Background: ATP synthase (F0F1) is a rotary motor enzyme.
Results: F1 with a short-sized helix-1 in β subunit rotates with half of the normal torque and supports reduced ATP synthesis activity.
Conclusion: Helix-1 acts as a “pushrod” to generate torque, and torque-reduced F0F1 retains the catalytic ability of ATP synthesis.
Significance: Generation and utilization of the torque are crucial for motor enzymes.
Keywords: ATP Synthase, ATPases, Bioenergetics, Enzyme Mechanisms, F1F0 ATPase, Motor Enzyme, Torque
Abstract
ATP synthase (F0F1) is made of two motors, a proton-driven motor (F0) and an ATP-driven motor (F1), connected by a common rotary shaft, and catalyzes proton flow-driven ATP synthesis and ATP-driven proton pumping. In F1, the central γ subunit rotates inside the α3β3 ring. Here we report structural features of F1 responsible for torque generation and the catalytic ability of the low-torque F0F1. (i) Deletion of one or two turns in the α-helix in the C-terminal domain of catalytic β subunit at the rotor/stator contact region generates mutant F1s, termed F1(1/2)s, that rotate with about half of the normal torque. This helix would support the helix-loop-helix structure acting as a solid “pushrod” to push the rotor γ subunit, but the short helix in F1(1/2)s would fail to accomplish this task. (ii) Three different half-torque F0F1(1/2)s were purified and reconstituted into proteoliposomes. They carry out ATP-driven proton pumping and build up the same small transmembrane ΔpH, indicating that the final ΔpH is directly related to the amount of torque. (iii) The half-torque F0F1(1/2)s can catalyze ATP synthesis, although slowly. The rate of synthesis varies widely among the three F0F1(1/2)s, which suggests that the rate reflects subtle conformational variations of individual mutants.
Introduction
F0F1-ATP synthase (F0F1) catalyzes synthesis of ATP in mitochondria, chloroplasts, and bacteria in oxidative and photo-phosphorylation, using the energy of proton translocation caused by the proton motive force (pmf)2 across membranes. Proton translocation through membrane-embedded F0 portion drives ATP synthesis in membrane-protruding F1 portion and, in the reverse reaction, ATP hydrolysis in F1 can drive pumping back protons through F0. These two reactions, the inward/outward proton translocation in F0 and synthesis/hydrolysis in F1, are coupled by mechanical rotation (1–10). Both F0 and F1 are rotary motors; F0 motor is driven by downhill proton translocation, F1 motor is driven by ATP hydrolysis, and the two motors share a common rotary shaft. When pmf is high, F0 drives rotation of the shaft that results in ATP synthesis in F1. When the chemical potential of ATP hydrolysis exceeds pmf, F1 rotates the shaft and F0 is forced to pump protons. Indeed, isolated F1 rotates when it hydrolyzes ATP (11), and the reverse rotation by an external force results in ATP synthesis (12, 13).
ATP-driven rotation of the isolated F1 has been extensively studied by using a minimum motor complex consisting of α3β3γ subunits from thermophilic Bacillus PS3 (14–16), which we also refer to as F1 in this study unless otherwise indicated. The γ subunit is a rotor subunit. Its N- and C-terminal helices form a coiled-coil and are deeply inserted into the cavity of the stator α3β3 ring. The globular domain of γ subunit resides outside of the cavity of the α3β3 ring (see Fig. 1, left panel). To visualize rotation, the α3β3 ring is immobilized on the glass surface, and a bead (or bead duplex) is attached to the globular domain of the γ subunit. A single cycle rotates the γ subunit 120°, consuming one ATP, meaning that F1 carries out three reaction cycles per revolution. The 120° cycle begins with a pause at 0°, then an ∼80°-step rotation is followed by another pause, and lastly, an ∼40°-step rotation occurs. F1 waits for a substrate ATP during the pause at 0° (“ATP-waiting dwell”) and rotates ∼80° upon ATP binding. A pause at ∼80° (“catalytic dwell”) is ∼2 ms, independent of ATP concentration, representing the period required for hydrolytic cleavage of ATP and release of Pi from the catalytic site.
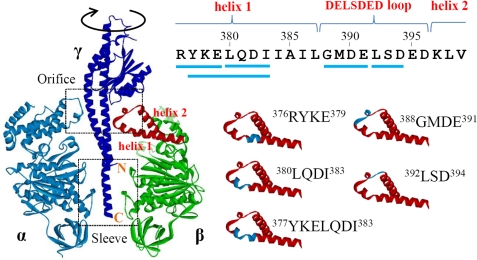
FIGURE 1.
Structure around the DELSDED loop. Left panel, structure of α, β, and γ subunits in bovine F1 (Protein Data Bank (PDB) ID: 2v7q). Orifice and sleeve regions and N and C termini of γ subunit are indicated. The region corresponding to the helix-1, the DELSDED loop, and the helix-2 of the β subunit are shown in red. Right panel, helix-1-DELSDED loop-helix-2 region of the β subunit of Bacillus PS3 F1. Sequences truncated in this work are shown by blue lines (upper panel) and blue stretches (lower panel).
The torque of ATP-driven F1 motor estimated from rotation speed, bead radius, rotation radius, and viscosity of aqueous medium amounts to ∼40 pN·nm (17). The energy required for a 120° rotation (∼40 pN·nm × 2π/3 radians) is roughly equal to the free energy liberated from hydrolysis of an ATP molecule (∼90 pN·nm) under physiological conditions and, therefore, energy conversion by F1 motor is very efficient. The torque of F1 is generated from sequential interactions between a rotor γ subunit and three catalytic β subunits in the stator α3β3 ring (18). According to the crystal structures (19, 20), β subunits can interact directly with the γ subunit at the two positions, the “sleeve” and the “orifice” regions (see Fig. 1, left panel). A mutant F1, whose γ subunit is truncated at the C terminus and loses interactions with the α3β3 ring at the sleeve region, rotates with half of the normal torque (21). Inversely, F1 devoid of the entire N-terminal helix, which preserves the sleeve interactions but loses many interactions at the orifice, also rotates with half of the normal torque (22). Therefore, the orifice interactions alone can produce half of the total torque, and the sleeve interactions, probably with the assistance of some of the orifice interactions, produce another half.
