Abstract
Inherited retinal degeneration in Drosophila has been explored for insights into similar processes in humans. Based on the mechanisms, I divide these mutations in Drosophila into three classes. The first consists of genes that control the specialization of photoreceptor cells including the morphogenesis of visual organelles (rhabdomeres) that house the visual signaling proteins. The second class contains genes that regulate the activity or level of the major rhodopsin, Rh1, which is the light sensor and also provides a structural role for the maintenance of rhabdomeres. Some mutations in Rh1 (NinaE) are dominant due to constitutive activity or folding defects, like autosomal dominant retinitis pigmentosa (ADRP) in humans. The third class consists of genes that control the Ca2+ influx directly or indirectly by promoting the turnover of the second messenger and regeneration of PIP2, or mediate the Ca2+-dependent regulation of the visual response. These gene products are critical for the increase in cytosolic Ca2+ following light stimulation to initiate negative regulatory events. Here I will focus on the signaling mechanisms underlying the degeneration in norpA, and in ADRP-type NinaE mutants that produce misfolded Rh1. Accumulation of misfolded Rh1 in the ER triggers the unfolded protein response (UPR), while endosomal accumulation of activated Rh1 may initiate autophagy in norpA. Both autophagy and the UPR are beneficial for relieving defective endosomal trafficking and the ER stress, respectively. However, when photoreceptors fail to cope with the persistence of these stresses, a cell death program is activated leading to retinal degeneration.
Keywords: Drosophila, autosomal dominant retinitis pigmentosa, rhodopsin, arrestin, rhabdomeres, vision, phospholipase Cβ, unfolded protein response, apoptosis, autophagy
Introduction
Drosophila is an excellent model organism to explore the molecular basis of retinal degeneration. Retinal degeneration in the fly is characterized by the initial loss of rhabdomeres, the visual organelles, followed by deterioration of photoreceptor cell bodies. The power of genetics allows the identification of mutants using a simple screening method, the loss of deep pseudopupil (dpp). Dpp represents the autofluorescence of the Rh1 rhodopsin in rhabdomeres of the major photoreceptor cells (R1–R6) from several adjacent unit eyes of the compound eye.1 The absence of dpp may be due to a loss of Rh1, which likely results from a loss of rhabdomeres contributed by degeneration of photoreceptor cells. Characterization of retinal degeneration mutants is greatly aided by our knowledge of rhabdomere morphogenesis.2 Moreover, the use of genetic tools including tissue-specific overexpression3,4 and RNAi-mediated knockdown5,6 provide additional avenues for uncovering mechanistic insights leading to photoreceptor death. In essence, degeneration of photoreceptor cells is often accompanied by defective development of rhabdomeres or aberrant visual signaling. As an introduction, a brief background concerning the visual signaling pathway, structure of the compound eye, and terminal differentiation of photoreceptor cells will be provided.
Structure of the Compound Eye
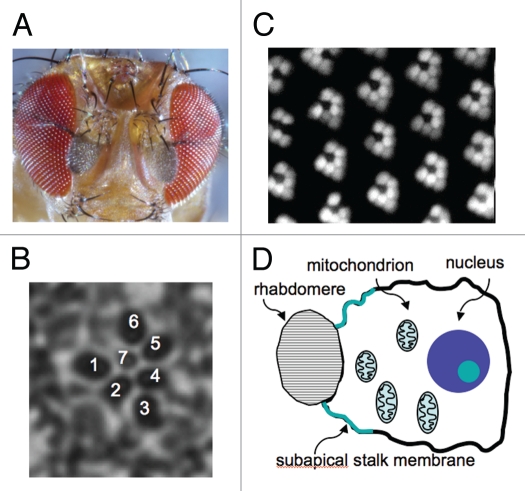
The major visual system of Drosophila and dipteran insects consists of two compound eyes, each of which contains approximately 800 unit eyes or ommatidia in Drosophila (Fig. 1). Each ommatidium is made up of 22 cells including eight photoreceptor cells, R1–R8, which are arranged in a cylindrical pattern with the outer photoreceptor cells, R1–R6, occupying the peripheral region (Fig. 1B), whereas the central photoreceptor cells (R7 and R8) reside in the central region with R7 distally located while R8 proximally. R1–R6 are the major photoreceptor cells in Drosophila. Similar to vertebrate rod photoreceptors in which rod outer segments house the signaling proteins involved in the visual cascade, Drosophila photoreceptor cells possess specialized visual organelles, rhabdomeres, in which visual signaling proteins reside (Fig. 1D). Each rhabdomere consists of densely stacked membrane structures of about 60,000 microvilli supported by an actin-based cytoskeleton (Fig. 1C).7
Figure 1.
