Abstract
Chronic beryllium disease (CBD) is a granulomatous lung disorder caused by beryllium (Be) exposure in the workplace. It is characterized by the accumulation of Be-specific CD4+ T cells in the lung as well as persistent lung inflammation, culminating in the development of lung fibrosis. CBD occurs in 2 to 16% of Be-exposed workers depending on the individuals' genetic susceptibility and the characteristics of the exposure. Genetic susceptibility to Be-induced disease has been linked to major histocompatibility complex class II molecules. In particular, HLA-DP alleles possessing a glutamic acid at the 69th position of the β-chain (βGlu69) are most strongly linked to disease susceptibility. The HLA-DP alleles that present Be to T cells match those implicated in the genetic susceptibility, suggesting that the HLA contribution to disease is based on the ability of those molecules to bind and present Be to T cells. However, the structural features of βGlu69-containing HLA-DP molecules that explain the disease association remain unknown. We have recently crystallized HLA-DP2, which is the most prevalent of the βGlu69-containing HLA-DP molecules. Its unique structure, which includes surface exposure of βGlu69, provides an explanation of the genetic linkage between βGlu69-containing HLA-DP alleles and Be-induced disease.
Keywords: beryllium, granuloma, human, lung, T cells
Beryllium (Be) is a lightweight metal with unique chemical and physical properties that make it ideally suited for use in high-technology industries, such as aerospace, ceramics, electronics, and defense (1). Beryllium exposure primarily occurs through inhalation in workers involved in machining Be-containing products. It is estimated that approximately 200,000 current and at least 1 million total workers have been exposed to Be in the United States alone (2, 3). Depending on the nature of the exposure and the genetic susceptibility of the individual, chronic beryllium disease (CBD) develops in 2 to 16% of these subjects (4–8). Thus, CBD remains an important public health concern.
Workplace screening of Be-exposed workers has identified individuals sensitized to Be who have no evidence of lung disease. These Be-sensitized subjects demonstrate a Be-specific immune response in peripheral blood but have no clinical, radiographic, or histopathologic features of CBD (1, 9). A subset of Be-sensitized subjects progress to CBD at a rate of 6 to 8% per year (9). CBD is characterized by the presence of noncaseating granulomatous inflammation that primarily affects the lung, although other organs may be involved (1, 10, 11). The diagnosis depends on the detection of a Be-specific immune response in blood and/or lung and the presence of noncaseating granulomas and/or mononuclear cell inflammation on a biopsy specimen (12). The histopathologic features of CBD are identical to those seen in sarcoidosis (13, 14). Due to the persistence of Be in the lung years after the cessation of exposure (15, 16), the natural history of CBD is characterized by a gradual decline in lung function, with one-third of untreated patients historically progressing to end-stage respiratory insufficiency (17).
Recently, major advances in our understanding of the immunopathogenesis of Be-induced disease have occurred. This review will focus on the link between major histocompatibility complex class II (MHCII) molecules, which have been strongly linked to disease susceptibility, and activation of Be-responsive CD4+ T cells.