At the orifice region, the helix-1-loop-helix-2 structure in the C-terminal domain of the β subunit makes contact with the γ subunit (19). The loop in the structure contains a well conserved DELSDED acidic cluster sequence, but its acidic nature is not essential for torque generation (23). Recently, it was shown that deletion of three or four residues in the loop does not impair ATP synthesis activity of F0F1 (24). If neither acidic residues in the loop nor the full-length loop itself are essential for rotation, what structural features of the orifice region are responsible for torque generation? Here, we report isolation of mutant F1s that exert half of the normal torque, indicating that the full-length helix-1 acts as a “pushrod” for the γ subunit to generate half of the total torque. Then, we addressed the next question. If the F1 portion of F0F1 wastes a large fraction of the energy transferred from the F0 motor, can it synthesize ATP? The results show that it can, although slowly.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Proteins
Escherichia coli strain JM109 was used for the genetic manipulation, and JM103 Δ(uncB-uncD), which does not express E. coli F0F1, was used for the expression of F1 and F0F1 from thermophilic Bacillus PS3. Mutations of Bacillus PS3 F1 were introduced into the plasmid pKAGB1/HC95, which expresses the α(C193S)3β(His10 tag at N terminus)3γ(S107C/I210C) complex of F1. Wild-type and mutant F1 were expressed and purified as described (25) except for two procedures. Heat treatment to precipitate heat-labile E. coli proteins was carried out at 64 °C for 20 min for wild-type F1 but at 60 °C for 10 min for the mutants. The procedure to remove endogenously bound nucleotides was omitted for the mutants because mutants without bound nucleotides tend to dissociate. The purified F1 was stored in 70% ammonium sulfate suspension containing 2 mm DTT at 4 °C. Mutations of F0F1 were introduced into the plasmid pTR19-ASDS-ϵΔc in which the F1-coding region is the same as pKAGB1/HC95 and the C-terminal domain of the ϵ subunit is deleted (26). F0F1 was expressed and purified as described (27, 28). The purified F0F1 preparation was frozen by liquid N2 and stored at −80 °C until use. The yields of purified mutant F1s and F0F1s were 20–50% of that of the wild type. The purity of the isolated preparations, exemplified by the three mutants that are the main focus of this study, is shown (supplemental Fig. S1). The concentrations of F1 and F0F1 were determined from the absorbance with the molar extinction coefficient at 280 nm of 154,000 and 253,000 m−1cm−1 and with the molecular mass as 356 and 530 kDa, respectively (25, 29).
Observation and Analysis of Rotation
Rotation was observed on the stage of an inverted microscope (IX70, Olympus) at 25 °C as described (21). A mercury lamp was used to observe rotation of 209-, 291-, and 350-nm polystyrene beads, and a 532-nm laser was used to observe rotation of 40-nm gold beads. Images of beads were captured with a CCD camera (ICL-B0620M-KC, IMPERX and GX-8, nac Image Technology Inc.) with 250 frames/s (fps) for polystyrene beads and with 4,000 fps for gold beads. The glass chamber was made of the nickel-nitrilotriacetic acid-coated bottom coverslip (21) and an uncoated top coverslip. The bottom coverslip and the top coverslip were separated by two strips of double-faced tape. F1 was biotinylated by incubation with biotin-PEAC5-maleimide for 90 min. The biotinylated F1 (10 nm) in 50 mm MOPS/KOH, pH 7.0, containing 50 mm KCl and 10 mg/ml bovine serum albumin was applied into the chamber, and overflowing solution was blotted with paper. After 2 min, the chamber was washed by 50 mm MOPS/KOH containing 50 mm KCl, and beads were suspended in 50 mm MOPS/KOH, 50 mm KCl, and 10 mg/ml bovine serum albumin were applied into the chamber. After waiting for 15 min and washing, the rotation buffer (50 mm MOPS/KOH, pH 7.0, 50 mm KCl, 2 mm MgCl2, 1 mm phosphoenolpyruvate (Sigma), 200 μg/ml pyruvate kinase (Roche Applied Science), and indicated concentration of ATP) was applied into the chamber. As commonly experienced for single molecule observations of this kind, only a few percentages, at most, of beads showed clear rotation due to surface denaturation, improper bead attachment, and other unknown reasons. Numbers of rotating beads of mutant F1 molecules were slightly fewer than that of wild-type F1. Rotating beads rotated at various speeds. In general, fast rotating beads always rotated quickly, whereas sluggish ones almost never showed fast rotation. The rotations at the fastest speed were analyzed because slower rotation can be explained by surface obstructions.
Torque of rotation was defined as
where ω and ξ are the rotation speed (radians/second) and viscous load, respectively. The value of ξ of duplex beads is given as
where η is the viscosity of a medium (η = 0.93 × 10−9 pN/nm2) and R and xi (i = 1, 2) are the radius and rotation radius of each bead, respectively (30). The beads rotating with about the same rotation radius were selected and analyzed. Torque was also estimated from fluctuation analysis (31) and defined as
where N is torque, kB is the Boltzmann constant, T is temperature (T = 298 K), Δθ = θ(t + Δt) − θ(t), Δt is 4 ms, and P(Δθ) is the probability distribution of Δθ. One can estimate N from this equation without the value of ξ.
