Background: ZFP57 is a maternal-zygotic effect gene that maintains genomic imprinting in mouse embryos.
Results: KAP1 facilitates the interaction between ZFP57 and DNA methyltransferases. The KRAB box of ZFP57 is required for maintaining DNA methylation imprint in ES cells.
Conclusion: ZFP57 recruits DNA methyltransferases and maintains DNA methylation imprint through KRAB box-mediated interaction.
Significance: This work implies that ZFP57 recruits DNA methyltransferases via KAP1 to maintain DNA methylation imprint.
Keywords: DNA Methyltransferase, Embryonic Stem Cell, Epigenetics, Homologous Recombination, Zinc Finger, DNA Methylation, Embryonic Stem Cells, Genomic Imprinting, KAP1/TRIM28/TIF1beta, ZFP57
Abstract
Previously, we discovered that ZFP57 is a maternal-zygotic effect gene, and it maintains DNA methylation genomic imprint at multiple imprinted regions in mouse embryos. Despite these findings, it remains elusive how DNA methyltransferases are targeted to the imprinting control regions to initiate and maintain DNA methylation imprint. To gain insights into these essential processes in genomic imprinting, we examined how ZFP57 maintains genomic DNA methylation imprint in mouse embryonic stem (ES) cells. Here we demonstrate that the loss of ZFP57 in mouse ES cells led to a complete loss of genomic DNA methylation imprint at multiple imprinted regions, similar to its role in mouse embryos. However, reintroduction of ZFP57 into Zfp57-null ES cells did not result in reacquisition of DNA methylation imprint, suggesting that the memory for genomic imprinting had been lost or altered in Zfp57-null ES cells in culture. Interestingly, ZFP57 and DNA methyltransferases could form complexes in the presence of KAP1/TRIM28/TIF1β when co-expressed in COS cells. We also found that the wild-type exogenous ZFP57 but not the mutant ZFP57 lacking the KRAB box that interacts with its co-factor KAP1/TRIM28/TIF1β could substitute for the endogenous ZFP57 in maintaining the DNA methylation imprint in ES cells. These results suggest that ZFP57 may recruit DNA methyltransferases to its target regions to maintain DNA methylation imprint, and this interaction is likely facilitated by KAP1/TRIM28/TIF1β.
Introduction
Genomic imprinting is a special kind of parental control over the progeny (1, 2). It was discovered more than three decades ago and is mainly observed in mammals, marsupials, and plants (3–8). In contrast to the normal bi-allelic expression of the vast majority of genes, imprinted genes are expressed either from the paternal chromosome or maternal chromosome. So far, ∼100 imprinted genes have been identified in mammals (9). Most of the imprinted genes are clustered, so that a few of them ranging from two to over a dozen imprinted genes are co-regulated by a cis-acting imprinting control region (10–13).
DNA methylation at the cytosine residue usually occurs at CpG dinucleotides in mammals (14, 15). Cytosine methylation is also frequently observed at non-CpG sites in plants as well as in ES cells (5, 16). This modification is catalyzed by DNA methyltransferases (17). DNA methyltransferase I (DNMT1)3 is the main maintenance DNA methyltransferase, whereas two related DNA methyltransferases DNMT3a and DNMT3b are involved in de novo DNA methylation (18–20). DNMT3L does not have its own catalytic enzyme ability, but it can enhance the enzymatic activities of DNMT3a and DNMT3b (21–24).
One hallmark of the imprinting control regions is that they contain a germ line-derived differentially methylated region (DMR) that is inherited either on the paternal chromosome or on the maternal chromosome (4, 10, 12, 25). Up until recently, it was largely unknown how this differential methylation is established in the germ line and stably maintained thereafter. Two maternal effect genes, PGC7/Stella and Zfp57, were found to play partially overlapping roles in the maintenance of differential DNA methylation at the imprinting control regions (26–29). Zfp57 is the first identified mammalian maternal-zygotic effect gene, and it has both maternal and zygotic functions (2, 26). Zfp57 is also required for the establishment of differential DNA methylation at the Snrpn imprinted region in the female germ line (26). Furthermore, ZFP57 associates with the Snrpn DMR based on a ChIP assay in ES cells (26). Therefore, we hypothesize that ZFP57 may target DNA methyltransferases to the imprinting control regions to establish and/or maintain differential DNA methylation imprint at the imprinting control regions.
In our previous study, ZFP57 was found to be an ES cell-specific gene that is highly expressed in undifferentiated ES cells but dramatically down-regulated during ES cell differentiation (30). ZFP57 is a member of KRAB zinc finger family of proteins, and it is estimated that there are over 300 members in the human genome (26, 31). KAP1/TRIM28/TIF1β is the obligate co-factor for KRAB zinc finger proteins, including ZFP57 (32, 33). Indeed, our previous study confirmed that ZFP57 binds to KAP1/TRIM28/TIF1β both in ES cells as well as in COS cells (26). KAP1/TRIM28/TIF1β contains multiple functional domains. It has a RING finger at the N terminus followed by B-Boxes and coiled coil domains (34, 35). It also has an HP1-binding motif in the middle (36–38). Its carboxyl end is composed of PhD and BRM domains that are essential for the interaction of KAP1/TRIM28/TIF1β with histones and other chromatin-associated proteins (39). The PhD domain also functions as an intramolecular E3 ligase for SUMO modification of the adjacent BRM domain (40). Indeed, sumoylation of the BRM domain facilitates the recruitment of the SETDB1 histone methyltransferase and the NuRD complex to initiate gene silencing (40, 41).
In this study, we carried out extensive biochemical interaction analyses to assess whether ZFP57 can interact with DNA methyltransferases either directly or indirectly via an intermediate protein. We found that ZFP57 does not appear to be able to bind DNA methyltransferases directly. By contrast, KAP1/TRIM28/TIF1β can bind multiple DNA methyltransferases and mediates the interactions between ZFP57 and DNA methyltransferases.
ES cells have been increasingly employed as a model system for studying genomic imprinting (9, 25, 42–46). Indeed, we found that ZFP57 maintains DNA methylation imprint at a large subset of imprinted regions in ES cells. This function of ZFP57 requires its KRAB box, suggesting that the interaction between ZFP57 and its co-factor KAP1/TRIM28/TIF1β is essential for the maintenance of DNA methylation imprint.
EXPERIMENTAL PROCEDURES
Plasmid Construction
KAP1/TRIM28/TIF1β deletion mutants were mostly constructed by PCR. Specifically, the cDNA encoding KAP1/TRIM28/TIF1β was subcloned into pBluescript. One internal primer with a HindIII site at the 5′ end was paired with an external primer in the vector backbone (T7 or M13Rev) to amplify the N- or C-terminal portion of the KAP1/TRIM28/TIF1β cDNA by PCR. Then these two portions of KAP1/TRIM28/TIF1β cDNA were linked by T4 DNA ligase-mediated ligation after HindIII digestion. The primer pairs used for construction of these KAP1/TRIM28/TIF1β deletion mutants are listed in Table 1.
TABLE 1.
Oligonucleotides used for PCR to generate deletion mutants of KAP1 and ZFP57
| Deletion mutant | Oligonucleotide name | Oligonucleotide sequence |
|---|---|---|
| KAP1ΔRING | KAP1-RING-FH | tgcaagcttcagtgctactccaaagac |
| KAP1ΔRING | huKAP1-RING-RH | caaagcttcagcagctccagcgcctcgg |
| KAP1ΔBCC | KAP1-RBCC-FH | gtgaagcttgtggagcctcatggtgagatg |
| KAP1ΔBCC | KAP1-BCC-RH | agaaagcttactgccactatcacg |
| KAP1ΔRBCC | KAP1-RBCC-FH | gtgaagcttgtggagcctcatggtgagatg |
| KAP1ΔRBCC | huKAP1-RING-RH | caaagcttcagcagctccagcgcctcgg |
| KAP1ΔHP1 | KAP1-HP1-FH | ctgaagcttacctctgacagccagccac |
| KAP1ΔHP1 | KAP1-HP1-RH2 | aaaagcttacttacctctccctcacc |
| KAP1ΔPhD | KAP1-PhD-FH | ctaaagcttgaagatggaagcctcagc |
| KAP1ΔPhD | KAP1-PhD-RH | ggtaagcttaggagccaccacctc |
| KAP1ΔBRM | KAP1-BRM-FH | accaagctttctgctgtgctggtag |
| KAP1ΔBRM | huKAP1-BRM-RH | gaaagcttggccaccacgccag |
| KAP1ΔPhD-BRM | KAP1-BRM-FH | accaagctttctgctgtgctggtag |
| KAP1ΔPhD-BRM | KAP1-PhD-RH | ggtaagcttaggagccaccacctc |
| ZFP57ΔKRAB | KRAB-SPHF | ttcgcatgcgaagcaagaagaaacctcaagaac |
| ZFP57ΔKRAB | KRAB-SPHR | acagcatgcgtgtcctggatggctgggaag |
The ZFP57 mutant lacking the KRAB domain was similarly constructed by PCR, and the primers used are listed in Table 1. An HindIII/PmeI cDNA fragment containing the Myc epitope tag and the six-histidine tag derived from the pcDNA3.1/Myc-His vector (Invitrogen) was fused to the C terminus of the cDNAs for the wild-type ZFP57 or the mutant ZFP57 lacking the KRAB box. This fusion was facilitated by an HindIII restriction site introduced into the Zfp57 cDNA at the C terminus. The pBluescript vector harboring Zfp57 or the mutant Zfp57 cDNA was digested with HindIII and EcoRV before ligation with the HindIII/PmeI cDNA fragment encoding the Myc-His tag. The tagged Zfp57 cDNA or the mutant Zfp57 cDNA was inserted into KpnI and NotI sites of an mammalian expression vector containing the chicken β-actin and CMV hybrid promoter (a gift from Dr. Jianrong Lu of University of Florida).