CBD IS A CD4+ T CELL–MEDIATED DISORDER
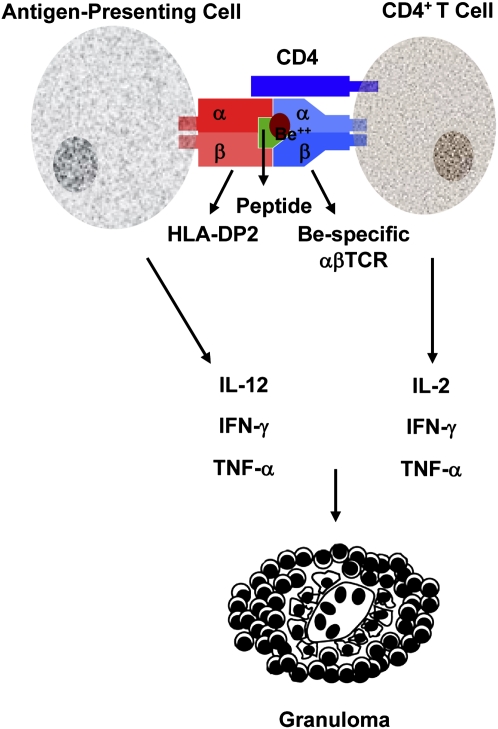
The healthy human lung contains few lymphocytes. In comparison, the lungs of patients with CBD are characterized by a CD4+ T cell alveolitis (18–22). Evidence suggests that these CD4+ T cells play a critical role in the immunopathogenesis of CBD, while CD8+ T cells play a minor, if any, role in the disease process (18, 19, 21, 22). Using an in vivo skin model of granulomatous inflammation, the development of granulomas was preceded by the influx into skin of CD4+ T cells expressing identical T cell receptors (TCRs) to those clones found in the bronchoalveolar lavage (BAL) of the same patients, confirming the importance of these antigen-specific CD4+ T cells in the initiation of the granulomatous response (23). Furthermore, in the lungs of patients with CBD, a remarkably large number of Be-responsive CD4+ T cells accumulate (24). Be-specific T cells are specific for patients with CBD (i.e., cells expressing these TCRs are not found in the lungs of patients with other granulomatous or autoimmune diseases) and persist at high frequency in patients with active disease (21). These cells also express markers of previous activation and exhibit an effector memory T cell phenotype (characterized by expression of CD45RO and the loss of the lymph node homing receptors, CD62L and CCR7) (20, 24, 25). After Be recognition, CD4+ T cells undergo clonal proliferation and secrete T helper 1 (Th1)-type cytokines such as interleukin-2 (IL-2), interferon-γ (IFN-γ), and tumor necrosis factor-α (TNF-α) (Figure 1) (24, 26). Th2-type cytokines are not detected in the lungs of patients with CBD (24, 26). Importantly, IFN-γ and TNF-α promote macrophage accumulation, activation, and aggregation, resulting in the initiation of the granulomatous response.
Figure 1.
Immunopathogenesis of chronic beryllium disease. Following the inhalation of Be-containing particulates, antigen-presenting cells expressing a βGlu69-containing HLA-DP molecule present Be to CD4+ T cells resulting in T cell activation, proliferation, and Th1-type cytokine production.
Although the vast majority of Be-specific T cells are compartmentalized to the lung, a greater frequency of antigen-specific cells in blood strongly correlates with the extent of lung inflammation, as measured by the severity of the CD4+ T cell alveolitis (27). These findings raise the possibility that determination of the number of circulating Be-specific T cells may provide a glimpse of the inflammatory response occurring in the lung without the need for invasive procedures.
GENETIC SUSCEPTIBILITY TO BERYLLIUM-INDUCED DISEASE
In addition to Be exposure in the workplace, genetic susceptibility plays a major role in the immunopathogenesis of CBD. Susceptibility to CBD has been linked to HLA-DP alleles that contain a glutamic acid at position 69 of the β-chain (βGlu69) (28–36). Richeldi and coworkers (28) showed that HLA-DPB1*0201 was associated with the development of CBD, whereas DPB1*0401 was protective. Sequence analysis showed that DPB1 alleles with βGlu69, which includes DPB1*0201, were strongly associated with disease susceptibility (28). Other studies have confirmed these findings, documenting the presence of βGlu69-containing DPB1 alleles in approximately 80% of patients with CBD compared with 30 to 48% of exposed controls (29–36). Recent studies have shown that βGlu69-containing DPB1 alleles are a risk factor for the development of Be sensitization and not simply a marker of progression from sensitization to disease (32–35). In addition, rare HLA-DP alleles carrying βGlu69 (i.e., non-HLA-DPB1*0201 alleles), more than the HLA-DP βGlu69 itself, were associated with CBD in some studies (30, 32, 33).
Approximately 20% of patients with CBD do not possess a βGlu69-containing HLA-DPB1 allele, suggesting the importance of other MHCII molecules in the genetic susceptibility to Be-induced disease. In this subset of patients with CBD, an increased frequency of HLA-DR13 alleles was observed (33, 34). These alleles possess a glutamic acid at position 71 of the DR β-chain (βGlu71), which corresponds to position 69 of HLA-DP. Another study showed that the presence of a phenylalanine at position 47 of the β-chain of HLA-DR was independently associated with Be sensitization and CBD in those subjects without a βGlu69-containing HLA-DP allele (36).