ATP-driven Proton Pumping and ATP Synthesis
Purified F0F1 was reconstituted into proteoliposomes by using BioBeads as described (28). Proteoliposome suspension (20 μl) was mixed with 1.2 ml of 10 mm HEPES/KOH, pH 7.5, 5 mm MgCl2, 100 mm KCl, and 0.3 μg/ml 9-amino-6-chloro-2-methoxyacridine (ACMA). The reaction was started by the addition of 12 μl of 100 mm ATP, and formation of a transmembrane pH gradient (ΔpH) was monitored with quenching of ACMA fluorescence (excitation 410 nm, emission 480 nm) at 25 °C. Final concentrations of F0F1 were 11.4 nm. At the end of measurement, nigericin (2 μm) was added to eliminate ΔpH. ATP synthesis was measured as described (27, 28) except for a long (6-h) incubation of proteoliposome with the acidic mixture (35 mm MES, 10 mm NaH2PO4, 2.5 mm MgCl2, 0.68 mm KCl, 340 mm sucrose, 0.5 mm ADP, 0.2 μm valinomycin, pH 6.05). A basic mixture was prepared by mixing 21 μl of the luciferin/luciferase mixture (2× concentration, CLS II ATP bioluminescence assay kit, Roche Applied Science), 870 μl of the basic buffer (300 mm HEPES, 10 mm NaH2PO4, 2.5 mm MgCl2, 240 mm KOH, pH adjusted with NaOH to 8.05), and 9 μl of 50 mm ADP and was incubated for 5 min at 30 °C. The acidic mixture containing proteoliposome (100 μl) was added into 900 μl of the basic mixture, and emission of luciferin was monitored. The calculated size of the imposed pmf was 260 mV (ΔpH = 2, Δψ = 140 mV). The final concentrations of F0F1, ADP, and Pi were 11.4 nm, 0.5 mm, and 10 mm, respectively. The final pH of the solution outside the proteoliposomes was 8.05. At the end of measurements, 10 μl of 10 μm ATP was added three times for calibration (28).
RESULTS
Torque Mutants of F1
F1 with decreased torque is expected to rotate beads slowly when compared with the wild-type F1. For the search for structural features responsible for the torque generation, various mutants of Bacillus PS3 F1 were expressed in E. coli. They are, in most cases, expressed in smaller amounts and are less heat-stable than the wild-type but purified without difficulty. The purified F1 was immobilized onto the nickel-nitrilotriacetic acid-coated glass surface through His-tagged β subunits and, 291-nm polystyrene beads were attached on the γ subunit as a rotation probe. We observed the rotation of duplex beads in 2 mm ATP under the microscopic field with a camera (250 fps). Under these experimental conditions, ATP-waiting dwell and catalytic dwell are too short to be observed, and continuous rotations with occasional pauses by ADP-Mg inhibition or for other reasons were observed. Among beads rotating at various speeds, the rotations at the fastest speed were taken to be the unobstructed rotation of native molecules.
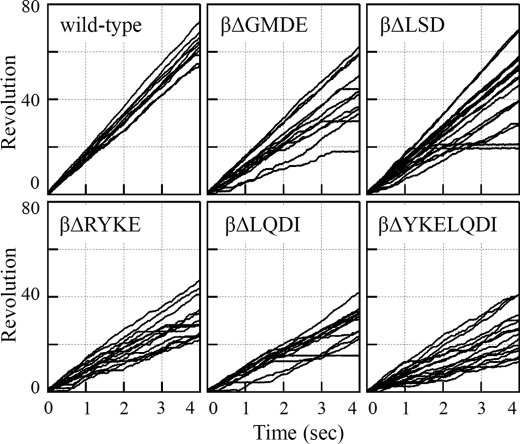
We at first focused on amino acid residues that are located at the rotor/stator interface narrower than 3.5 Å at the orifice region. Except for the residues in the 390DELSDED396 sequence of the β subunit that were already demonstrated not to be critical for torque generation, we replaced these residues with alanine and made F1 variants containing mutations αG398A/S399A/D400A, αD400A/L401A/D402A, γD83A/R84A/G85A, γG85A/L86A, γG88A, γY90A/N91A, or γN91A/S92A/N93A. We found that these mutants rotated beads at speeds similar to the wild-type F1, ∼20 revolutions/s (data not shown), indicating that specific interactions between side chains of these residues do not significantly contribute to torque generation. Next, we made two mutant F1s, βΔ388GMDE391 and βΔ392LSD394, that have a deletion in the 388GMDELSDED396 sequence of the β subunit (Fig. 1, right panel), but again, they showed rotation at a normal speed (Fig. 2, upper panels). Then, we deleted one or two turns of the helix-1 that precedes the 390DELSDED396 sequence, and βΔ380LQDI383, βΔ376RYKE379, and βΔ377YKELQDI383 were made (Fig. 1, right panel) (superscripts are abbreviated hereafter). These three mutant F1s rotated more slowly than the wild-type F1 (Fig. 2, lower panels), and we focused on these mutants.
FIGURE 2.
Rotation trajectories of rotating duplex beads in 2 mm ATP. Bead size is 291 nm. Each line represents rotation of each F1 molecule. Experimental details are described under “Experimental Procedures.”