Transfection
COS cells were split the day before transfection. Plasmid DNA was mixed at appropriate ratios and co-transfected into COS cells with FuGENE 6 (Roche Applied Science). After 2 days of culture at 37 °C, transiently transfected cells were harvested for co-immunoprecipitation (co-IP) interaction assays.
Isolation of Doxycycline-inducible ES Clones
ES cells were cultured as previously described (26). The A2lox ES cell line was used for isolation of doxycycline-inducible ES clones that express FLAG epitope-tagged DNMT3L or DNMT3a (47). The hybrid cDNA encoding FLAG-tagged DNMT3L or DNMT3a was subcloned into the p2Lox targeting construct (47). The resultant constructs were transfected, together with pCAGGS-Cre that constitutively expresses Cre recombinase, into A2lox ES cells by electroporation. The transfected ES cells were plated on 10-cm plates seeded with irradiated feeder fibroblast cells. The following day, ES cell culture was switched to growth medium containing 260 μg/ml of G-418. After G-418 selection for 1 week to 10 days, stably transfected colonies were picked individually and grown on 24-well plates seeded with irradiated feeder fibroblast cells. ES cells derived from these colonies were induced with 1 μg/ml of doxycycline for 3 days, and protein lysate was subjected to Western blot analysis with monoclonal antibody against the FLAG epitope (Sigma) to examine whether the FLAG-tagged DNMT3L or DNMT3a was expressed after doxycycline induction. Candidate ES clones with doxycycline-inducible expression of FLAG-tagged DNMT3L or DNMT3a were used for co-immunoprecipitation interaction assays to detect the interaction between endogenous KAP1 and FLAG-tagged DNMT3L or DNMT3a.
Co-immunoprecipitation Interaction Assay
The cells were lysed in buffer containing 20 mm of Tris-HCl (pH 7.3), 150 mm of NaCl, 0.5% of Nonidet P-40, 10% of glycerol, 1 mm of PMSF, as well as the mixture of protease inhibitors (Roche Applied Science). After brief sonication to rupture the cell membrane, the cell lysate was centrifuged, and the supernatant was transferred to a new Eppendorf tube. Mouse monoclonal antibody against the FLAG epitope was added to the supernatant, and protein A/G beads were added thereafter to precipitate the immunocomplexes.
Bisulfite Sequencing
Genomic DNA was isolated from ES cells grown on the 6-well plates coated with 0.1% gelatin for one passage after being cultured in the presence of feeder fibroblasts. These genomic DNA samples were subjected to bisulfite mutagenesis with the EpiTect bisulfite kit (Qiagen) or the EZ DNA methylation kit (Zymo Research). After the bisulfite treatment, genomic DNA was purified and amplified by PCR with the primers corresponding to the imprinted DMR regions and nonimprinted repeat regions. The amplified PCR product was cloned into the pGEM-T vector system and transformed into competent DH5α cells. Single bacterial colonies were picked and grown on 96-well plates before being sent for sequencing. PCR primers are listed in Table 2 with the exception of the primers for IG-DMR in Fig. 6A.
TABLE 2.
Oligonucleotides for PCR amplification of the bisulfite-treated DNA
| Imprinted region | Inside/outside nested primer | Oligonucleotide name | Oligonucleotide sequence |
|---|---|---|---|
| Snrpn DMR | Outside | gSn-F642 | GGGCTTCATGTTTGATTGTGTG |
| Snrpn DMR | Outside | SnR1204 | AATCAAATAAAATACACTTTCACTACT |
| Snrpn DMR | Inside | Sn-F780 | TGTGTGATGTTTGTAATTATTTGGGAG |
| Snrpn DMR | Inside | Sn-R1180 | ACTAAAATCCACAAACCCAACTAAC |
| IG-DMR | Outside | IG-nF1 | TGTGGATCCTAGAGATGTTTTTGTTGA |
| IG-DMR | Outside | IG-nR1 | TTCGGATCCCTACAACTTAAAAATTTCTCCAACC |
| IG-DMR | Inside | IG-F2416 | TTTTAGTTTTTTGGGTTTTAGAGAA |
| IG-DMR | Inside | IG-A2799 | AATAATCACCCTAACCCAAC |
| H19 DMR | Outside | H19-F1172 | GAAAGAAAAAGGTTGGTGAGAAAA |
| H19 DMR | Outside | H19A1745 | AACTAACATAAACCCCTAACCTCA |
| H19 DMR | Inside | H19-F1278 | GAGTATTTAGGAGGTATAAGAATTTTG |
| H19 DMR | Inside | H19 R1738 | ATAAACCCCTAACCTCATAAAACC |
| Line-1 | Outside | Line OF | GTTAGAGAATTTGATAGTTTTTGGAATAGG |
| Line-1 | Outside | Line OR | CCAAAACAAAACCTTTCTCAAACACTATAT |
| Line-1 | Inside | Line IF | TAGGAAATTAGTTTGAATAGGTGAGAGGGT |
| Line-1 | Inside | Line IR | TCAAACACTATATTACTTTAACAATTCCCA |
| IAP | Outside | IAP-OF | TTGATAGTTGTGTTTTAAGTGGTAAATAAA |
| IAP | Outside | IAP-OR | CAAAAAAAACACCACAAACCAAAAT |
| IAP | Inside | IAP-IF | TTGTGTTTTAAGTGGTAAATAAATAATTTG |
| IAP | Inside | IAP-IR | AAAACACCACAAACCAAAATCTTCTAC |
| Peg1 | Outside | Pe1 F504 | TTGGGATATAAAAGGTTAATGAGA |
| Peg1 | Outside | Pe1 R1190 | TCATTAAAAACACAAACCTCCTTTAC |
| Peg1 | Inside | Pe1 F533 | TTTTAGATTTTGAGGGTTTTAGGTTG |
| Peg1 | Inside | Pe1 R1096 | AATCCCTTAAAAATCATCTTTCACAC |
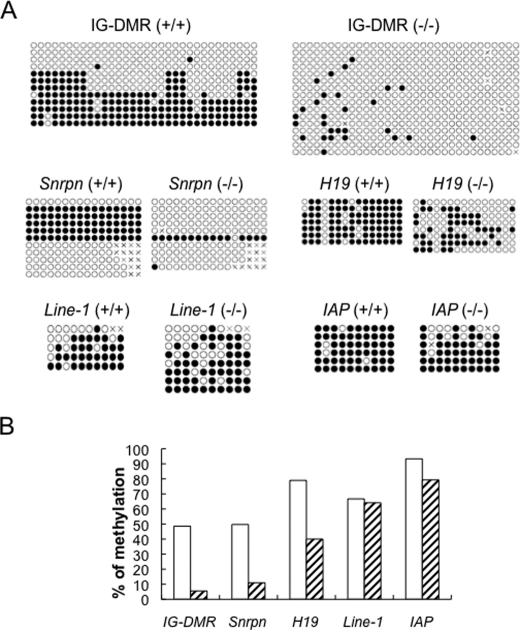
FIGURE 6.
DNA methylation imprint is lost at multiple imprinted regions in Zfp57-null ES cells. Zfp57-null ES cells were generated from the ES clones containing one floxed allele and one targeted allele of Zfp57 after Cre recombinase-mediated excision (26). Genomic DNA derived from the wild-type TC1 ES cells (+/+) and Zfp57-null ES cells (−/−) were subjected to bisulfite sequencing. A web-based software QUMA was used for DNA methylation analysis. A, bisulfite sequencing data are shown for the imprinted regions (IG-DMR, Snrpn DMR, H19 DMR) and nonimprinted repeat regions (Line-1 and IAP). ●, methylated CpG. ○, unmethylated CpG. ×, definitive sequencing result is not available for that particular CpG site. Each row indicates a unique clone based on bisulfite sequencing results. The outside nested PCR primers for the IG-DMR are: IG-F770, 5′-TTTGTAGTATTTTGTGTAGTTGTG and IG-A1588, 5′-CCAACTAACCTAAACTCCATACTA. The inside nested PCR primers for the IG-DMR are: IG-F941, 5′-ATATTATGTTAGTGTTAGGAAGGATTGTG and IG-A1371, 5′-TACAACCCTTCCCTCACTCCAAAAATT. B, a diagram is shown for the percentage of methylated CpG sites at the imprinted regions (IG-DMR, Snrpn DMR, and H19 DMR) and nonimprinted regions (Line-1 and IAP) in ES cells. Open bar, wild-type ES cells. Hatched bar, Zfp57-null ES cells.