BERYLLIUM PRESENTATION TO CD4+ T CELLS
Human MHCII molecules have been implicated in susceptibility to various immune-mediated diseases. Due to the unknown initiating autoantigen(s) in the majority of these disorders, the mechanism for the association is poorly defined. Conversely, CBD allowed a determination of whether the association of HLA-DP molecules and disease susceptibility occurs at the level of antigen presentation. Using Be-specific CD4+ T cells and either lymphoblastoid cells or human DNA-engineered mouse fibroblasts as antigen-presenting cells, investigators have shown that most Be presentation occurs through HLA-DP (37, 38). However, only certain HLA-DP molecules are capable of presenting Be to pathogenic CD4+ T cells (37). The DPB1 alleles that mediate Be presentation match those implicated in disease susceptibility, providing a mechanism for the association of these HLA-DP alleles and disease susceptibility (37, 38). In addition, Be recognition required the presence of βGlu69, raising the possibility that a single polymorphic amino acid might dictate Be presentation and, more importantly, disease susceptibility. Studies using soluble HLA-DP molecules expressing βGlu69, but not HLA-DP molecules with a lysine at that position, corroborate these findings (39, 40).
In subjects without a βGlu69-containing HLA-DP allele, recent studies have shown that HLA-DR assumes the predominant role in Be presentation (36, 41). For example, HLA-DR13 molecules are capable of presenting Be to Be-responsive T cells (41), and mutational analysis revealed that the critical polymorphic amino acid in the DR13 molecules is βGlu71 (41). Thus, a hierarchy of Be presentation to CD4+ T cells has been established: HLA-DP assuming the predominant role, with a minor role for HLA-DR and no known role for HLA-DQ.
STRUCTURAL BASIS FOR THE ASSOCIATION OF βGlu69-CONTAINING HLA-DP ALLELES AND DISEASE SUSCEPTIBILITY
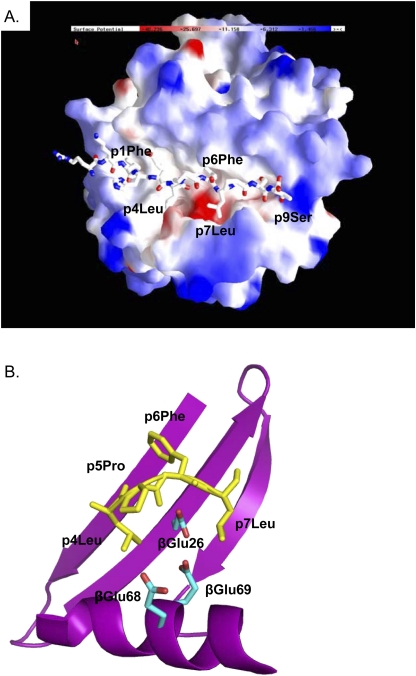
The structures of at least 10 disease-associated HLA-DR and -DQ molecules have been reported. Conversely, the HLA-DP molecule has been recalcitrant to attempts at crystallization. As a result, the unique structural features of βGlu69-containing HLA-DP molecules that explain the disease association have remained unknown. Recently, we crystallized HLA-DP2 (DPA1*0103, DPB1*0201) in complex with a self-peptide derived from the HLA-DR α-chain (pDRA) (42) and solved its structure to a resolution of 3.25 Å (43). Although the overall structure of the DP2-pDRA complex was similar to that of other MHCII/peptide complexes, two unique structural features were identified that may explain the ability of HLA-DP2 to present Be to T cells. The first was a widening of the gap between the peptide and the β-chain α-helix, with DP2 having the largest gap of any of the 29 published MHCII structures (43). This region of the β-chain α-helix may be quite flexible (44), explaining the large variation in the width of this part of the binding groove. The second distinct feature of the DP2 peptide binding groove was an upward shift in the DR α-chain peptide such that the leucine at the p4 position of the peptide was solvent-exposed. As a result of these structural changes, an acidic pocket was solvent-exposed (Figure 2A) (43). This pocket was flanked by leucine residues at the p4 and p7 positions of the pDRA and the β-chain α-helix and was composed of three DP2 β-chain amino acids that contribute to the net negative surface charge of this pocket: βGlu68 and βGlu69 from the β-chain α-helix and βGlu26 from the floor of the peptide binding groove (Figure 2B) (43).