Half-torque F1
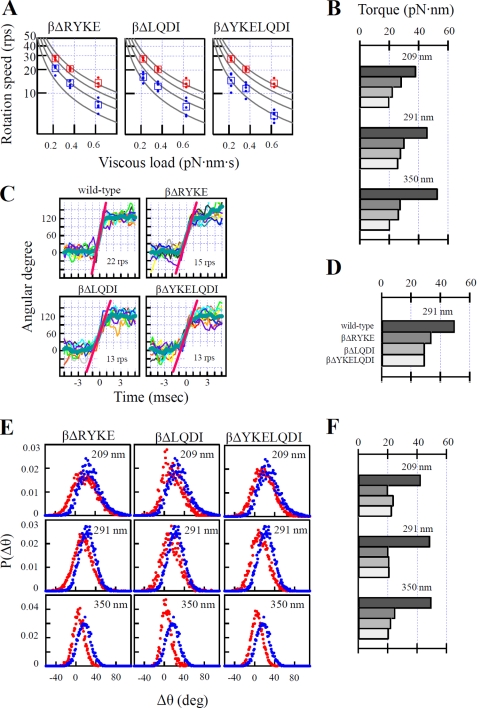
Torque of the three mutants relative to that of wild-type F1 was estimated from three different analyses. First, continuous rotation of 209-, 291-, and 350-nm duplex beads in 2 mm ATP was analyzed. The rotation speed of each mutant F1 was obtained from the consecutive, smooth 10 revolutions taken from the five fastest-rotating molecules. As the size of bead increases, viscous load increases and rotation slows down (Fig. 3A). With beads of the same size, viscous load is constant, and torque is obtained from the equation (torque = viscous load × rotation speed) (Fig. 3B). The averaged torque with three different sizes of beads is 46 pN·nm (wild-type), 29 pN·nm (βΔRYKE), 26 pN·nm (βΔLQDI), and 22 pN·nm (βΔYKELQDI). Second, rotation of 291-nm duplex beads in 200 nm ATP was observed. At this low ATP concentration, F1 repeats a pause of ATP-waiting dwell and a 120°-step rotation. The consecutive 10 120°-step rotations were overlaid and averaged (Fig. 3C). Torque was estimated from the rotation speed of the middle part of the averaged 120°-step rotation trajectory. As seen (Fig. 3D), torques of mutants obtained by this analysis are slightly larger than those obtained from continuous rotation. Third, bead fluctuation during rotational motion was analyzed by the recently developed fluctuation theorem (Fig. 3E). By this analysis, the magnitude of torque is directly obtained from fluctuation of rotating beads without knowledge of the values of viscous load and rotation speed. This is advantageous because the estimation of viscous load needs information of exact size of individual beads, rotational radius, and viscosity of the medium near the surface of glass and may contain errors (32). The averaged torque with three different sizes of beads is 47 pN·nm (wild type), 22 pN·nm (βΔRYKE), 23 pN·nm (βΔLQDI), and 22 pN·nm(βΔYKELQDI). Altogether, three independent methods give similar torque values for all three mutant F1s, that is, about half of the normal torques of wild-type F1. Thus, shortening of the helix-1 results in the loss of half of the torque. Out of the energy supplied by hydrolysis of one ATP molecule for 120° rotation, these mutant F1s can utilize only ∼45 pN·nm (=torque × 2π/3), whereas wild-type F1 can utilize ∼90 pN·nm.
FIGURE 3.
Torque measurement by three methods. A, continuous rotation of three different beads in 2 mm ATP. For each size of bead, 10 consecutive, smooth revolutions were selected from the five fastest rotation trajectories out of several tens of observed rotations, and the time-averaged slope of the trajectory was obtained as rotation speed (shown by circles). The average of five circles is shown by the square symbols. Wild-type F1 is shown in red, and mutants are shown in blue. Viscous load was calculated from bead size, rotation radius, and viscous coefficient. The lines of constant torque (50, 40, 30, 20 pN·nm from the top) are shown. rps indicates revolutions per second. B, torques calculated from A. Labels of bars are wild-type, βΔRYKE, βΔLQDI, and βΔYKELQDI from the top as in D. C, 120°-step rotation of 291-nm beads in 200 nm ATP. Ten consecutive 120°-step rotations were overlaid and averaged (blue, thick line), and then the slope between 30 and 90° (red line) was obtained as the rotation speed. D, torques obtained from C. E, fluctuation analysis of rotation of three differently sized beads in 2 mm ATP. Δθ is the difference between rotary angles after 4 ms. P(Δθ) is a distribution function of Δθ. Red, mutant F1; blue, wild-type F1. F, torques obtained from E. Labels of bars are the same as D. Experimental details are described under “Experimental Procedures.”
Kinetic Constants of Half-torque F1s
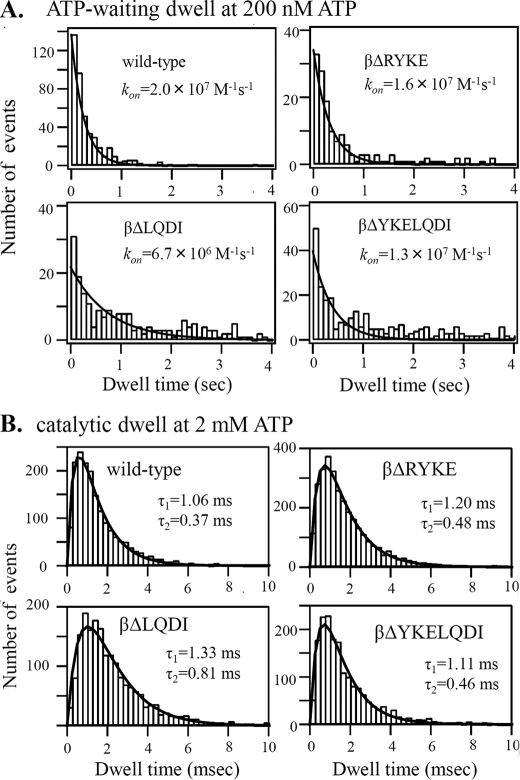
Kinetic characteristics of half-torque F1, termed F1(1/2) hereafter, were examined by analysis of rotation. First, durations of ATP-waiting dwell in rotation in 200 nm ATP were collected and plotted as histograms (Fig. 4A). The histograms are fitted by an exponential function that contains the rate of ATP binding (kon) to the catalytic site. The kon values that give the best fit are 2.0 × 107 m−1s−1 (wild-type), 1.6 × 107 m−1s−1 (βΔRYKE), 0.7 × 107 m−1s−1 (βΔLQDI), and 1.3 × 107 m−1s−1 (βΔYKELQDI). Taking into account that the values of βΔLQDI and βΔYKELQDI might be underestimated because fitting curves are dragged to the right by the presence of dwells longer than 2 s of unknown origin, it appears that kon values of F1(1/2)s are not significantly changed from that of wild-type F1.