Structure of the compound eye in Drosophila. (A) Each compound eye consists of 600–800 unit eyes (ommatidia). (B) Each ommatidium contains eight photoreceptors, R1–R8. Shown is a cross section of the distal region of an ommatidium, which reveals the R1–R7 photoreceptors with the stereotypical arrangement of rhabdomeres. Rhabdomere is the visual organelle in photoreceptor cells, and consists of densely packed membranes supported by the actin cytoskeleton (C and D). Shown is the rhabdomere of the R1–R6 photoreceptors revealed by epifluorescence of actin tagged with the green fluorescent protein (C). (D) A diagrammatic depiction of a photoreceptor cell (cross section).
Six Rhodopsins, Rh1–Rh6, are Expressed in Drosophila Photoreceptors
The sensitivity of photoreceptor cells to a particular wavelength of light is governed by the nature of the rhodopsin it expresses.8,9 Drosophila expresses six distinct rhodopsins, Rh1–Rh6, and senses a wide spectrum of light including ultraviolet (UV) light. Rhodopsin 1 (Rh1) is the major rhodopsin with absorption maximum at 480 nM, and is present in R1–R6 photoreceptor cells.10,11 The abundance of Rh1 rhodopsin in the compound eye is parallel to that of rod rhodopsin in vertebrate retinas. In Drosophila, two UV-absorbing rhodopsins, rhodopsin 3 and rhodopsin 4, are found in non-overlapping R7 photoreceptor cells.12,13 In contrast, R8 photoreceptor cells express either rhodopsin 5,14 or rhodopsin 6 15 conferring the sensitivity of these cells to either blue or green light, respectively. Lastly, rhodopsin 2 is found in the ocelli,16 the three simple eyes located on the vertex of the head, and is critical for visual guidance. All Drosophila rhodopsins belong to the class A subfamily17 of G-protein coupled receptors (GPCRs), and are the most abundant membrane proteins in the rhabdomere of the photoreceptor cell.
Morphogenesis of Rhabdomeres
The shape and content of the rhabdomere support the critical elements of photoreceptor cell's function and survival. Without the completion of the morphogenesis program, photoreceptor cells will not achieve the desired shape. In particular, malformation of the rhabdomere renders it unable to accommodate the signaling machinery, affecting both visual transduction and maintenance of photoreceptor cells, defects in which eventually bring about retinal degeneration.