Figure 2.
HLA-DP2 possesses a solvent-exposed acidic pocket which includes βGlu69. (A) The electrostatic surface charge of the HLA-DP2 molecule (with bound pDRA) is shown colored by the relative charge of the surface atoms (red, negative; blue, positive) (program GRASP). A wireframe representation of pDRA is shown with CPK coloring. (B) DP2 molecule is shown in the area between p4Leu and the DP2 β-chain α-helix with ribbon and wireframe representations of the DP2 β-chain (magenta) and pDRA (yellow coloring), respectively. Wireframe representations of the side chains of βGlu26, βGlu68, and βGlu69 are also shown.
In addition to βGlu69, these findings raised the possibility that βGlu26 and βGlu68 of HLA-DP2 may be involved in Be coordination and presentation. Since these two amino acids are invariant among HLA-DP alleles (Table 1), their presence is not sufficient for Be presentation in the absence of βGlu69. Site-directed mutagenesis of βGlu26 and βGlu68 and expression of the mutated HLA-DP2 molecules on the surface of fibroblasts did not activate beryllium-specific T cells, confirming the importance of these additional glutamic acid residues in Be coordination and presentation (43). Of note, the HLA-DR13 alleles, which are linked to disease susceptibility and can present Be to Be-responsive T cells, also possess an acidic cluster composed of βAsp28, βAsp70, and βGlu71. Together, these findings add further support for the critical nature of this acidic pocket in Be coordination and subsequent T cell activation.
TABLE 1.
AMINO ACID RESIDUES OF HLA-DPB1 ALLELES INVOLVED IN CHRONIC BERYLLIUM DISEASE SUSCEPTIBILITY
| HLA-DPB1 Allele | Ability to present Be | Amino Acid Position* |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 9 | 11 | 26 | 35 | 36 | 55 | 56 | 57 | 67 | 68 | 69 | 76 | 84 | 85 | 86 | 87 | ||
| DPB1*0101 | — | V | Y | G | E | Y | A | A | A | E | E | E | K | V | D | E | A | V |
| DPB1*0401 | — | L | F | G | E | F | A | A | A | E | E | E | K | M | G | G | P | M |
| DPB1*0402 | — | L | F | G | E | F | V | D | E | E | E | E | K | M | G | G | P | M |
| DPB1*0501 | — | L | F | G | E | F | V | E | A | E | E | E | K | V | D | E | A | V |
| DPB1*0201 | + | L | F | G | E | F | V | D | E | E | E | E | E | M | G | G | P | M |
| DPB1*0601 | + | V | Y | L | E | F | V | D | E | D | E | E | E | M | D | E | A | V |
| DPB1*1001 | + | V | H | L | E | F | V | D | E | E | E | E | E | V | D | E | A | V |
| DPB1*1301 | + | V | Y | L | E | Y | A | A | A | E | E | E | E | M | D | E | A | V |
| DPB1*1701 | + | V | H | L | E | F | V | D | E | D | E | E | E | M | D | E | A | V |
Sequences for the different HLA-DP alleles are presented based on homology to DPB1*0101. The bolded amino acid residues at positions 26, 68, and 69 indicate the three glutamic acid residues that comprise the solvent-exposed acidic cluster of HLA-DP2 and are shared between the presenting HLA-DP molecules.
SUMMARY
The combination of genetic susceptibility studies, along with functional and structural research, strongly suggest that the acidic pocket of the HLA-DP2 binding groove is the Be binding site and provides an explanation for the genetic linkage of HLA-DP2 to the development of granulomatous inflammation in the Be-exposed worker. However, despite these recent advances in our understanding of the immunopathogenesis of Be-induced disease, important unanswered questions remain. For example, which MHCII-bound peptides are capable of completing the Be-responsive αβTCR ligand? Are Be-responsive T cells recognizing Be alone or conformational changes in the self-peptide induced by Be? Hopefully, answers to these and other questions will allow disease prevention in susceptible individuals as well as the development of new therapeutic strategies to prevent the development of fibrotic lung disease in patients with CBD.