FIGURE 4.
Kinetic constants of F1(1/2)s. A, histogram of ATP-waiting dwell of rotation of 291-nm duplex beads in 200 nm ATP recorded by 250 fps. The ATP binding rate constant (kon) is obtained from a fitting curve (solid line), constant × exp(−kon × [ATP] × t). B, histogram of the catalytic dwell of rotation of a 40-nm gold bead in 2 mm ATP recorded by 4,000 fps. The data are fitted by a double exponential function (solid line), constant × (exp(−(1/τ1) × t) − exp(−(1/τ2) × t). τ1 and τ2 represent the time constants of two successive reactions, ATP hydrolysis and Pi release, that occur in the catalytic dwell (16). Experimental details are described under “Experimental Procedures.”
Next, rotation of a 40-nm single gold bead attached to the γ subunit was observed in 2 mm ATP with a high speed camera (4,000 fps). Under these conditions, viscous load of bead rotation is negligible, ATP-waiting dwell is not observed, and only catalytic dwell is observed at each 120° interval. The histogram of durations of the catalytic dwells is fitted by a double exponential function defined by two time constants (Fig. 4B). Previous studies assigned two time constants to the two sequential catalytic events on the enzyme, ATP hydrolysis and Pi release (16). Two of the F1(1/2)s have time constants (βΔRYKE, 0.5 and 1.2 ms; βΔYKELQD, 0.5 and 1.1 ms) that are very similar to those of the wild-type F1 (0.4 and 1.4 ms). The βΔLQDI mutant has slightly longer time constants (0.8 and 1.3 ms). Enzyme turnover rates calculated from the above values are 560 s−1 (wild-type), 590 s−1 (βΔRYKE), 480 s−1 (βΔLQDI), and 630 s−1 (βΔYKELQDI). ATP hydrolysis activities measured in the bulk solution containing 2 mm ATP are 330 s−1 (wild-type), 270 s−1 (βΔRYKE), 110 s−1 (βΔLQDI), and 330 s−1 (βΔYKELQDI) in the presence of lauryldimethylamine-N-oxide, which is thought to relieve ADP-Mg inhibition. Taking into account that purified F1s may contain some fraction of inactive molecules, these values are compatible with values obtained from rotation. To summarize the above results, enzyme kinetics of F1(1/2)s are not changed much from those of wild-type F1.
Proton Pumping and ATP Synthesis
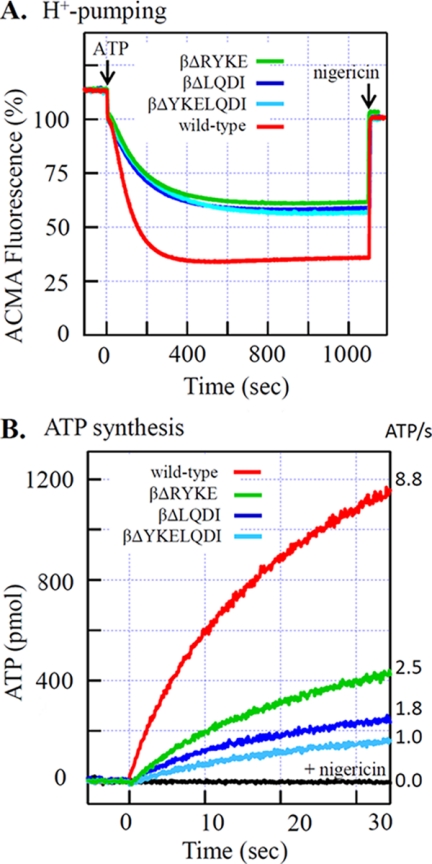
Bacillus PS3 F0F1 containing F1(1/2) was expressed in E. coli and purified. The purified mutant F0F1s are as stable as the wild type. To avoid complexity caused by regulatory function of the ϵ subunit, the C-terminal domain of the ϵ subunit (ϵΔC) was deleted and will be termed wild-type F0F1 hereafter (26, 33, 34). The purified F0F1 was reconstituted into proteoliposomes, and ATP-driven proton pumping was monitored with the quenching of ACMA fluorescence that reflects pH gradient across membranes (ΔpH). Upon the addition of ATP, quenching started with a short lag, progressed at a maximum rate, and leveled off at the stationary phase (Fig. 5A). The established ΔpH was completely abolished by the addition of nigericin, an H+-K+ antiporter that can dissipate ΔpH. In general, quenching time courses of the three F0F1(1/2)s are very similar to each other. The fastest quenching progress rates of F0F1(1/2)s are about 40% of that of wild-type F0F1, and the maximum quenching level at the stationary phase with F0F1(1/2)s was ∼60% of that of wild-type F0F1. Thus, F0F1(1/2)s can pump protons, but the final ΔpH built up is much smaller than that of wild-type F0F1.
FIGURE 5.