Combined Bisulfite Restriction Analysis (COBRA)
Genomic DNA samples were harvested and subjected to the similar bisulfite mutagenesis as above. PCR product amplified from the bisulfite-treated samples was digested with the restriction enzymes that recognize the CpG sites present in the imprinted DMR regions and nonimprinted repeat regions. These CpG sites when unmethylated were sensitive to bisulfite mutagenesis, and the restriction enzyme sites were lost, but the restriction enzyme sites were preserved if these CpG sites were methylated. After gel electrophoresis, the methylated product and the unmethylated DNA product displayed different patterns of restriction enzyme digestion. For PCR primers, see Table 2.
RESULTS
Binding of ZFP57 to KAP1/TRIM28/TIF1β via Its KRAB Box
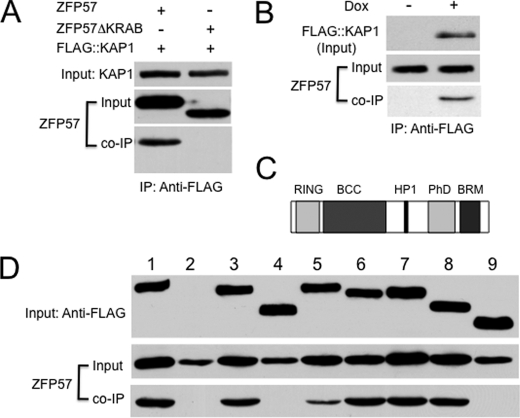
ZFP57 contains putative KRAB Box (26). Previously, we found that ZFP57 bound to KAP1 in COS cells when co-expressed (26). As expected, this interaction appears to be mediated by the KRAB box. When the ZFP57 mutant lacking the KRAB box was co-expressed with KAP1/TRIM28/TIF1β tagged with the FLAG epitope in COS cells, no detectable ZFP57 product was present in the immunoprecipitate when the monoclonal antibody against the FLAG epitope tag was used for immunoprecipitation (Fig. 1A). By contrast, wild-type ZFP57 was present in the immunoprecipitate when it was co-expressed with FLAG-tagged KAP1/TRIM28/TIF1β. These data suggests that the KRAB box is essential for binding of ZFP57 to KAP1/TRIM28/TIF1β, similar to what has been observed in other KRAB zinc finger proteins.
FIGURE 1.
ZFP57 binds to the BCC domain of KAP1 via its KRAB box. FLAG-tagged KAP1 or KAP1 deletion mutants were co-expressed in COS cells with ZFP57 or the ZFP57 deletion mutant without the KRAB box (ZFP57ΔKRAB). The monoclonal antibody against the FLAG epitope was used to precipitate KAP1-associated proteins, and the polyclonal antibodies against ZFP57 were used to probe the immunoprecipitate. A, PCR was used to create the cDNA for ZFP57ΔKRAB. FLAG-tagged KAP1 was co-expressed with ZFP57 or ZFP57ΔKRAB in COS cells. B, doxycycline-inducible ES clones that can express FLAG-tagged KAP1 were isolated from A2lox ES cells (47). Co-IP was performed for ES cells grown in the presence (+) or absence (−) of 1 μg/ml of doxycycline (Dox). C, a diagram for the functional domains of KAP1. HP1, HP1-binding motif. The RING and BCC domains together form the RBCC domain. D, PCR was used to generate the cDNAs encoding KAP1 deletion mutants that lack one or two functional domains of KAP1. Lane 1, wild-type KAP1; lane 2, vector only (pcDNA3-FLAG); lane 3, KAP1ΔRING; lane 4, KAP1ΔBCC; lane 5, KAP1ΔHP1; lane 6, KAP1ΔPhD; lane 7, KAP1ΔBRM; lane 8, KAP1ΔPhD-BRM (lacking both PhD and BRM domains); lane 9, KAP1ΔRBCC.
Binding of Endogenous ZFP57 to FLAG-tagged KAP1 in ES Cells
Previously, we demonstrated that endogenous ZFP57 binds to endogenous KAP1 based on the co-IP result with affinity-purified polyclonal antibodies against ZFP57 (26). To test whether endogenous ZFP57 can also interact with FLAG-tagged KAP1 in a co-IP assay in ES cells, we isolated ES clones that can inducibly express FLAG-tagged KAP1 in response to doxycycline in A2lox ES cells (47) (Fig. 1B). Indeed, we found that endogenous ZFP57 was present in the immunoprecipitate when monoclonal antibody against the FLAG epitope was used for immunoprecipitation (Fig. 1B), suggesting that endogenous ZFP57 can bind to FLAG-tagged KAP1 in ES cells.
The B-Box Coiled Coil (BCC) Domain of KAP1 Is Essential for Its Binding to ZFP57
KAP1/TRIM28/TIF1β is a scaffold protein and has multiple functional domains including the RING, B-Boxes, coiled coil, HP1-binding, PhD, and BRM domains or motifs (39). The B-Boxes and the coiled coil domains form the BCC domain. Together with the RING domain, they form the functional RBCC domain. To determine which domain of KAP1/TRIM28/TIF1β is required for its interaction with ZFP57, we have generated a series of KAP1/TRIM28/TIF1β deletion mutants that lack one or two functional domains of KAP1/TRIM28/TIF1β (Fig. 1C). We found that KAP1/TRIM28/TIF1β mutants lacking a single functional domain of the RING (Fig. 1D, lane 3), HP1-binding (Fig. 1D, lane 5), PhD (Fig. 1D, lane 6), or BRM (Fig. 1D, lane 7) domain or both PhD and BRM domains (Fig. 1D, lane 8) still retained their binding capability. In contrast, the KAP1/TRIM28/TIF1β deletion mutant lacking the BCC domain (Fig. 1D, lane 4) or the RBCC domain (Fig. 1D, lane 9) did not bind ZFP57 in COS cells when co-expressed. These results suggest that the BCC domain acts as the binding domain of KAP1/TRIM28/TIF1β for ZFP57.
Binding of DNMT3a to RBCC Domain of KAP1
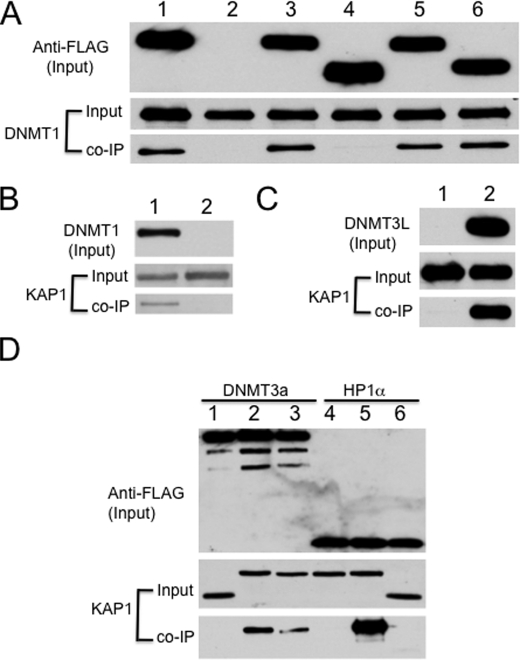
In our previous study, we found that ZFP57 is involved in the maintenance of DNA methylation imprints at multiple imprinted regions (26). In addition, ZFP57 is required for the acquisition of DNA methylation imprint at the Snrpn imprinted domain. These findings suggest that ZFP57 and its associated complexes may recruit DNA methyltransferases to the imprinting control regions of the affected imprinted domains. Our initial test results indicated that ZFP57 does not bind to DNA methyltransferases directly (see Fig. 4). To examine whether KAP1 may mediate the interactions between ZFP57 and DNA methyltransferases, we performed co-IP assays between KAP1 and DNMT3a when they were co-expressed in COS cells. Interestingly, we found that KAP1 did bind to DNMT3a when co-expressed (Fig. 2A, lane 1). Then we used various KAP1 deletion mutants to determine the functional interaction domain of KAP1 that is essential for its interaction with DNMT3a (Fig. 2A). Similar to the results obtained between ZFP57 and KAP1, the BCC domain is the essential interaction domain of KAP1 for DNMT3a. As shown in Fig. 2A, KAP1 deletion mutants lacking either the BCC (lane 3) or the RBCC domain (lane 7) were defective in binding to DNMT3a in COS cells when co-expressed. In contrast, those KAP1 mutant proteins lacking the RING (Fig. 2A, lane 2), HP1-binding (Fig. 2A, lane 4), PhD (Fig. 2A, lane 5), or BRM (Fig. 2A, lane 6) domain or both PhD and BRM domains (Fig. 2A, lane 8) displayed similar DNMT3a binding ability to the wild-type KAP1. Furthermore, the truncated protein containing just the RBCC domain was sufficient in binding to DNMT3a (Fig. 2B, lane 3). We also found that KAP1 was present in the immunoprecipitate when the monoclonal antibody against the FLAG epitope was used to precipitate DNMT3a-associated proteins (Fig. 2C, lane 1). These results suggest that the RBCC domain of KAP1 is both necessary and sufficient for its interaction with DNMT3a.