Supported by the following NIH grants: HL62410, HL92997, and ES011810 (to A.P.F.), and by the Clinical Translational Research Center (UL1 RR025780) from the National Center for Research Resources.
Conflict of Interest Statement: M.T.F. received grant support from the Foundation for Sarcoidosis Research ($50,001–$100,000). N.A.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.W.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.P.F. received lecture fees from Amgen (up to $1,000) and grant support ($50,001–$100,000). He receives royalties from UpToDate (up to $1,000) and received grant support from the National Institutes of Health ($100,001 or more).
References
- 1.Newman LS, Maier LA. Beryllium disease. In: Schwarz MI, King TE Jr, editors. Interstitial lung disease., 3rd ed. Hamilton: B.C. Decker Inc.; 2003. pp. 435–451.
- 2.Henneberger PK, Goe SK, Miller WE, Doney B, Groce DW. Industries in the United States with airborne beryllium exposure and estimates of the number of current workers potentially exposed. J Occup Environ Hyg 2004;1:648–659. [DOI] [PubMed] [Google Scholar]
- 3.Newman LS, Rose CS, Bresnitz EA, Rossman MD, Barnard J, Frederick M, Terrin ML, Weinberger SE, Moller DR, McLennan G, et al. A case control etiologic study of sarcoidosis: environmental and occupational risk factors. Am J Respir Crit Care Med 2004;170:1324–1330. [DOI] [PubMed] [Google Scholar]
- 4.Kreiss K, Newman LS, Mroz M, Campbell PA. Screening blood test identifies subclinical beryllium disease. J Occup Med 1989;31:603–608. [DOI] [PubMed] [Google Scholar]
- 5.Kreiss K, Wasserman S, Mroz MM, Newman LS. Beryllium disease screening in the ceramics industry: blood test performance and exposure-disease relations. J Occup Med 1993;35:267–274. [PubMed] [Google Scholar]
- 6.Kreiss K, Mroz MM, Zhen B, Martyny JW, Newman LS. Epidemiology of beryllium sensitization and disease in nuclear workers. Am Rev Respir Dis 1993;148:985–991. [DOI] [PubMed] [Google Scholar]
- 7.Kreiss K, Mroz MM, Newman LS, Martyny J, Zhen B. Machining risk of beryllium disease and sensitization with median exposures below 2 μg/m3. Am J Ind Med 1996;30:16–25. [DOI] [PubMed] [Google Scholar]
- 8.Henneberger PK, Cumro D, Deubner DD, Kent MS, McCawley M, Kreiss K. Beryllium sensitization and disease among long-term and short-term workers in a beryllium ceramics plant. Int Arch Occup Environ Health 2001;74:167–176. [DOI] [PubMed] [Google Scholar]
- 9.Newman LS, Mroz MM, Balkissoon R, Maier LA. Beryllium sensitization progresses to chronic beryllium disease: a longitudinal study of disease risk. Am J Respir Crit Care Med 2004;171:54–60. [DOI] [PubMed] [Google Scholar]
- 10.Fontenot AP, Newman LS, Kotzin BL. Chronic beryllium disease: T cell recognition of a metal presented by HLA-DP. Clin Immunol 2001;100:4–14. [DOI] [PubMed] [Google Scholar]
- 11.Fontenot AP, Kotzin BL. Chronic beryllium disease: immune-mediated destruction with implications for organ-specific autoimmunity. Tissue Antigens 2003;62:449–458. [DOI] [PubMed] [Google Scholar]
- 12.Newman LS, Kreiss K, King TE Jr, Seay S, Campbell PA. Pathologic and immunologic alterations in early stages of beryllium disease: re-examination of disease definition and natural history. Am Rev Respir Dis 1989;139:1479–1486. [DOI] [PubMed] [Google Scholar]
- 13.Newman LS, Kreiss K. Nonoccupational beryllium disease masquerading as sarcoidosis: identification by blood lymphocyte proliferation response to beryllium. Am Rev Respir Dis 1992;145:1212–1214. [DOI] [PubMed] [Google Scholar]