Activities of F0F1(1/2)s. A, ATP-driven proton pump. Acidification of the inside lumen of F0F1(1/2)-containing proteoliposomes by proton pumping was monitored with fluorescence quenching of ACMA. The reaction was started by the addition of 1 mm ATP. B, ATP synthesis activity. The F0F1(1/2)-containing proteoliposomes were pre-equilibrated with the low K+, acidic mixture and then injected into a 10-fold volume of the high K+ basic mixture. The acidic mixture contained ADP, Pi, and valinomycin. The basic mixture contained ADP, Pi, and luciferin/luciferase. Final concentrations of F0F1, ADP, and Pi were 11.4 nm, 0.5 mm, and 10 mm, respectively. ATP produced was monitored with emission of luciferin, and the initial slope of exponential fitting curve between 0 and 6 s was taken to be ATP synthesis activity as shown in the right side of the figure. Experimental details are described under “Experimental Procedures.”
Using the same proteoliposomes, we observed ATP synthesis driven by acid-base transition and valinomycin-induced K+ diffusion potential (Fig. 5B). The reaction was started by the addition of acidified proteoliposome mixture into the basic mixture, and generation of ATP was monitored by luciferin/luciferase. In the presence of nigericin, which acts as an uncoupler in combination with valinomycin, ATP synthesis did not occur. In contrast to proton pumping, activities of ATP synthesis of the F0F1(1/2)s are different from each other, and initial rates of synthesis are 28% (βΔRYKE), 20% (βΔLQDI), and 11% (βΔYKELQDI) of that of wild-type F0F1.
DISCUSSION
Short-sized Helix-1 Impairs Energy Conversion but Not Chemical Catalysis
Properties of the half-torque mutants are summarized in Table 1. As seen, when the mutant enzymes hydrolyze ATP without the burden of the viscous load, their enzymatic kinetics are not very different from those of the wild-type enzyme. However, once they have to do work such as rotating large beads and pumping protons, their defects become obvious. Thus, shortening of helix-1 does not impair chemical catalysis but does impair mechanochemical energy conversion.
TABLE 1.
Properties of mutant F1s and F0F1s with short-sized pushrod helix
Torque Generation in F1(1/2)
The half-torque mutants we reported here retain the sleeve interactions. In addition, they do not lose all orifice interactions but retain the interactions at the conical entrance of the orifice of α3β3 ring that touches the globular portion of the γ subunit (Fig. 1). F1 with the truncated γ subunit, which apparently has only the interactions with the conical entrance region of the α3β3 ring, can generate a small torque that barely supports unidirectional slow rotation of a load-negligible gold bead (35). It appears, therefore, that the sleeve interactions coordinated with the interactions at the conical entrance would be responsible for the generation of torque in F1(1/2)s.
Helix-1 with Sufficient Length Is Necessary for Torque Generation, Acting as Pushrod
The orifice interactions by themselves produce at least half of the torque of the ATP-driven F1 motor (21, 35). Given that the interactions at the conical entrance region contribute only a small fraction of the torque, the majority of the torque produced at the orifice is attributed to the interactions of the helix-1-loop-helix-2 with the convex coiled-coil region of the γ subunit. Concerning the central loop, the conserved DELSDED sequence is not necessary for the torque generation (23). Even the mutants with the shortened loop can synthesize ATP (24) and produce normal torque (Fig. 2). Instead, this work reveals a critical role of full-length helix-1 in the generation of torque. When helix-1 is shortened by one helical turn (5.4 Å) by deletion of either the RYKE or the LQDI segments, torque is reduced by half. When the segment RYKELQDI, which forms two turns (10.8 Å) in helix-1, is truncated, torque is again reduced by no more than half. It is clear from these results that the physical length, rather than residue-specific interactions, of helix-1 is important for torque generation. In addition, the fact that deletion of one turn and of two turns gives the same result implies the presence of a critical length for helix-1 function. Crystal structure (36), single molecule study (37), and rapid scan atomic force microscopy (38) show that C-terminal domains of three β subunits in F1 undergo bending motion sequentially and alternately during the catalytic cycle. The helix-1-loop-helix-2 is a protruding body of the C-terminal domain toward the γ subunit (Fig. 1), and it can protrude more toward the γ subunit by a bending motion (36). It appears that a protruding solid body of the helix-1-loop-helix-2 physically pushes the convex part of the coiled-coil of the γ subunit and assists the γ subunit to rotate (39). To transmit the force, helix-1, and probably helix-2 as well, must play the role of pushrod with sufficient length and stiffness.
Critical Length of Helix-1
Very recently, more research on the helix-1-loop-helix-2 structure was published by Mnatsakanyan et al. (40). They found that a mutant of Bacillus PS3 F0F1, Δ7 (βΔ(LQDI+LSD)), can catalyze ATP synthesis, although slowly, but Δ10 (βΔ(QDIIAIL+LSD)) cannot. From the difference between Δ7 and Δ10, Mnatsakanyan et al. (40) concluded that the critical length of the helix-1-loop-helix-2 structure is around ∼10 Å shorter than the native length. Our results clearly show that shortening one turn (∼5 Å) of helix-1 (βΔLQDI and βΔRYKE) is enough to abolish half of the torque. Therefore, we agree on the presence of a critical length but propose that the critical length of the helix as a solid body is ∼5 Å shorter than the native length. Most likely, Δ10 is also a half-torque mutant and has the catalytic ability of ATP synthesis, although it might have been very weak and escaped the assay employed in Ref. 40. Actually, Δ10 retains proton-pumping activity, although weak (40), which should not happen without torque.