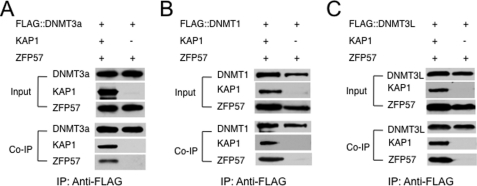
FIGURE 4.
KAP1 mediates the interaction between ZFP57 and DNA methyltransferases. ZFP57 and FLAG-tagged DNMT3a, DNMT1, or DNMT3L were co-transfected into COS cells with or without KAP1. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and polyclonal antibodies against ZFP57 or KAP1 were used to probe the immunoprecipitate. In addition, the monoclonal antibody against the FLAG epitope was used to confirm that an equal amount of FLAG-tagged DNMT3a, DNMT1, or DNMT3L was present in the immunoprecipitate. A, FLAG-DNMT3a was co-transfected with ZFP57 into COS cells with or without KAP1. B, FLAG-DNMT1 was co-transfected with ZFP57 into COS cells with or without KAP1. C, FLAG-DNMT3L was co-transfected with ZFP57 into COS cells with or without KAP1.
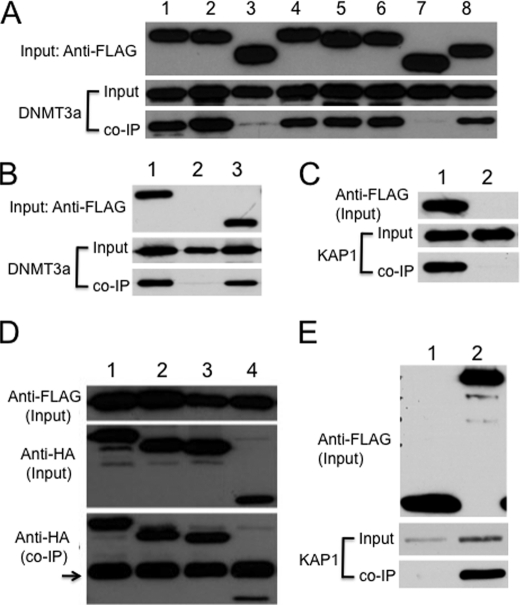
FIGURE 2.
KAP1 binds to DNMT3a via its RBCC domain. A, HA-tagged DNMT3a was co-expressed with FLAG-tagged KAP1 or KAP1 deletion mutants in COS cells. The monoclonal antibody against the FLAG epitope was used to precipitate KAP1-associated proteins, and the monoclonal antibody against the HA epitope was used to probe the immunoprecipitate. Lane 1, wild-type KAP1; lane 2, KAP1ΔRING; lane 3, KAP1ΔBCC; lane 4, KAP1ΔHP1; lane 5, KAP1ΔPhD; lane 6, KAP1ΔBRM; lane 7, KAP1ΔRBCC; lane 8, KAP1ΔPhD-BRM (lacking both PhD and BRM domains). B, HA-tagged DNMT3a was co-expressed with FLAG-tagged KAP1 or FLAG-tagged RBCC in COS cells. The monoclonal antibody against the FLAG epitope was used to precipitate KAP1- or RBCC-associated proteins, and the monoclonal antibody against the HA epitope was used to probe the immunoprecipitate. Lane 1, FLAG::KAP1; lane 2, vector only; lane 3, FLAG::RBCC. C, FLAG epitope-tagged DNMT3a was co-expressed with Myc epitope-tagged KAP1 in COS cells. The monoclonal antibody against the FLAG epitope was used to precipitate DNMT3a-associated proteins, and the monoclonal antibody against the Myc epitope was used to probe the immunoprecipitate. Lane 1, FLAG::DNMT3a; lane 2, vector only. D, FLAG-tagged KAP1 was co-expressed with HA-tagged DNMT3a or DNMT3a truncation mutants in COS cells. The monoclonal antibody against the FLAG epitope was used to precipitate KAP1-associated proteins, and the monoclonal antibody against the HA epitope was used to probe the immunoprecipitate. Arrow, IgG heavy chain. Lane 1, wild-type DNMT3a; lane 2, N1 mutant of DNMT3a; lane 3, N2 mutant of DNMT3a; lane 4, C mutant of DNMT3a. These truncation mutants had been used previously in another study (48). E, Myc-tagged KAP1 was co-expressed with FLAG-tagged EGFP or FLAG-tagged DNMT3a in COS cells. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and the monoclonal antibody against the Myc epitope was used to probe the immunoprecipitate. Lane 1, FLAG::EGFP; lane 2, FLAG::DNMT3a.
To examine whether there is a distinct domain of DNMT3a that is responsible for this interaction, we employed previously constructed DNMT3a truncation proteins that contain either the C-terminal catalytic domain or the N-terminal noncatalytic domains with or without the PWWP region (48). Interestingly, these truncated DNMT3a proteins all bound to KAP1 in co-IP when co-expressed in COS cells, although the interaction of the N-terminal noncatalytic domains (Fig. 2D, lanes 2 and 3) with KAP1 appeared to be stronger than that of the C-terminal catalytic domain (Fig. 2D, lane 4). These results suggest that the N-terminal noncatalytic domain of DNMT3a is the major interaction site for KAP1, and the C-terminal catalytic domain of DNMT3a plays a minor role as well.
The interaction between DNMT3a and KAP1 appears to be specific because a negative control involving the FLAG-tagged EGFP protein did not bind to KAP1 in co-IP when they were co-expressed in COS cells (Fig. 2E, lane 1). By contrast, KAP1 was present in the immunoprecipitate when the monoclonal antibody against the FLAG epitope was used for immunoprecipitation to pull down FLAG::DNMT3a-associated proteins in COS cells co-transfected with KAP1 and FLAG::DNMT3a (Fig. 2E, lane 2).
Binding of Other DNA Methyltransferases to KAP1
Because ZFP57 is involved in both acquisition and maintenance of DNA methylation imprints, it is possible that KAP1 will bind to both maintenance as well as de novo DNA methyltransferases. Indeed, we found that KAP1 binds to DNMT1, the major maintenance DNA methyltransferase, in co-IP when co-expressed in COS cells (Fig. 3A, lanes 1 and 3). Similarly, this interaction is mediated by the BCC domain of KAP1 because the KAP1 deletion mutant lacking the BCC domain was defective in binding of DNMT1 (Fig. 3A, lane 4), whereas all other KAP1 deletion mutants lacking just the HP1-binding domain (lane 5) or both the PhD and BRM domains (lane 6) were capable of binding to DNMT1. Consistent with this, KAP1 was also present in the immunoprecipitate when the monoclonal antibody against the FLAG epitope was used for co-IP against FLAG-tagged DNMT1 when Myc-tagged KAP1 and FLAG-tagged DNMT1 were co-expressed in COS cells (Fig. 3B, lane 1). Furthermore, DNMT3L bound to KAP1 as well when they were co-expressed in COS cells (Fig. 3C).
FIGURE 3.
KAP1 binds to other DNA methyltransferases. A, Myc-tagged DNMT1 was co-expressed with FLAG-tagged KAP1 or KAP1 deletion mutants in COS cells. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and the monoclonal antibody against the MYC epitope was used to probe the immunoprecipitate. Lane 1, wild-type KAP1; lane 2, vector only; lane 3, wild-type KAP1; lane 4, KAP1ΔBCC; lane 5, KAP1ΔHP1; lane 6, KAP1ΔPhD-BRM. B, FLAG-tagged DNMT1 was co-expressed with Myc-tagged KAP1 in COS cells. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and the monoclonal antibody against the Myc epitope was used to probe the immunoprecipitate. Lane 1, wild-type DNMT1; lane 2, vector only. C, FLAG-tagged DNMT3L was co-expressed with Myc-tagged KAP1 in COS cells. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and the monoclonal antibody against the Myc epitope was used to probe the immunoprecipitate. Lane 1, vector only; lane 2, FLAG::DNMT3L. D, FLAG-tagged DNMT3a (lanes 1–3) or HP1a (lanes 4–6) was co-expressed with Myc-tagged KAP1 or KAP1 deletion mutants. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and the monoclonal antibody against the Myc epitope was used to probe the immunoprecipitate. Lane 1, KAP1ΔRBCC; lane 2, wild-type KAP1; lane 3, KAP1ΔHP1; lane 4, KAP1ΔHP1; lane 5, wild-type KAP1; lane 6, KAP1ΔRBCC.
Interactions between RBCC Domain of KAP1 and DNA Methyltransferases Appear to Be Specific
Based on the above co-IP results, the RBCC domain appears to be both necessary and sufficient for the interaction between KAP1 and DNA methyltransferases in COS cells when co-expressed. To further test whether this interaction involving the RBCC domain is specific, we also examined the binding between KAP1 and the previously identified interaction protein HP1a. As shown in Fig. 3D, wild-type KAP1 did indeed bind to HP1a (lane 5). In contrast, KAP1 mutants lacking either the HP1-binding (lane 4) or the RBCC domain (lane 6) were defective in binding to HP1a. These data suggest that the interaction between KAP1 and HP1a requires both the RBCC domain and the HP1-binding motif of KAP1. This is distinct from those involving KAP1 and DNA methyltransferases in which only the RBCC domain of KAP1 is required. The KAP1 deletion mutant lacking just the HP1-binding motif retained its ability to bind to DNMT3a (Fig. 3D, lane 3), whereas KAP1 without the RBCC domain was completely defective in its binding to DNMT3a (Fig. 3D, lane 1). Therefore, the interaction between the RBCC domain and DNMT3a appears to be specific because the KAP1 deletion mutant lacking just the HP1-binding motif did not bind to HP1a, even though it contains an intact RBCC domain (Fig. 3D, lane 4).