- 14.Newman LS, Rose CS, Maier LA. Sarcoidosis. N Engl J Med 1997;336:1224–1234. [DOI] [PubMed] [Google Scholar]
- 15.Jones-Williams W, Kelland D. New aid for diagnosing chronic beryllium disease (CBD): laser ion mass analysis (LIMA). J Clin Pathol 1986;39:900–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones-Williams W, Wallach ER. Laser microprobe mass spectrometry (LAMMS) analysis of beryllium, sarcoidosis, and other granulomatous diseases. Sarcoidosis 1989;6:111–117. [PubMed] [Google Scholar]
- 17.Newman LS, Lloyd J, Daniloff E. The natural history of beryllium sensitization and chronic beryllium disease. Environ Health Perspect 1996;104:937S–943S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossman MD, Kern JA, Elias JA, Cullen MR, Epstein PE, Preuss OP, Markham TN, Daniele RP. Proliferative response of bronchoalveolar lymphocytes to beryllium. Ann Intern Med 1988;108:687–693. [DOI] [PubMed] [Google Scholar]
- 19.Saltini C, Winestock K, Kirby M, Pinkston P, Crystal RG. Maintenance of alveolitis in patients with chronic beryllium disease by beryllium-specific helper T cells. N Engl J Med 1989;320:1103–1109. [DOI] [PubMed] [Google Scholar]
- 20.Saltini C, Kirby M, Trapnell BC, Tamura N, Crystal RG. Biased accumulation of T lymphocytes with “memory”-type CD45 leukocyte common antigen gene expression on the epithelial surface of the human lung. J Exp Med 1990;171:1123–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontenot AP, Falta MT, Freed BM, Newman LS, Kotzin BL. Identification of pathogenic T cells in patients with beryllium-induced lung disease. J Immunol 1999;163:1019–1026. [PubMed] [Google Scholar]
- 22.Fontenot AP, Kotzin BL, Comment CE, Newman LS. Expansions of T-cell subsets expressing particular T cell receptor variable regions in chronic beryllium disease. Am J Respir Cell Mol Biol 1998;18:581–589. [DOI] [PubMed] [Google Scholar]
- 23.Fontenot AP, Maier LA, Canavera SJ, Hendry-Hofer TB, Boguniewicz M, Barker EA, Newman LS, Kotzin BL. Beryllium skin patch testing to analyze T cell stimulation and granulomatous inflammation in the lung. J Immunol 2002;168:3627–3634. [DOI] [PubMed] [Google Scholar]
- 24.Fontenot AP, Canavera SJ, Gharavi L, Newman LS, Kotzin BL. Target organ localization of memory CD4+ T cells in patients with chronic beryllium disease. J Clin Invest 2002;110:1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontenot AP, Palmer BE, Sullivan AK, Joslin FG, Wilson CC, Maier LA, Newman LS, Kotzin BL. Frequency of beryllium-specific, central memory CD4+ T cells in blood determines proliferative response. J Clin Invest 2005;115:2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tinkle SS, Kittle LA, Schumacher BA, Newman LS. Beryllium induces IL-2 and IFN-γ in berylliosis. J Immunol 1997;158:518–526. [PubMed] [Google Scholar]
- 27.Pott GB, Palmer BE, Sullivan AK, Silviera L, Maier LA, Newman LS, Kotzin BL, Fontenot AP. Frequency of beryllium-specific, TH1-type cytokine-expressing CD4+ T cells in patients with beryllium-induced disease. J Allergy Clin Immunol 2005;115:1036–1042. [DOI] [PubMed] [Google Scholar]
- 28.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science 1993;262:242–244. [DOI] [PubMed] [Google Scholar]
- 29.Richeldi L, Kreiss K, Mroz MM, Zhen B, Tartoni P, Saltini C. Interaction of genetic and exposure factors in the prevalence of berylliosis. Am J Ind Med 1997;32:337–340. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, White PS, Petrovic M, Tatum OL, Newman LS, Maier LA, Marrone BL. Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and -DPA1 alleles. J Immunol 1999;163:1647–1653. [PubMed] [Google Scholar]