Low-torque F0F1 Builds Up a Small ΔpH
Given that F1(1/2) can use only a fraction of the energy of ATP hydrolysis for driving the motor, it is reasonable to assume that F0F1(1/2) can also use only a fraction of the energy for proton pumping. Indeed, the final ΔpH built up by the proton-pumping action of F0F1(1/2) is significantly smaller than ΔpH built up by wild-type F0F1. Final ΔpH could be determined by equilibrium between proton-pumping activity and back pressure of pmf and/or equilibrium between proton-pumping activity and passive leakage of protons through membranes. In any case, however, the fact that three kinds of F0F1(1/2)s build up the same final ΔpH indicates that the final ΔpH is directly determined by the magnitude of the torque, that is, by the energy conversion yield. This agrees with the thermodynamic prediction that ΔpH is directly related to the torque (n × ZΔpH = torque × 2π/3; where n is the number of transported protons per ATP, and Z is a constant).
Low-torque F0F1 Can Synthesize ATP Slowly under Thermodynamically Favorable Conditions
If the same inefficient energy conversion occurs in the ATP synthesis reaction of F0F1(1/2), the F1(1/2) portion of F0F1(1/2) can use only a fraction (∼45 pN·nm per 120° rotation) of the total energy provided by the F0 portion. Despite this defect, F0F1(1/2) can synthesize ATP (Fig. 5B). This might be because of the small free energy required for ATP synthesis under actual experimental conditions. The reaction mixtures contained 0.5 mm ADP and 10 mm Pi. The ADP we used contained about 0.05% ATP as measured by luciferase assay, and the free energy required for ATP synthesis would amount to ∼40 pN·nm, which the F1(1/2) portion of F0F1(1/2) can manage to exert. Therefore, low-torque F0F1 has catalytic ability and can synthesize ATP under optimum thermodynamic situations.
Because the free energy defines the direction and equilibrium but not the rate of the reaction, it is not surprising that each of the three F0F1(1/2)s catalyzes ATP synthesis at a different rate. Subtle variations in the enzyme can affect the rate, and the reason for the slow ATP synthesis rate of F0F1(1/2)s cannot be explained with certainty. During the ATP synthesis reaction, the F0 portion provides energy to the F1 portion via rotation of the γ subunit. The F1 portion uses this energy to induce the otherwise thermodynamically unfavorable conformational changes to β subunits that lead to ATP synthesis. We speculate that with insufficient ability to utilize energy, F0F1(1/2) would fail frequently and take a long time to induce such changes, or even make uncoupled rotations frequently without inducing such changes. We observed that the F0F1(1/2)s have larger uncoupled ATP hydrolysis activity than wild-type F0F1, which is probed by resistance to inactivation by dicyclohexylcarbodiimide, a reagent that prevents proton translocation and hence rotation of F0. This observation favors the scenario of the uncoupled rotation but does not exclude other interpretations.
Finally, it should be noted that although F0F1(1/2) can synthesize ATP, the synthesis reaction reaches equilibrium when ATP concentration is 4 or 5 orders of magnitude lower than in the case of wild-type F0F1. This means that cellular ATP concentration supported by F0F1(1/2) would be very low and could hardly sustain normal cell activities. Thus, natural selection would not allow field organisms to have a low-torque version of F0F1, even if it had the catalytic ability of ATP synthesis.
Supplementary Material
Acknowledgment
We thank Daniel Xu for critically reading and polishing the manuscript.
This work was supported by the ATP synthesis Regulation Project (ICORP) through Japan Science and Technology Agency (to M. Y.).

This article contains supplemental Fig. S1.
- pmf
- proton motive force
- fps
- frames per second
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- F1(1/2)
- mutant F1 that rotates with half-torque of the wild-type F1
- pN·nm
- piconewtons nanometers.
REFERENCES
- 1. Yoshida M., Muneyuki E., Hisabori T. (2001) ATP synthase–a marvelous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 2. Boyer P. D. (2002) A research journey with ATP synthase. J. Biol. Chem. 277, 39045–39061 [DOI] [PubMed] [Google Scholar]
- 3. Senior A. E., Nadanaciva S., Weber J. (2002) The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta 1553, 188–211 [DOI] [PubMed] [Google Scholar]
- 4. Kinosita K., Jr., Adachi K., Itoh H. (2004) Rotation of F1-ATPase: how an ATP-driven molecular machine may work. Annu. Rev. Biophys. Biomol. Struct. 33, 245–268 [DOI] [PubMed] [Google Scholar]
- 5. Weber J. (2006) ATP synthase: subunit-subunit interactions in the stator stalk. Biochim. Biophys. Acta 1757, 1162–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakamoto R. K., Baylis Scanlon J. A., Al-Shawi M. K. (2008) The rotary mechanism of the ATP synthase. Arch. Biochem. Biophys. 476, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Junge W., Sielaff H., Engelbrecht S. (2009) Torque generation and elastic power transmission in the rotary F0F1-ATPase. Nature 459, 364–370 [DOI] [PubMed] [Google Scholar]
- 8. Düser M. G., Zarrabi N., Cipriano D. J., Ernst S., Glick G. D., Dunn S. D., Börsch M. (2009) 36° step size of proton-driven c-ring rotation in F0F1-ATP synthase. EMBO J. 28, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. von Ballmoos C., Wiedenmann A., Dimroth P. (2009) Essentials for ATP synthesis by F1F0 ATP synthases. Annu. Rev. Biochem. 78, 649–672 [DOI] [PubMed] [Google Scholar]
- 10. Nakanishi-Matsui M., Sekiya M., Nakamoto R. K., Futai M. (2010) The mechanism of rotating proton pumping ATPases. Biochim. Biophys. Acta 1797, 1343–1352 [DOI] [PubMed] [Google Scholar]