In Drosophila, morphogenesis and terminal differentiation of photoreceptors start at the mid-pupal stage, resulting in the elaboration of rhabdomeres (reviewed in ref. 2). These events are orchestrated by the Crumbs complex [Crumbs, Stardust and DPATJ (PALS1-associated tight junction protein)], which first initiates the subdivision of the apical membrane into two distinct regions, and subsequently controls the specialization of these regions into subapical stalk membranes and rhabdomeres (Fig. 1D). This initial division of the apical domain is mediated by Bazooka (Baz),18 a scaffolding protein containing three PDZ (PSD95, discs-large, ZO-1) domains. Baz participates in the recruitment of PTEN (Phosphatase and Tensin Homolog deleted on chromosome 10), a lipid phosphatase that converts phosphatidylinositol-3,4,5-trisphosphate (PIP3) into phosphatidylinositol-4,5-bisphosphate (PIP2).19 PIP2 is critical for actin polymerization20 whereas PIP3 activates AKT/protein kinase B.21 Mutants missing PTEN display defects in rhabdomeres and stalk membranes, indicating that specialization of photoreceptor cells is sensitive to the relative level of these two phosphoinositides.22
As indicated, the Crumbs protein complex is implicated in the expansion of the subapical stalk membrane, and the elongation of rhabdomeres. Indeed, mutants devoid of components of the Crumbs complex display a reduction of the stalk membrane and a failure in the rhabdomere elongation.18,23–25 The elaboration of rhabdomeres enables photoreceptor cells to increase the amount of the membrane in order to accommodate about 108 molecules of Rh1 rhodopsin.26 This high density of rhodopsin in rhabdomeres makes the visual signaling exquisitely sensitive. Moreover, compartmentalization of the signaling molecules within the rhabdomere facilitates fast kinetics of the visual signaling by promoting protein-protein interactions, thus enhancing the kinetics of the photoresponse.
Visual Signaling in Drosophila
Photoreceptors in the eye are highly specialized neurons that provide a window for the brain to see the world. These neurons utilize a set of signaling proteins to detect the light signal and convert it into an electrical impulse, which then propagates to neurons in the visual brain. Specifically, activation of the visual signaling cascade generates a second messenger that regulates the ion flux, resulting in a change of membrane potential of photoreceptors. In essence, the visual signaling mechanism operates in a similar way to that mediated by a prototypical GPCR.
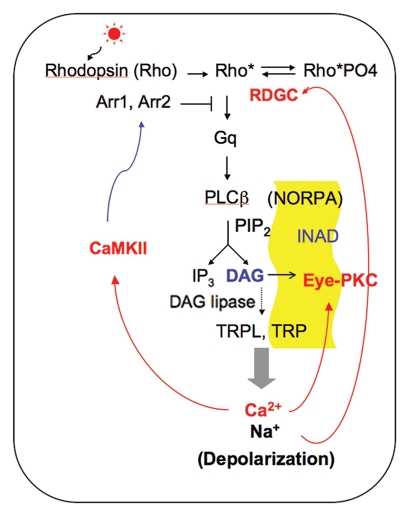
In the visual cascade, rhodopsin is the critical GPCR that senses the light signal by the covalently bound chromophore, 11-cis retinal (11-cis 3-hydroxy-retinal in the fly rhodopsin). The energy of light causes the isomerization of 11-cis retinal to an all-trans configuration, which activates rhodopsins (reviewed in ref. 33). In Drosophila, activated rhodopsin promotes the exchange of GTP for GDP in the α-subunit of the heterotrimeric Gq protein,27 which switches on NORPA (no-receptor-potential A). NORPA28 is a phospholipase Cβ that catalyzes the hydrolysis of phospholipids PIP2 to generate inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG). It appears that DAG and its metabolites including polyunsaturated fatty acids29,30 are critical for the gating of the downstream TRP (transient receptor potential) and TRPL (TRP-like) cation channels, whereas IP3 is not required for the light-mediated membrane depolarization (Fig. 2) (reviewed in refs. 31–33).
Figure 2.
Visual signaling in Drosophila. Light stimulation results in depolarization of Drosophila photoreceptors, as activated rhodopsin (Rho*) couples to Gq leading to the activation of PLCβ (NORPA), and the eventual opening of the TRP and TRPL channels. The INAD complex, highlighted in yellow, consists of INAD, eye-PKC, NORPA and TRP. The Ca2+-dependent regulatory events employ CaMKII, RDGC and eye-PKC partly to regulate Arr2, rhodopsin and TRP for the deactivation of the visual response.