KAP1 Mediates Interaction between ZFP57 and DNMT3a
KAP1 binds to ZFP57 via its BCC domain. Similarly, the BCC domain is also the interaction domain between KAP1 and DNMT3a. It is possible that ZFP57 and DNMT3a may not be able to bind to the BCC domain simultaneously because of potential competition for the same binding domain. Alternatively, they may form a ternary complex, with KAP1 as the scaffold protein. To distinguish these possibilities, we co-transfected ZFP57 and FLAG::DNMT3a with or without KAP1 into COS cells and then immunoprecipitated DNMT3a-associated proteins with the monoclonal antibody against the FLAG epitope. Detectable ZFP57 was present in the immunoprecipitate only when KAP1 was included as one of the co-transfected proteins (Fig. 4A). Consistent with this, KAP1 was also present as one of the FLAG::DNMT3a-associated proteins immunoprecipitated with the monoclonal antibody against the FLAG epitope when ZFP57, KAP1, and DNMT3a were co-transfected into COS cells (Fig. 4A). By contrast, no detectable ZFP57 was observed in the immunoprecipitate from the cell lysate co-transfected with ZFP57 and FLAG::DNMT3a when the antibody against the FLAG epitope was used to precipitate the DNMT3a-associated proteins (Fig. 4A). These results suggest that ZFP57 does not bind to DNMT3a directly, and KAP1 forms a bridge between ZFP57 and DNMT3a (see Fig. 10).
FIGURE 10.
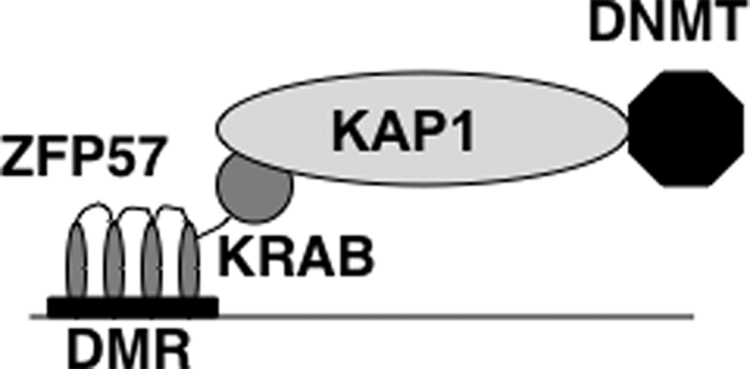
A hypothetical model for recruitment of DNA methyltransferases by ZFP57 via KAP1. As shown in the diagram, the zinc finger domain of ZFP57 presumably binds to its target regions such as a differentially methylated region (DMR) in an imprinted domain. The KRAB box of ZFP57 can bind to KAP1, which in turn recruits DNA methyltransferases (DNMT). DNA methyltransferases then can either initiate or maintain DNA methylation at the DMR.
Indirect Interaction between ZFP57 and Other DNA Methyltransferases
Besides DNMT3a, KAP1 bound to DNMT1 via the BCC domain when co-expressed in COS cells (Fig. 3A). KAP1 also interacted with DNMT3L in a co-IP assay in COS cells (Fig. 3C). To test whether KAP1 also mediates the interaction between ZFP57 and these two DNA methyltransferases, we performed similar co-IP assays for interaction of ZFP57 with DNMT1 or DNMT3L, with or without KAP1 co-transfected into COS cells. Similar to what had been observed between ZFP57 and DNMT3a, ZFP57 was detected in the immunoprecipitate pulled down with the monoclonal antibody against the FLAG epitope only when KAP1 was included as a co-transfected protein, together with the FLAG::DNMT1 or FLAG::DNMT3L (Fig. 4, B and C). These results suggest that KAP1 also mediates the interaction between ZFP57 and other DNA methyltransferases, similar to what had been observed between ZFP57 and DNMT3a (Fig. 4A).
Endogenous KAP1 Binds to FLAG-tagged DNMT3a in Mouse ES Cells
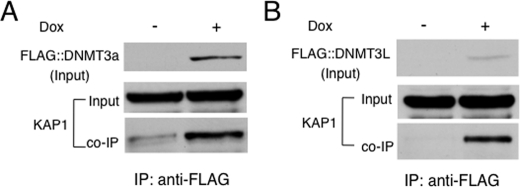
To test whether KAP1 binds to DNMT3a in ES cells as well, we isolated doxycycline-inducible ES clones that can express FLAG-tagged DNMT3a in response to doxycycline (Fig. 5A). Then we used the monoclonal antibody against the FLAG epitope to precipitate FLAG::DNMT3a-associated proteins in doxycycline-induced ES cells. Indeed, KAP1 was detected in the immunoprecipitate from the ES cells in the presence of 1 μg/ml of doxycycline but not from the ES cells without the addition of doxycycline (Fig. 5A), suggesting that endogenous KAP1 binds to FLAG::DNMT3a in mouse ES cells.
FIGURE 5.
Endogenous KAP1 binds to exogenous DNMT3a or DNMT3L in mouse ES cells. Doxycycline-inducible ES clones that can express FLAG-tagged DNMT3a or DNMT3L were used for co-IP to examine potential interactions between endogenous KAP1 and FLAG::DNMT3a or FLAG::DNMT3L. The monoclonal antibody against the FLAG epitope was used for immunoprecipitation, and polyclonal antibodies against KAP1 were used to probe the immunoprecipitate. Dox, doxycycline. FLAG-tagged proteins were induced by 1 μg/ml of doxycycline added to the ES cell culture. A, FLAG::DNMT3a. B, FLAG::DNMT3L.
DNMT3L Binds to Endogenous KAP1 in ES Cells
To test whether KAP1 binds to DNMT3L in ES cells, we isolated doxycycline-inducible ES clones that can express FLAG-tagged DNMT3L in A2lox ES cells in response to doxycycline induction (Fig. 5B). Then we performed co-IP with the monoclonal antibody against the FLAG epitope to precipitate FLAG::DNMT3L-associated proteins. We found that KAP1 was present in the immunoprecipitate derived from the ES cells in the presence of 1 μg/ml of doxycycline, but not in that from the ES cells without doxycycline (Fig. 5B), suggesting that endogenous KAP1 binds to FLAG::DNMT3L in ES cells as well.
ZFP57 Maintains DNA Methylation Imprint in ES Cells
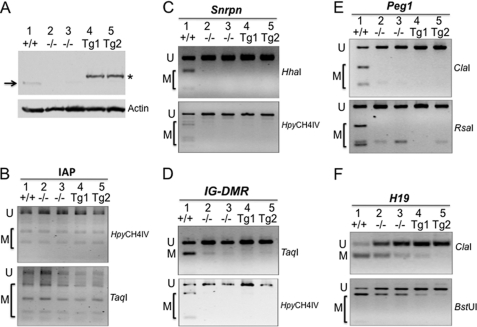
We constructed Zfp57-null ES cells by transiently expressing Cre recombinase in ES cells containing one floxed allele and one targeted allele of Zfp57 (26). We harvested genomic DNA samples from the original wild-type ES cells, as well as from two independent Zfp57-null ES clones. Then we subjected these genomic DNA samples to COBRA and bisulfite sequencing to analyze the DNA methylation status at the imprinted control regions, as well as nonimprinted repeat regions (Fig. 6, A and B). We found that DNA methylation imprint at Snrpn DMR and IG-DMR was lost in Zfp57-null ES cells but not in the wild-type ES cells (Fig. 6, A and B). For the H19 DMR, DNA methylation imprint was partially missing in the Zfp57-null ES cells in comparison with the wild-type ES cells (Fig. 6, A and B). By contrast, DNA methylation was stably maintained in nonimprinted repeat regions such as Line-1 elements and IAP repeats (Fig. 6, A and B). These results suggests that ZFP57 is required for the maintenance of DNA methylation imprint at a subset of imprinted regions in ES cells, similar to what we had observed previously in mouse embryos (26).