- 31.Saltini C, Richeldi L, Losi M, Amicosante M, Voorter C, van den Berg-Loonen E, Dweik RA, Wiedemann HP, Deubner DC, Tinelli C. Major histocompatibility locus genetic markers of beryllium sensitization and disease. Eur Respir J 2001;18:677–684. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Farris GM, Newman LS, Shou Y, Maier LA, Smith HN, Marrone BL. Beryllium sensitivity is linked to HLA-DP genotype. Toxicology 2001;165:27–38. [DOI] [PubMed] [Google Scholar]
- 33.Rossman MD, Stubbs J, Lee CW, Argyris E, Magira E, Monos D. Human leukocyte antigen Class II amino acid epitopes: susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med 2002;165:788–794. [DOI] [PubMed] [Google Scholar]
- 34.Maier LA, McGrath DS, Sato H, Lympany P, Welsh K, Du Bois R, Silveira L, Fontenot AP, Sawyer RT, Wilcox E, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol 2003;171:6910–6918. [DOI] [PubMed] [Google Scholar]
- 35.McCanlies EC, Ensey JS, Schuler CR, Kreiss K, Weston A. The association between HLA-DPB1Glu69 and chronic beryllium disease and beryllium sensitization. Am J Ind Med 2004;46:95–103. [DOI] [PubMed] [Google Scholar]
- 36.Amicosante M, Berretta F, Rossman M, Butler RH, Rogliani P, van den Berg-Loonen E, Saltini C. Identification of HLA-DRPheβ47 as the susceptibility marker of hypersensitivity to beryllium in individuals lacking the berylliosis-associated supratypic marker HLA-DPGlubeta69. Respir Res 2005;6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontenot AP, Torres M, Marshall WH, Newman LS, Kotzin BL. Beryllium presentation to CD4+ T cells underlies disease susceptibility HLA-DP alleles in chronic beryllium disease. Proc Natl Acad Sci USA 2000;97:12717–12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lombardi G, Germain C, Uren J, Fiorillo MT, du Bois RM, Jones-Williams W, Saltini C, Sorrentino R, Lechler R. HLA-DP allele-specific T cell responses to beryllium account for DP-associated susceptibility to chronic beryllium disease. J Immunol 2001;166:3549–3555. [DOI] [PubMed] [Google Scholar]
- 39.Amicosante M, Sanarico N, Berretta F, Arroyo J, Lombardi G, Lechler R, Colizzi V, Saltini C. Beryllium binding to HLA-DP molecule carrying the marker of susceptibility to berylliosis glutamate beta 69. Hum Immunol 2001;62:686–693. [DOI] [PubMed] [Google Scholar]
- 40.Fontenot AP, Keizer TS, McCleskey M, Mack DG, Meza-Romero R, Huan J, Edwards DM, Chou YK, Vandenbark AA, Scott B, et al. Recombinant HLA-DP2 binds beryllium and tolerizes beryllium-specific pathogenic CD4+ T cells. J Immunol 2006;177:3874–3883. [DOI] [PubMed] [Google Scholar]
- 41.Bill JR, Mack DG, Falta MT, Maier LA, Sullivan AK, Joslin FG, Martin AK, Freed BM, Kotzin BL, Fontenot AP. Beryllium presentation to CD4+ T cells is dependent on a single amino acid residue of the MHC class II β-chain. J Immunol 2005;175:7029–7037. [DOI] [PubMed] [Google Scholar]
- 42.Diaz G, Canas B, Vazquez J, Nombela C, Arroyo J. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Immunogenetics 2005;56:754–759. [DOI] [PubMed] [Google Scholar]
- 43.Dai S, Murphy GA, Crawford F, Mack DG, Falta MT, Marrack P, Kappler JW, Fontenot AP. Crystal strucutre of HLA-DP2: implications for chronic beryllium disease. Proc Natl Acad Sci USA (In press). [DOI] [PMC free article] [PubMed]
- 44.Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS ONE 2008;3:e2403. [DOI] [PMC free article] [PubMed] [Google Scholar]