The light-dependent depolarization of Drosophila photoreceptor cells is one of the fastest GPCR-mediated signaling events. The fast kinetics is partly contributed by the compartmentalization of the signaling molecules in rhabdomeres. Moreover, several signaling proteins including TRP, NORPA and eye-PKC (eye-specific protein kinase C) are organized into a macromolecular signaling complex (transduciosome or signalplex) (Fig. 2) through the association with INAD (inactivation-no-afterpotential D),34–39 a multivalent scaffolding protein containing five PDZ domains. INAD is critical for the subcellular localization as well as the stability of its interacting proteins in vivo, and it promotes fast activation and deactivation of the visual response.37
In vertebrates, by contrast, light hyperpolarizes photoreceptor cells (rods and cones) by closing cGMP channels (reviewed in ref. 40). In the dark, the cGMP level is higher, which opens the cGMP channel to generate the ‘dark currents’. Upon light stimulation, activated phosphodiesterase lowers the cGMP level, allowing fewer cGMP channels to remain open leading to hyperpolarization of rods or cones. In short, light switches on phosphodiesterase via the heterotrimeric G-protein transducin, resulting in a reduction of cGMP and hyperpolarization of vertebrate photoreceptor cells.
Modulation of the Visual Signaling by Ca2+
Photoreceptor cells are capable of responding to a wide-range of light intensities by adjusting degrees of amplification in the visual response. This is accomplished by several feedback mechanisms. In Drosophila, for example, the influx of Ca2+ via the TRP and TRPL channels generates localized Ca2+ transients which activate several Ca2+-dependent regulatory events including those initiated by Ca2+/calmodulin-dependent protein kinase II (CaMKII), eye-PKC and RDGC (retinal degeneration C) (Fig. 2). RDGC catalyzes the dephosphorylation of rhodopsin,41,42 whereas CaMKII is involved in the phosphorylation of arrestin 2 (Arr2),43,44 the major visual arrestin critical for the inactivation of activated rhodopsin.45 Eye-PKC is a conventional PKC46 activated by both DAG and Ca2+. Importantly, eye-PKC is tethered to the INAD signaling complex to regulate INAD and TRP;47 inaC mutants devoid of eye-PKC display defective light adaptation and slow deactivation of the visual response.48,49
Genes Affecting Retinal Degeneration in Drosophila can be Divided into Three Classes
The class I genes regulate the specialization of rhabdomeres.
Class I encompasses genes involved in the terminal differentiation of photoreceptor cells including those controlling the morphogenesis of rhabdomeres (Table 1). As detailed earlier, rhabdomeres are the visual organelles that house most of the visual signaling machinery. The specialization of rhabdomeres is orchestrated by the Crumbs protein complex including Crumb, Stardust and dPATJ: mutations in each component of this protein complex, and their interacting proteins result in defective morphogenesis with shortened stalk and aberrant rhabdomeres.2 Photoreceptors with abnormal rhabdomeres undergo light-dependent retinal degeneration.2 Significantly, the degeneration phenotype can be prevented when rhodopsin synthesis is greatly decreased by feeding flies with vitamin A-deficient food, presumably because it reduces the demand for active elongation of rhabdomeres to accommodate massive Rh1 rhodopsin.23
Table 1.