Reintroduction of Exogenous ZFP57 into Zfp57-null ES Cells
In our previous study, we found that DNA methylation imprint at the Snrpn DMR was regained in approximately half of the mouse embryos in the presence of zygotic ZFP57 (26). We wondered whether reintroduction of ZFP57 into Zfp57-null ES cells would lead to reacquisition of DNA methylation imprint at the Snrpn DMR as well. For this, we targeted an expression construct to the hprt locus of Zfp57-null ES cells via homologous recombination, which can constitutively express the exogenous ZFP57 driven by a chicken β-actin and CMV hybrid promoter (Fig. 7A, lanes 4 and 5). Despite the fact that exogenous ZFP57 is highly expressed in two independent ES clones that lack endogenous ZFP57, the DNA methylation imprint was not reacquired at the Snrpn DMR region (Fig. 7C). With the exception of the H19 DMR (Fig. 7F), we also observed no effect on other imprinted regions or nonimprinted IAP repeats (Fig. 7, B, D, and E). Methylation was in fact lost at the H19 DMR whose methylation imprint has been shown previously to be relatively unstable in ES cells before (49, 50). These results suggest that the imprinting memory at the Snrpn DMR and other imprinted regions have been lost in Zfp57-null ES cells, and reintroduction of exogenous ZFP57 is not sufficient to reinitiate the acquisition of DNA methylation imprint in ES cells.
FIGURE 7.
Reintroduction of the exogenous wild-type ZFP57 does not restore the DNA methylation imprint in ES cells. A mammalian expression construct harboring the Zfp57 cDNA under the control of chicken β-actin and CMV hybrid promoter was inserted into the hprt locus Zfp57-null ES cells via homologous recombination. Genomic DNA was isolated from the wild-type TC1 ES cells (+/+), two independent Zfp57-null ES clones (−/−) and two independent ES clones (Tg1 and Tg2) that constitutively express exogenous ZFP57 in Zfp57-null ES cells lacking the endogenous ZFP57. A, Western blot analysis of the ES clones with purified polyclonal antibodies against ZFP57 (top panel) or with an monoclonal antibody against β-actin (bottom panel). Arrow, position of the endogenous ZFP57. Asterisk, exogenous ZFP57 with an Myc epitope tag and a six-histidine tag at the C terminus. COBRA analysis was performed for the nonimprinted IAP repeat region with HpyCH4IV (B), the imprinted Snrpn DMR with HhaI and HpyCH4IV (C), the imprinted IG-DMR of the Dlk1-Dio3 imprinted region with TaqI and HpyCH4IV (D), the imprinted Peg1 DMR with ClaI and RsaI (E), and the imprinted H19 DMR with ClaI and BstUI (F). U, restriction enzyme digestion fragment of unmethylated DNA. M, restriction enzyme digestion fragment of methylated DNA.
The KRAB Box of ZFP57 Is Indispensible for Maintaining DNA Methylation Imprint
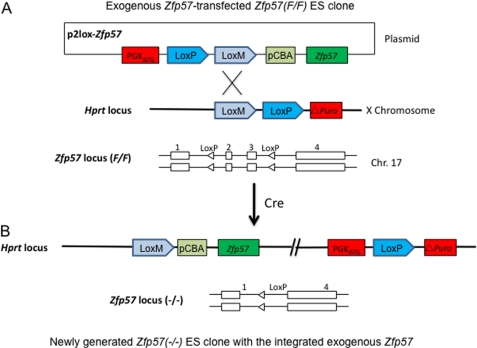
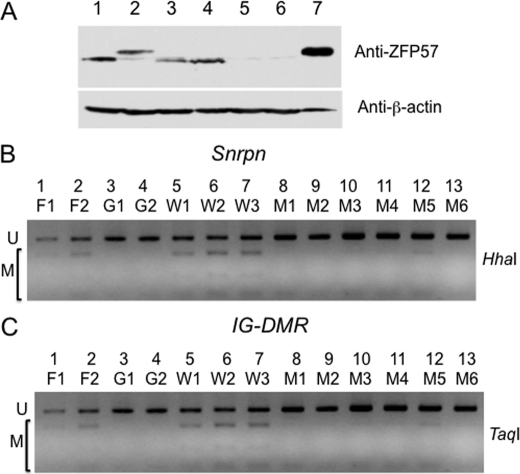
Without ZFP57, DNA methylation imprint is lost at a subset of imprinted regions in both mouse embryos, as well as in ES cells (26) (Fig. 6). ZFP57 binds to its co-factor KAP1/TRIM28/TIF1β via its KRAB box (26) (Fig. 1A). To test whether this interaction is essential for the role of ZFP57 in maintenance of DNA methylation imprint, we had constructed an ES cell system that can replace the endogenous ZFP57 with the exogenous ZFP57 or its variants constitutively expressed from the chicken β-actin and CMV hybrid promoter inserted into the hprt locus (47) (Fig. 8B). First, we obtained the ES clone (F/F) containing two floxed alleles at the Zfp57 locus that retains DNA methylation imprint at the Snrpn DMR and IG-DMR imprinted regions (Figs. 8A and Fig. 9B, lanes 1 and 2). Then a puromycin cassette that contains a canonical LoxP site and a mutant LoxP site (LoxM) as well as a puromycin-resistance gene lacking the initiation codon ATG was inserted into the hprt locus of the ES clone (F/F) through homologous recombination with a targeting vector containing a fragment of the hprt gene (Fig. 8A and data not shown). Next, the plasmid containing Zfp57 (p2lox-Zfp57 in Fig. 8A) or its variants or GFP cDNA was electroporated, together with the plasmid pCAGGS-Cre that can constitutively express Cre recombinase, into the cells of the above ES clone (F/F) carrying the puromycin cassette. Upon transient expression of Cre recombinase from pCAGGS-Cre, both floxed alleles of Zfp57 were excised, and expression for the endogenous ZFP57 was terminated (Figs. 8B and 9A, lanes 2–6). Simultaneously, expression constructs carrying the cDNAs for the GFP reporter, the wild-type ZFP57, or the mutant ZFP57 lacking the KRAB box were integrated into the LoxP and LoxM sites at the hprt locus upon Cre recombinase-mediated insertion of the plasmid (p2lox-Zfp57) or its derivatives (Fig. 8, A and B). Multiple independent ES clones for each transfected construct were picked individually. As expected, these ES clones express the integrated transgenes (Fig. 9A, lane 1, wild-type ZFP57 with a tag; lanes 3 and 4, mutant ZFP57 with a tag and no KRAB box; lanes 5 and 6, EGFP). Green fluorescence was observed for the ES clones expressing EGFP under fluorescence microscope (data not shown). Genomic DNA samples harvested from these ES clones were subjected to COBRA analysis. Indeed, the ES clones expressing GFP had all lost DNA methylation imprint at the Snrpn DMR and IG-DMR in the absence of endogenous ZFP57 upon Cre recombinase-mediated excision (Fig. 9, B and C, lanes 3 and 4). Methylaton imprint was maintained at these imprinted regions in the ES clones that express the wild-type ZFP57 (Fig. 9, B and C, lanes 5–7). By contrast, the DNA methylation imprint was lost at the Snrpn DMR and IG-DMR in the ES clones that constitutively express the mutant ZFP57 lacking KRAB box (Fig. 9, B and C, lanes 8–13). These results indicate that exogenous wild-type ZFP57 but not the mutant ZFP57 lacking the KRAB box can substitute for the endogenous ZFP57 in maintaining DNA methylation imprint in ES cells.
FIGURE 8.
A diagram is shown for the strategy for creating an ES cell system to test the maintenance function of exogenous ZFP57 and its variants. Mouse Zfp57 gene has four exons (exons 1, 2, 3, and 4) and is located on mouse chromosome 17 (26). ES clones containing two floxed alleles of Zfp57 (F/F) were generated by Flp recombinase-mediated excision of the Neo and Zeo cassette flanked by two FRT sites from the targeted allele in the ES clone containing one targeted allele and one floxed allele of Zfp57 (F/T) (26). Homologous recombination was used to insert a puromycin cassette into the hprt locus containing a canonical LoxP site and a mutant LoxM site, as well as a puromycin-resistant gene lacking the initiation codon ATG (△Puro). Similar to the strategy described in the previous study (47), Cre recombinase-mediated recombination was used to generate the ES cell clones for testing whether exogenous ZFP57 or its variants could substitute for the endogenous ZFP57. Diagrams were shown for the ES clone transfected with the plasmid containing the exogenous Zfp57 cDNA before (A) and after (B) Cre recombinase-mediated recombination. A, the cDNA for the wild-type Zfp57 or the mutant Zfp57 under the control of chicken β-actin and CMV promoter was cloned into the p2Lox vector. The resultant plasmid was co-transfected with a mammalian expression plasmid pCAGGS-Cre that constitutively expresses Cre recombinase into ES cells containing two floxed alleles of Zfp57 and a puromycin cassette at the hprt locus located on mouse × chromosome. B, upon transient expression of Cre recombinase, both floxed alleles of Zfp57 were excised. Simultaneously, the constitutive PGK promoter with the initiator codon ATG, together with the cDNA for Zfp57 or its mutants under the control of chicken β-actin and CMV promoter was integrated onto the LoxP and LoxM sites of hprt locus. The subsequent recombination between two LoxP sites results in in-frame fusion of the constitutive PGK promoter containing the initiator codon ATG and the puromycin-resistance gene lacking the initiator codon ATG, allowing expression of the full-length puromycin-resistant gene (47). These puromycin-resistant ES clones will also express the exogenous ZFP57 or its variants under the control of the constitutive chicken β-actin and CMV promoter (pCBA).
FIGURE 9.