- 11. Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 12. Itoh H., Takahashi A., Adachi K., Noji H., Yasuda R., Yoshida M., Kinosita K. (2004) Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 [DOI] [PubMed] [Google Scholar]
- 13. Rondelez Y., Tresset G., Nakashima T., Kato-Yamada Y., Fujita H., Takeuchi S., Noji H. (2005) Highly coupled ATP synthesis by F1-ATPase single molecules. Nature 433, 773–777 [DOI] [PubMed] [Google Scholar]
- 14. Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 15. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40 degree substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 17. Yasuda R., Noji H., Kinosita K., Jr., Yoshida M. (1998) F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 18. Ariga T., Muneyuki E., Yoshida M. (2007) F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat. Struct. Mol. Biol. 14, 841–846 [DOI] [PubMed] [Google Scholar]
- 19. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 20. Bowler M. W., Montgomery M. G., Leslie A. G., Walker J. E. (2007) Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 282, 14238–14242 [DOI] [PubMed] [Google Scholar]
- 21. Hossain M. D., Furuike S., Maki Y., Adachi K., Suzuki T., Kohori A., Itoh H., Yoshida M., Kinosita K., Jr. (2008) Neither helix in the coiled coil region of the axle of F1-ATPase plays a significant role in torque production. Biophys. J. 95, 4837–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kohori A., Chiwata R., Hossain M. D., Furuike S., Shiroguchi K., Adachi K., Yoshida M., Kinosita K., Jr. (2011) Torque generation in F1-ATPase devoid of the entire amino-terminal helix of the rotor that fills half of the stator orifice. Biophys. J. 101, 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hara K. Y., Noji H., Bald D., Yasuda R., Kinosita K., Jr., Yoshida M. (2000) The role of the DELSEED motif of the beta subunit in rotation of F1-ATPase. J. Biol. Chem. 275, 14260–14263 [DOI] [PubMed] [Google Scholar]
- 24. Mnatsakanyan N., Krishnakumar A. M., Suzuki T., Weber J. (2009) The role of the βDELSEED-loop of ATP synthase. J. Biol. Chem. 284, 11336–11345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuki T., Ueno H., Mitome N., Suzuki J., Yoshida M. (2002) F0 of ATP synthase is a rotary proton channel. Obligatory coupling of proton translocation with rotation of c-subunit ring. J. Biol. Chem. 277, 13281–13285 [DOI] [PubMed] [Google Scholar]
- 26. Konno H., Suzuki T., Bald D., Yoshida M., Hisabori T. (2004) Significance of the ϵ subunit in the thiol modulation of chloroplast ATP synthase. Biochem. Biophys. Res. Commun. 318, 17–24 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki T., Murakami T., Iino R., Suzuki J., Ono S., Shirakihara Y., Yoshida M. (2003) F0F1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of epsilon subunit in response to proton motive force and ADP/ATP balance. J. Biol. Chem. 278, 46840–46846 [DOI] [PubMed] [Google Scholar]
- 28. Soga N., Kinosita K., Jr., Yoshida M., Suzuki T. (2011) Efficient ATP synthesis by thermophilic Bacillus F0F1-ATP synthase. FEBS J. 278, 2647–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furuike S., Adachi K., Sakaki N., Shimo-Kon R., Itoh H., Muneyuki E., Yoshida M., Kinosita K., Jr. (2008) Temperature dependence of the rotation and hydrolysis activities of F1-ATPase. Biophys. J. 95, 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakaki N., Shimo-Kon R., Adachi K., Itoh H., Furuike S., Muneyuki E., Yoshida M., Kinosita K., Jr. (2005) One rotary mechanism for F1-ATPase over ATP concentrations from millimolar down to nanomolar. Biophys. J. 88, 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayashi K., Ueno H., Iino R., Noji H. (2010) Fluctuation theorem applied to F1-ATPase. Phys. Rev. Lett. 104, 218103. [DOI] [PubMed] [Google Scholar]
- 32. Pänke O., Cherepanov D. A., Gumbiowski K., Engelbrecht S., Junge W. (2001) Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 81, 1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feniouk B. A., Suzuki T., Yoshida M. (2007) Regulatory interplay between proton motive force, ADP, phosphate, and subunit ϵ in bacterial ATP synthase. J. Biol. Chem. 282, 764–772 [DOI] [PubMed] [Google Scholar]
- 34. Masaike T., Suzuki T., Tsunoda S. P., Konno H., Yoshida M. (2006) Probing conformations of the beta subunit of F0F1-ATP synthase in catalysis. Biochem. Biophys. Res. Commun. 342, 800–807 [DOI] [PubMed] [Google Scholar]
- 35. Furuike S., Hossain M. D., Maki Y., Adachi K., Suzuki T., Kohori A., Itoh H., Yoshida M., Kinosita K., Jr. (2008) Axle-less F1-ATPase rotates in the correct direction. Science 319, 955–958 [DOI] [PubMed] [Google Scholar]
- 36. Bowler M. W., Montgomery M. G., Leslie A. G., Walker J. E. (2006) How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl. Acad. Sci. U.S.A. 103, 8646–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masaike T., Koyama-Horibe F., Oiwa K., Yoshida M., Nishizaka T. (2008) Cooperative three-step motions in catalytic subunits of F1-ATPase correlate with 80 degrees and 40 degrees substep rotations. Nat. Struct. Mol. Biol. 15, 1326–1333 [DOI] [PubMed] [Google Scholar]
- 38. Uchihashi T., Iino R., Ando T., Noji H. (2011) High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 [DOI] [PubMed] [Google Scholar]
- 39. Oster G., Wang H. (2003) Rotary protein motors. Trends Cell Biol. 13, 114–121 [DOI] [PubMed] [Google Scholar]
- 40. Mnatsakanyan N., Kemboi S.K., Salas J., Weber J. (2011) The β subunit loop that couples catalysis and rotation in ATP synthase has a critical length. J. Biol. Chem. 286, 29788–29796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.