Three major classes of retinal degeneration mutations in Drosophila
| Class | Genes | Proteins (Activities) | Mechanisms# | References | |||
| I | Regulate terminal differentiation of photoreceptors | ||||||
| Crumbs | transmembrane protein with EGF-like and laminin G repeats | 1 | 24, 25 | ||||
| Stardust | membrane associated guanylate kinase (MAGUK) | 1 | 18, 23 | ||||
| Dpatj | cytosolic protein with PDZ and L27 domains | 1 | 18 | ||||
| Baz | scaffolding protein | 1 | 18 | ||||
| WASp | rhabdomere morphogenesis (actin polymerization) | 1 | 7 | ||||
| II | Control the level or activity of the Rh1 rhodopsin | ||||||
| Structural gene for Rh1 | |||||||
| NinaE (type I) | Rh1 rhodopsin | 2 | 75 | ||||
| NinaE (type II, LOF) | Rh1 with folding defects | 3 | 84–87 | ||||
| NinaE (type II, GOF) | Rh1 with constitutive activity | 4, 5 | 77 | ||||
| Rh1 folding and maturation | |||||||
| ninaA | cyclophilin-like; chaperone | 3 | 65, 99 | ||||
| cnx99A | calreticulin-like, lectin chaperone (Rh1 maturation) | 3 | 50 | ||||
| Trafficking of Rh1 | |||||||
| rab6 | small GTPase (trafficking of Rh1) | 2 | 52 | ||||
| rab11 | small GTPase (post-Golgi trafficking of Rh1) | 2 | 53 | ||||
| ninaC | unconventional myosin III (anchoring Rh1 in rhabdomeres) | 2 | 54 | ||||
| Inactivation of Rh1 | |||||||
| Arr2 | arrestin 2 (inactivation of Rh1) | 5 | 45, 55 | ||||
| Arr1 | arrestin 1 (inactivation and internalization of Rh1) | 5 | 55 | ||||
| rdgC | rhodopsin phosphatase | 4 | 41, 56 | ||||
| Tsp42Ej (sun) | tetraspanin (degradation of Rh1) | 4 | 57 | ||||
| TADR | Rh1 interacting protein | 4, 5 | 149 | ||||
| Endocytosis of Rh1 | |||||||
| car | sec-1 like protein (endosome to lysosome trafficking) | 4 | 58, 128, 129 | ||||
| lt | vacuolar protein sorting 41 (endosome to lysosome trafficking) | 4 | 58, 130 | ||||
| cm | clathrin adaptor mu subunit (endosome to lysosome trafficking) | 4 | 58 | ||||
| III | Modulate Ca2+ level or control Ca2+ influx | ||||||
| Modulate cytosolic Ca2+ level | |||||||
| cnx99A | calreticulin-like (cytoplasmic Ca2+ buffering) | 4, 5 | 50 | ||||
| calx | Na+/Ca2+ exchanger (cytoplasmic Ca2+ homeostasis) | 5 | 104 | ||||
| Participate in the synthesis/recycling of PIP2 | |||||||
| rdgA | diacylglycerol kinase (inactivation of second messenger) | 5 | 29, 109 | ||||
| cdsA | CDP-DAG synthase (regeneration of PIP2) | 4 | 111 | ||||
| pis | phosphatidylinositol synthase (regeneration of PIP2) | 4 | 111 | ||||
| rdgB | phosphatidylinositol transfer protein (regeneration of PIP2) | 4 | 107, 111 | ||||
| cerk | ceramide kinase (regulates norpA, PIP2) | 4 | 108 | ||||
| Regulate TRP Ca2+ channel | |||||||
| trp | Ca2+ channel | 4 | 120 | ||||
| Trp | constitutive TRP activity | 5 | 118 | ||||
| inaC | photoreceptor-specific protein kinase C | 4 | 46 | ||||
| inaD | scaffolding protein (regulates the stability of TRP) | 4 | 109 | ||||
| norpA | phospholipase Cβ (generates second messenger) | 4 | 113, 127 | ||||
Five mechanisms that contribute to retinal degeneration are (1) defective rhabdomere formation, (2) reduced Rh1 levels, (3) misfolded Rh1, (4) trafficking defects associated with internalization of activated Rh1 and (5) the Ca2+-mediated excitatoxicity due to overactive signaling. Abbreviations: LOF, loss-of-function; GOF, gain-of-function.
The class II genes are involved in the biosynthetic pathway and/or regulating Rh1 rhodopsin.