Exogenous wild-type ZFP57 can substitute for the endogenous ZFP57 in maintaining genomic imprinting. ES clones expressing the exogenous wild-type ZFP57 or the mutant ZFP57 or EGFP from the hprt locus in place of the endogenous ZFP57 were generated as depicted in Fig. 8. Total cell lysate was harvested from these clones for Western blot analysis with affinity-purified polyclonal antibodies against ZFP57 (A, top panel). Western blot with an monoclonal antibody against β-actin was also performed for these total cell lysate samples (A, bottom panel). ES clones in lanes 2–6 lack endogenous ZFP57 after Cre recombinase-mediated excision. Lane 1, wild-type TC1 ES cells; lane 2, an ES clone expressing the wild-type ZFP57 with an Myc epitope tag and a six-histidine tag; lanes 3 and 4, two independent Zfp57(−/−) ES clones expressing the mutant ZFP57 with an Myc epitope tag and a six-histidine tag but without the KRAB box; lanes 5 and 6, two independent Zfp57(−/−) ES clones expressing EGFP; lane 7, COS cells expressing the wild-type ZFP57 with an Myc epitope tag and a six-histidine tag. Genomic DNA was isolated from these clones as well as from two ES clones containing two floxed alleles of Zfp57. COBRA analysis was performed at the Snrpn DMR with HhaI (B) and at the IG-DMR of the Dlk1-Dio3 imprinted region with TaqI (C). U, restriction enzyme digestion fragment of unmethylated DNA. M, restriction enzyme digestion fragment of methylated DNA. F1 and F2, two independent ES clones containing two floxed alleles of Zfp57. G1 and G2, two independent Zfp57(−/−) ES clones expressing EGFP. W1-W3, three independent Zfp57(−/−) ES clones expressing the wild-type ZFP57 with an Myc epitope tag and a six-histidine tag. M1–M6, six independent Zfp57(−/−) ES clones expressing the mutant ZFP57 with an Myc epitope tag and a six-histidine tag but without the KRAB box.
DISCUSSION
It is interesting to find that KAP1 can bind multiple DNA methyltransferases when co-expressed in COS cells. This interaction is mediated by the BCC domain of KAP1. We employed two different strategies to rule out the possibility of nonspecific interactions caused by the BCC domain. First, a negative control with a FLAG-tagged EGFP protein was used to test whether the full-length KAP1 containing the BCC domain will bind an unrelated protein such as EGFP. As expected, no interaction was detected between KAP1 and FLAG::EGFP. Second, we employed a known interaction protein HP1a that binds to another domain (HP1-binding motif) of KAP1 to rule out the possibility that the presence of the BCC domain is sufficient to cause nonspecific interactions with other proteins. As shown in Fig. 3D, a KAP1 mutant lacking the HP1-binding motif but with an intact BCC domain still did not bind to HP1a. This result suggests that the BCC domain of KAP1 does not bind to other proteins promiscuously, and its interactions with DNA methyltransferases appear to be specific.
Based on previously published literature, KAP1 acts as a scaffold protein with multiple functional domains. Thus, we tested the possibility that it can mediate the interaction between ZFP57 and DNA methyltransferases. Indeed, we found that ZFP57 and DNA methyltransferases bound to KAP1 simultaneously when co-expressed in COS cells, suggesting that they can form a complex. This raises an interesting possibility that ZFP57 can recruit DNA methyltransferases to its target regions via KAP1 (Fig. 10). Consistent with this hypothesis, it was shown that an artificial KRAB box-containing protein can initiate de novo DNA methylation in early mouse embryos (51). If that is the case, ZFP57 will be the first known transcription factor that can potentially provide sequence-specific targeting activity for DNA methyltransferases to initiate and/or maintain DNA methylation. Intriguingly, KAP1 can bind to both the maintenance DNA methyltransferase and de novo DNA methyltransferases. This indicates that KAP1, like ZFP57, may be also involved in establishment as well as maintenance of DNA methylation imprint (26). More study is needed to dissect the roles of ZFP57 and KAP1 in these processes.
To test whether these interactions originally found in COS cells are physiologically relevant, we employed ES cells as a model system in which ZFP57, KAP1, and DNA methyltransferases are all expressed (30, 52–55). KAP1 has been shown to be important for the maintenance and differentiation of embryonic stem cells (53, 56, 57). In addition, DNA methylation genomic imprint is maintained in ES cells, and de novo methylation is established during ES cell differentiation (58). In our previous study, we found endogenous ZFP57 interacts with endogenous KAP1 (26). However, we do not have good antibodies against DNA methyltransferases. Therefore, we established doxycycline-inducible ES clones in which FLAG epitope-tagged DNMT3a or DNMT1 can be expressed in response to doxycycline induction. Indeed, they can also bind to the endogenous KAP1 present in ES cells. These results indirectly suggest that endogenous KAP1 interacts with DNA methyltransferases in ES cells, and they may form a ternary complex with ZFP57 (Fig. 10).
To further test whether these interactions among ZFP57, KAP1, and DNA methyltransferases are functionally relevant, we have examined the roles of ZFP57 and its KRAB box in DNA methylation genomic imprint in ES cells. Similar to what had been observed in mouse embryos, ZFP57 is also required for the maintenance of genomic DNA methylation imprint at multiple imprinted regions in ES cells (Fig. 6). In addition, we found that exogenous wild-type ZFP57 but not the mutant ZFP57 lacking the KRAB box could substitute for the endogenous ZFP57 in maintaining genomic imprinting in ES cells (Fig. 9). Because KAP1 is the obligate co-factor for KRAB zinc finger proteins including ZFP57 and the KRAB box of ZFP57 is the interaction domain for KAP1, these results imply that the interaction between ZFP57 and KAP1 is essential for maintaining DNA methylation imprint. This also supports our hypothesis that KAP1 facilitates the interaction between ZFP57 and DNA methyltransferases.
The loss of ZFP57 causes loss of DNA methylation imprint at multiple imprinted regions in ES cells (Fig. 6). However, reintroduction of the exogenous wild-type ZFP57 into Zfp57-null ES cells did not lead to reacquisition of DNA methylation imprint at these imprinted regions (Fig. 7). This suggests that the imprinting memory may have been lost because of the lack of continued presence of ZFP57 in cultured Zfp57-null ES cells that were generated from the ES clones containing one floxed allele and one targeted allele of Zfp57 after Cre recombinase-mediated excision (26). By contrast, DNA methylation imprint was maintained when the exogenous wild-type ZFP57 was expressed presumably before the endogenous ZFP57 disappeared after introduction of Cre recombinase into the ES cells harboring two floxed alleles of Zfp57 (Figs. 8 and 9). These results suggest that ZFP57 is continuously required for maintaining DNA methylation imprint in ES cells, and maintenance of genomic imprinting may be an active process. Similar findings have been discovered previously for the mechanisms of the maintenance DNA methyltransferase DNMT1 in DNA methylation imprint (45, 59, 60).
Although loss of ZFP57 had no effect on the H19 DMR in mouse embryos (26), DNA methylation imprint at the H19 DMR region was partially lost in Zfp57-null ES cells, as well as in the Zfp57-null ES cells transfected with the wild-type ZFP57 (Figs. 6 and 7F). This is consistent with the published literature that DNA methylation imprint at the H19 DMR region is relatively unstable in ES cells (49, 50).
Acknowledgments
We thank Dr. Stephen Goff and Dr. Daniel Wolf (Columbia University) for providing us the mouse KAP1 cDNA construct. We appreciate the help from Miriam Merzel and Lai Chan during the course of this work. We also want to thank other members of the Li lab for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM093335 (to X. L.). This work was also supported in part by the March of Dimes Basil O'Connor Starter Research Award (to X. L.) and New York State NYSTEM Contract C026434 (to X. L.).
- DNMT
- DNA methyltransferase
- ES
- embryonic stem
- DMR
- differentially methylated region
- CMV
- cytomegalovirus
- co-IP
- co-immunoprecipitation
- COBRA
- combined bisulfite restriction analysis
- BCC
- B-Box coiled coil
- RBCC
- RING B-Box coiled coil
- BRM
- bromodomain.
