study. Diabetes 59, 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lukic M. L., Mensah-Brown E., Wei X., Shahin A., Liew F. Y. (2003) Lack of the mediators of innate immunity attenuate the development of autoimmune diabetes in mice. J. Autoimmun. 21, 239–246 [DOI] [PubMed] [Google Scholar]
- 31. Feng F., Wang L., Albanese N., Holmes A., Xia P. (2008) Tumor necrosis factor-like weak inducer of apoptosis attenuates the action of insulin in hepatocytes. Endocrinology 149, 1505–1513 [DOI] [PubMed] [Google Scholar]
- 32. Steinkasserer A., Estaller C., Weiss E. H., Sim R. B. (1992) Human interleukin-1 receptor antagonist is expressed in liver. FEBS Lett. 310, 60–62 [DOI] [PubMed] [Google Scholar]
- 33. Cantó C., Auwerx J. (2009) PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaw R. J., Lamia K. A., Vasquez D., Koo S. H., Bardeesy N., Depinho R. A., Montminy M., Cantley L. C. (2005) The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 310, 1642–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Handschin C., Spiegelman B. M. (2008) The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Im S. S., Kim M. Y., Kwon S. K., Kim T. H., Bae J. S., Kim H., Kim K. S., Oh G. T., Ahn Y. H. (2011) Peroxisome proliferator-activated receptor {alpha} is responsible for the up-regulation of hepatic glucose-6-phosphatase gene expression in fasting and db/db mice. J. Biol. Chem. 286, 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perrier S., Darakhshan F., Hajduch E. (2006) IL-1 receptor antagonist in metabolic diseases. Dr Jekyll or Mr Hyde? FEBS Lett. 580, 6289–6294 [DOI] [PubMed] [Google Scholar]
- 38. Ehses J. A., Lacraz G., Giroix M. H., Schmidlin F., Coulaud J., Kassis N., Irminger J. C., Kergoat M., Portha B., Homo-Delarche F., Donath M. Y. (2009) IL-1 antagonism reduces hyperglycemia and tissue inflammation in the type 2 diabetic GK rat. Proc. Natl. Acad. Sci. U.S.A. 106, 13998–14003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsen C. M., Faulenbach M., Vaag A., Vølund A., Ehses J. A., Seifert B., Mandrup-Poulsen T., Donath M. Y. (2007) Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N. Engl. J. Med. 356, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 40. Lamacchia C., Palmer G., Bischoff L., Rodriguez E., Talabot-Ayer D., Gabay C. (2010) Distinct roles of hepatocyte- and myeloid cell-derived IL-1 receptor antagonist during endotoxemia and sterile inflammation in mice. J. Immunol. 185, 2516–2524 [DOI] [PubMed] [Google Scholar]
- 41. Corton J. C., Brown-Borg H. M. (2005) Peroxisome proliferator-activated receptor gamma coactivator 1 in caloric restriction and other models of longevity. J. Gerontol. A Biol. Sci. Med. Sci. 60, 1494–1509 [DOI] [PubMed] [Google Scholar]
- 42. Longuet C., Sinclair E. M., Maida A., Baggio L. L., Maziarz M., Charron M. J., Drucker D. J. (2008) The glucagon receptor is required for the adaptive metabolic response to fasting. Cell. Metab. 8, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berglund E. D., Kang L., Lee-Young R. S., Hasenour C. M., Lustig D. G., Lynes S. E., Donahue E. P., Swift L. L., Charron M. J., Wasserman D. H. (2010) Glucagon and lipid interactions in the regulation of hepatic AMPK signaling and expression of PPARα and FGF21 transcripts in vivo. Am. J. Physiol. Endocrinol. Metab. 299, E607–E614 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Kuczyński S., Winiarska H., Abramczyk M., Szczawińska K., Wierusz-Wysocka B., Dworacka M. (2005) IL-15 is elevated in serum patients with type 1 diabetes mellitus. Diabetes Res. Clin. Pract. 69, 231–236 [DOI] [PubMed] [Google Scholar]
- 45. Budagian V., Bulanova E., Paus R., Bulfone-Paus S. (2006) IL-15/IL-15 receptor biology. A guided tour through an expanding universe. Cytokine Growth Factor Rev. 17, 259–280 [DOI] [PubMed] [Google Scholar]
- 46. Smith X. G., Bolton E. M., Ruchatz H., Wei X., Liew F. Y., Bradley J. A. (2000) Selective blockade of IL-15 by soluble IL-15 receptor α-chain enhances cardiac allograft survival. J. Immunol. 165, 3444–3450 [DOI] [PubMed] [Google Scholar]
- 47. Ruchatz H., Leung B. P., Wei X. Q., McInnes I. B., Liew F. Y. (1998) Soluble IL-15 receptor α-chain administration prevents murine collagen-induced arthritis. A role for IL-15 in development of antigen-induced immunopathology. J. Immunol. 160, 5654–5660 [PubMed] [Google Scholar]
- 48. Mortier E., Bernard J., Plet A., Jacques Y. (2004) Natural, proteolytic release of a soluble form of human IL-15 receptor α-chain that behaves as a specific, high affinity IL-15 antagonist. J. Immunol. 173, 1681–1688 [DOI] [PubMed] [Google Scholar]
- 49. Mortier E., Quéméner A., Vusio P., Lorenzen I., Boublik Y., Grötzinger J., Plet A., Jacques Y. (2006) Soluble interleukin-15 receptor α (IL-15R α)-sushi as a selective and potent agonist of IL-15 action through IL-15R β/γ. Hyperagonist IL-15 x IL-15R α fusion proteins. J. Biol. Chem. 281, 1612–1619 [DOI] [PubMed] [Google Scholar]
- 50. Nielsen A. R., Mounier R., Plomgaard P., Mortensen O. H., Penkowa M., Speerschneider T., Pilegaard H., Pedersen B. K. (2007) Expression of interleukin-15 in human skeletal muscle. Effect of exercise and muscle fibre type composition. J. Physiol. 584, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Riechman S. E., Balasekaran G., Roth S. M., Ferrell R. E. (2004) Association of interleukin-15 protein and interleukin-15 receptor genetic variation with resistance exercise training responses. J. Appl. Physiol. 97, 2214–2219 [DOI] [PubMed] [Google Scholar]
- 52. Estall J. L., Ruas J. L., Choi C. S., Laznik D., Badman M., Maratos-Flier E., Shulman G. I., Spiegelman B. M. (2009) PGC-1α negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erbα axis. Proc. Natl. Acad. Sci. U.S.A. 106, 22510–22515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hardie D. G. (2011) AMP-activated protein kinase. An energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bulcão C., Ribeiro-Filho F. F., Sañudo A., Roberta Ferreira S. G. (2007) Effects of simvastatin and metformin on inflammation and insulin resistance in individuals with mild metabolic syndrome. Am. J. Cardiovasc. Drugs 7, 219–224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.