The Class II gene products participate in the biosynthesis or inactivation of the Rh1 rhodopsin (Table 1). Mutants affecting the biogenesis result in a reduced Rh1 level, which impacts the elaboration of rhabdomeres (see below, Structural Role of Rhodopsin). These gene products include CNX (calnexin),50 and NINAA (neither-inactivation-nor-afterpotential A),51 that controls the maturation of Rh1, rab6,52 and rab1153 that take part in the trafficking and delivery of Rh1 following its de novo synthesis, NINAC that anchors Rh1 in the rhabdomere,54 and ninaE (neither-inactivation-nor-after-potential E), the structural gene for Rh1.10,11 Mutations in ninaE can be further subdivided into two types, based on whether it is recessive (type I) or dominant (type II) in promoting retinal degeneration (see below).
In contrast, mutations that cause a delay in the inactivation of activated Rh1 resulting in either prolonged visual signaling and/or defective trafficking/degradation. These include arrestin 1 (Arr1),55 Arr2,45 rdgC,41,56 sun (sunglass),57 and the ‘granule group genes’.58 While both Arr1 and Arr2 contribute to the inactivation of Rh1,45 Arr1 also is critical for promoting Rh1 endocytosis.55 The internalized Rh1 can be detected in multivesicular bodies and appears targeted for degradation.55 The ‘granule group gene’ products play a role in the endosome to lysosome trafficking,58 while the sun gene encodes a novel tetraspanin that may modulate the degradation of Rh1 in the lysosome.57 Conceivably, these class II mutations result in light-dependent retinal degeneration because activation of Rh1 is required.57,58 The light-dependent degeneration of these mutants can be prevented by a reduction of Rh1 through dietary vitamin A restriction.
Biosynthesis of Rh1 Rhodopsin: Opsin and the Retinal Chromophore
Functional rhodopsin consists of a protein moiety, opsin and a retinal chromophore. The opsin serves to fine-tune the absorption spectrum of the covalently linked retinal chromophore resulting in a unique spectral sensitivity for each rhodopsin.8,9 Opsin, like other integral membrane proteins, is translated in the ribosome of the rough endoplasmic reticulum (ER). Following translation, Rh1 undergoes N-linked glycosylation at Asp(20),59,60 rendering it sensitive to endoglycosidase H. However, the carbohydrate moiety is subsequently removed when Rh1 exits the ER, resulting in a mature Rh1 without glycosylation.61 In the ER, the carbohydrate moiety is required for folding and maturation of Rh1,61 which is aided by two chaperones, NINAA51 and CNX.50 CNX is a resident protein in the ER involved in the quality control of protein synthesis, and it interacts with both the sugar moiety and the peptide chain of newly glycosylated polypeptides.62 A lack of CNX results in defects in Rh1 maturation.50 In contrast, NINAA is a chaperone with sequence homology to prolyl cistrans isomerase,63,64 and it regulates the export of Rh1 out of the ER.51 Mutations in ninaA lead to ER retention of Rh1 in R1–6 photoreceptors.65
The chromophore of rhodopsin in the fly, 11-cis 3-hydroxy-retinal, is derived from vitamin A of the diet. Vitamin A is taken up and processed to 11-cis retinal through multiple biochemical reactions, which are catalyzed by the gene products of ninaB,66 ninaD,67,68 ninaG69 and santa maria (scavenger receptor acting in neural tissue and majority of rhodopsin is absent).70 The stability of Drosophila Rh1 rhodopsin is dependent on its incorporation of the retinal chromophore:71 flies defective in proteins that control the uptake or processing of vitamin A, contain a reduced level of Rh1. Despite having a low Rh1 level, both ninaB and ninaD flies do not undergo retinal degeneration.72
Mature Rh1 is transported through the Golgi. The post-Golgi trafficking is orchestrated by monomeric G-proteins including Rab 652 and Rab11.53 Finally, Rh1 is delivered to and inserted into the rhabdomeric membrane. The rhabdomeric localization of Rh1 appears dependent on the helix 8 in its carboxyl terminal sequence.73 Moreover, the carboxyl tail of Rh1 is required for the maintenance of rhabdomere structure.74
Role of Rh1 in the Morphogenesis of Photoreceptors