REFERENCES
- 1. Cattanach B. M. (1986) Parental origin effects in mice. J. Embryol. Exp. Morphol. 97, 137–150 [PubMed] [Google Scholar]
- 2. Li X. (2010) Extending the maternal-zygotic effect with genomic imprinting. Mol. Hum. Reprod. 16, 695–703 [DOI] [PubMed] [Google Scholar]
- 3. Tilghman S. M. (1999) The sins of the fathers and mothers. Genomic imprinting in mammalian development. Cell 96, 185–193 [DOI] [PubMed] [Google Scholar]
- 4. Bartolomei M. S. (2009) Genomic imprinting. Employing and avoiding epigenetic processes. Genes Dev. 23, 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Law J. A., Jacobsen S. E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jullien P. E., Berger F. (2009) Gamete-specific epigenetic mechanisms shape genomic imprinting. Curr. Opin. Plant Biol. 12, 637–642 [DOI] [PubMed] [Google Scholar]
- 7. Feil R., Berger F. (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 23, 192–199 [DOI] [PubMed] [Google Scholar]
- 8. Renfree M. B., Hore T. A., Shaw G., Graves J. A., Pask A. J. (2009) Evolution of genomic imprinting. Insights from marsupials and monotremes. Annu. Rev. Genomics Hum. Genet. 10, 241–262 [DOI] [PubMed] [Google Scholar]
- 9. Koerner M. V., Pauler F. M., Huang R., Barlow D. P. (2009) The function of non-coding RNAs in genomic imprinting. Development 136, 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards C. A., Ferguson-Smith A. C. (2007) Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19, 281–289 [DOI] [PubMed] [Google Scholar]
- 11. Ben-Porath I., Cedar H. (2000) Imprinting. Focusing on the center. Curr. Opin. Genet. Dev. 10, 550–554 [DOI] [PubMed] [Google Scholar]
- 12. Reik W., Murrell A., Lewis A., Mitsuya K., Umlauf D., Dean W., Higgins M., Feil R. (2004) Chromosome loops, insulators, and histone methylation. New insights into regulation of imprinting in clusters. Cold Spring Harb. Symp. Quant. Biol. 69, 29–37 [DOI] [PubMed] [Google Scholar]
- 13. Verona R. I., Mann M. R., Bartolomei M. S. (2003) Genomic imprinting. Intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19, 237–259 [DOI] [PubMed] [Google Scholar]
- 14. Suzuki M. M., Bird A. (2008) DNA methylation landscapes. Provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 15. Ooi S. K., O'Donnell A. H., Bestor T. H. (2009) Mammalian cytosine methylation at a glance. J. Cell Sci. 122, 2787–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., Thomson J. A., Ren B., Ecker J. R. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goll M. G., Bestor T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 18. Klose R. J., Bird A. P. (2006) Genomic DNA methylation. The mark and its mediators. Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 19. Li E., Bestor T. H., Jaenisch R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 [DOI] [PubMed] [Google Scholar]
- 20. Okano M., Bell D. W., Haber D. A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 21. Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. (2001) Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 [DOI] [PubMed] [Google Scholar]
- 22. Kareta M. S., Botello Z. M., Ennis J. J., Chou C., Chédin F. (2006) Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 281, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 23. Chedin F., Lieber M. R., Hsieh C. L. (2002) The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. U.S.A. 99, 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hata K., Okano M., Lei H., Li E. (2002) Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 25. Li E., Beard C., Jaenisch R. (1993) Role for DNA methylation in genomic imprinting. Nature 366, 362–365 [DOI] [PubMed] [Google Scholar]
- 26. Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A. C. (2008) A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mackay D. J., Callaway J. L., Marks S. M., White H. E., Acerini C. L., Boonen S. E., Dayanikli P., Firth H. V., Goodship J. A., Haemers A. P., Hahnemann J. M., Kordonouri O., Masoud A. F., Oestergaard E., Storr J., Ellard S., Hattersley A. T., Robinson D. O., Temple I. K. (2008) Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 40, 949–951 [DOI] [PubMed] [Google Scholar]
- 28. Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Tanaka S., Shiota K., Nakano T. (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71 [DOI] [PubMed] [Google Scholar]
- 29. Payer B., Saitou M., Barton S. C., Thresher R., Dixon J. P., Zahn D., Colledge W. H., Carlton M. B., Nakano T., Surani M. A. (2003) Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13, 2110–2117 [DOI] [PubMed] [Google Scholar]
- 30. Li X., Leder P. (2007) Identifying genes preferentially expressed in undifferentiated embryonic stem cells. BMC Cell Biol. 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Looman C., Abrink M., Mark C., Hellman L. (2002) KRAB zinc finger proteins. An analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 19, 2118–2130 [DOI] [PubMed] [Google Scholar]
- 32. Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 33. Peng H., Begg G. E., Harper S. L., Friedman J. R., Speicher D. W., Rauscher F. J., 3rd (2000) Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J. Biol. Chem. 275, 18000–18010 [DOI] [PubMed] [Google Scholar]
- 34. Borden K. L. (2000) RING domains. Master builders of molecular scaffolds? J. Mol. Biol. 295, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 35. Peng H., Begg G. E., Schultz D. C., Friedman J. R., Jensen D. E., Speicher D. W., Rauscher F. J., 3rd (2000) Reconstitution of the KRAB-KAP-1 repressor complex. A model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 295, 1139–1162 [DOI] [PubMed] [Google Scholar]
- 36. Cammas F., Herzog M., Lerouge T., Chambon P., Losson R. (2004) Association of the transcriptional corepressor TIF1β with heterochromatin protein 1 (HP1). An essential role for progression through differentiation. Genes Dev. 18, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsuda E., Agata Y., Sugai M., Katakai T., Gonda H., Shimizu A. (2001) Targeting of Krüppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J. Biol. Chem. 276, 14222–14229 [DOI] [PubMed] [Google Scholar]
- 38. Ryan R. F., Schultz D. C., Ayyanathan K., Singh P. B., Friedman J. R., Fredericks W. J., Rauscher F. J., 3rd (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins. A potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell Biol. 19, 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng J., Wysocka J. (2008) It takes a PHD to SUMO. Trends Biochem. Sci. 33, 191–194 [DOI] [PubMed] [Google Scholar]
- 40. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeng L., Yap K. L., Ivanov A. V., Wang X., Mujtaba S., Plotnikova O., Rauscher F. J., 3rd, Zhou M. M. (2008) Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 15, 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones M. J., Bogutz A. B., Lefebvre L. (2011) An extended domain of Kcnq1ot1 silencing revealed by an imprinted fluorescent reporter. Mol. Cell Biol. 31, 2827–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maclean J. A., Bettegowda A., Kim B. J., Lou C. H., Yang S. M., Bhardwaj A., Shanker S., Hu Z., Fan Y., Eckardt S., McLaughlin K. J., Skoultchi A. I., Wilkinson M. F. (2011) The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol. Cell Biol. 31, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murakami K., Araki K., Ohtsuka S., Wakayama T., Niwa H. (2011) Choice of random rather than imprinted × inactivation in female embryonic stem cell-derived extra-embryonic cells. Development 138, 197–202 [DOI] [PubMed] [Google Scholar]
- 45. Borowczyk E., Mohan K. N., D'Aiuto L., Cirio M. C., Chaillet J. R. (2009) Identification of a region of the DNMT1 methyltransferase that regulates the maintenance of genomic imprints. Proc. Natl. Acad. Sci. U.S.A. 106, 20806–20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lewis A., Green K., Dawson C., Redrup L., Huynh K. D., Lee J. T., Hemberger M., Reik W. (2006) Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development 133, 4203–4210 [DOI] [PubMed] [Google Scholar]
- 47. Iacovino M., Hernandez C., Xu Z., Bajwa G., Prather M., Kyba M. (2009) A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 18, 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ge Y. Z., Pu M. T., Gowher H., Wu H. P., Ding J. P., Jeltsch A., Xu G. L. (2004) Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279, 25447–25454 [DOI] [PubMed] [Google Scholar]
- 49. Humpherys D., Eggan K., Akutsu H., Hochedlinger K., Rideout W. M., 3rd, Biniszkiewicz D., Yanagimachi R., Jaenisch R. (2001) Epigenetic instability in ES cells and cloned mice. Science 293, 95–97 [DOI] [PubMed] [Google Scholar]
- 50. Schumacher A., Doerfler W. (2004) Influence of in vitro manipulation on the stability of methylation patterns in the Snurf/Snrpn-imprinting region in mouse embryonic stem cells. Nucleic Acids Res. 32, 1566–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiznerowicz M., Jakobsson J., Szulc J., Liao S., Quazzola A., Beermann F., Aebischer P., Trono D. (2007) The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 282, 34535–34541 [DOI] [PubMed] [Google Scholar]
- 52. Wolf D., Goff S. P. (2007) TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 [DOI] [PubMed] [Google Scholar]
- 53. Hu G., Kim J., Xu Q., Leng Y., Orkin S. H., Elledge S. J. (2009) A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23, 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolf D., Goff S. P. (2009) Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li J. Y., Pu M. T., Hirasawa R., Li B. Z., Huang Y. N., Zeng R., Jing N. H., Chen T., Li E., Sasaki H., Xu G. L. (2007) Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell Biol. 27, 8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fazzio T. G., Huff J. T., Panning B. (2008) An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seki Y., Kurisaki A., Watanabe-Susaki K., Nakajima Y., Nakanishi M., Arai Y., Shiota K., Sugino H., Asashima M. (2010) TIF1β regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 107, 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Latos P. A., Stricker S. H., Steenpass L., Pauler F. M., Huang R., Senergin B. H., Regha K., Koerner M. V., Warczok K. E., Unger C., Barlow D. P. (2009) An in vitro ES cell imprinting model shows that imprinted expression of the Igf2r gene arises from an allele-specific expression bias. Development 136, 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tucker K. L., Talbot D., Lee M. A., Leonhardt H., Jaenisch R. (1996) Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc. Natl. Acad. Sci. U.S.A. 93, 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tucker K. L., Beard C., Dausmann J., Jackson-Grusby L., Laird P. W., Lei H., Li E., Jaenisch R. (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10, 1008–1020 [DOI] [PubMed] [Google Scholar]