Beside functions as a light detection molecule, Rh1 is also critical for the differentiation of R1–R6 photoreceptor cells. The incorporation of newly synthesized Rh1 into the rhabdomeric membrane coincides with its rapid elongation during terminal differentiation of R1–R6 cells. Consistently, the expansion of rhabdomeres is significantly compromised in ninaE flies unable to produce sufficient amount of Rh1. Leonard et al. demonstrated that a drastically reduced level of Rh1 resulted in smaller rhabdomeres at the eclosion when flies emerge. Moreover, photoreceptor cells that lack Rh1 display age-dependent degeneration with concomitant deterioration and disappearance of rhabdomeres.75 It appears that the severity of retinal degeneration is in proportion to the degree of reduction in the Rh1 level, indicating a structural role of Rh1 in the morphogenesis of photoreceptor cells.76 Interestingly, the size of rhabdomere is not solely dependent on the level of Rh1, as ninaEP332 flies with about 1% of the wild-type level actually contain about 65% of the normal rhabdomeric membrane area. It is likely that insertion of other membrane proteins also contributes to the expansion of rhabdomeres. Surprisingly, rhabdomeres are barely formed in a null allele (ninaEI17),75 suggesting that Rh1 also serves to initiate the morphogenesis of rhabdomeres. Thus Rh1 may have dual roles in orchestrating the elongation of rhabdomeres by serving as a signal as well as by providing the Rh1-enriched membrane. Similarly, mutations in the class II genes that greatly reduce either the biosynthetic capacity or the transport of Rh1 may lead to degeneration of photoreceptor cells because of a decreased Rh1 content that restricts the rhabdomeric expansion.
Mutations in Rh1 (ninaE) Elicit Either Dominant or Recessive Retinal Degeneration
In general, mutations in ninaE that trigger retinal degeneration fall into two categories. The first (type I) consists of recessive ninaE alleles in which degeneration is observed only in homozygotes containing two mutant alleles. Type I ninaE mutations include hypomorphic (e.g., ninaEP332) and null alleles (e.g., ninaEI17), which generally produce unstable Rh1 rhodopsin resulting in a drastically reduced level of Rh1. In contrast, the second group (type II) displays dominant degeneration as heterozygotes also exhibit a degeneration phenotype. There are two mechanistically distinct type II NinaE mutations: one that produces active Rh1 (e.g., NinaEpp100) leading to constitutive signaling and persistent Arr2 binding,77 and the other that produces misfolded Rh1 (e.g., NinaED1) that negatively impacts the expression of the wild-type ninaE allele.78 The degeneration phenotype of NinaED1 is lessened when the expression of ninaE is reduced by dietary vitamin A restriction, while that of constitutively active NinaEpp100 can be eliminated only when both Arr2 and Gqa are absent.77 Similar dominant mutations are found in human rod rhodopsin gene, which cause either stationary congenital night blindness or ADRP (autosomal dominant retinitis pigmentosa).79,80
Defect in Human Rod Rhodopsin Is One of the Major Causes of ADRP
ADRP is primarily characterized by the progressive loss of rods in the retina. Mutations in human rod rhodopsin, the visual pigment expressed in rod photoreceptors, constitute to 25–30% of ADRP.80 Importantly, these mutant rod rhodopsins display either an increased activity or defective protein folding, similar to dominant NinaE alleles. Moreover, some mutant rod rhodopsins exhibit defective transport to the outer segment or abnormal endocytosis (reviewed in ref. 81).
In short, most mutations of rod rhodopsin implicated in ADRP affect post-translational events including folding and trafficking, which result in the accumulation of mutant rhodopsin in the ER. Pharmacological chaperones such as 11-cis 7-ring retinal have been shown to rescue misfolded rhodopsin for its trafficking to the cell surface.82 However, misfolded opsins may form aggregates83 causing dysfunction of the ER. The ER stress triggered by the expression of mutant rhodopsin may initiate the unfolded protein response (UPR) in order to relieve the accumulation of misfolded proteins (see below).
Dominant NinaE Mutations: ADRP in Drosophila
As indicated before, dominant NinaE mutations can be caused by either dominant active Rh1 possessing constitutive activity, or dominant negative Rh1 that interferes with the biosynthesis of wild-type Rh1. A great number of dominant negative NinaE alleles (e.g., NinaEG69D, NinaED1) were originally recovered by genetic screens as suppressors of rdgB (retinal degeneration B),84 or rdgC.85 These NinaE mutants having either mis-sense mutations or premature stop codons, were shown to produce predominantly immature forms of Rh1 that are glycosylated and sensitive to endoglycosidase H. The mutant Rh1 appears to be retained in the ER.84
Interestingly, misfolded Rh1 tends to oligomerize, and mostly exists as dimers.78,84 It is very likely that dimers represent the conformation of the newly translated Rh1, because dimers can be detected following the heat-shock induced transcription in flies expressing Rh1 under the control of the hsp70 promoter.78,84 Importantly, NinaE heterozygotes, but not homozygotes, display over-proliferation of the ER cisternae,84 strongly suggesting defects associated with the ER accumulation of the non-productive Rh1 dimers containing both wild-type and mutant Rh1. Consistently, retinal degeneration is more severe in NinaE heterozygotes than in homozygotes,78 supporting the notion that the non-productive Rh1 dimers are more stable and exert a more profound effect than those containing only mutant Rh1. Similarly, folding defects associated with the expression of modified Rh1 (ninaEN20I) that lacks N-linked glycosylation,62 also lead to ER retention of non-glycosylated Rh1, and consequently dominant retinal degeneration.59
Mutations in NinaE that correspond to human ADRP also promote dominant retinal degeneration. This occurs, for example, in transgenic flies expressing modified Rh1 (Rh1P37H) containing a His at Pro(37),86,87 which is equivalent to human rod rhodopsin(P23H), the most common ADRP mutation in North America.88 In several fly models of ADRP, misfolded Rh1 is localized in the ER, and appears to trigger the ER stress that progresses to cell death. Besides folding defects, some ADRP rhodopsin mutations are shown to display constitutive activities,89 which may promote internalization of rhodopsin-arrestin complexes, similar to that occurring in gain-of-function NinaE alleles.
ER Stress Induces the Unfolded Protein Response
A folding deficiency often results in accumulation of misfolded proteins in the ER, which triggers ER stress or the unfolded protein response (UPR).90,91 The UPR is initiated by sensing misfolded proteins by three type I transmembrane proteins including ATF6 (activating transcription factor 6), IRE1 (inositol requiring 1), and PERK (PKR-like ER kinase), which are kept in inactive states by an ER chaperone Grp78 (BiP).91 As the level of misfolded proteins increases, which occupy Grp78, the inhibition of these three proteins is removed.92,93 Specifically, ATF6 becomes active following the protease cleavage in the Golgi.93 Protease-cleaved ATF6 is translocated to the nucleus and regulates the transcription of XBP1 (X box protein 1),94 a transcription factor that controls the expression of several genes involved in the retrograde transport and the degradation of misfolded proteins.95 The expression of XBP1 is also regulated by mRNA splicing, which requires IRE94 that contains an endoribonuclease domain and is activated following oligomerization.96 Similarly, PERK is switched on upon oligomerization and autophosphorylation. Activated PERK phosphorylates and inactivates eIF2a (eukaryotic initiation factor 2α),97 thereby shutting down protein synthesis, and reducing the protein load in the ER. In short, the UPR leads to a reduction of protein synthesis, yet an increased transcription and translation of chaperones that promote re-folding or degradation of misfolded proteins.
Protective Effect of the UPR Towards ADRP-Like NinaE Mutations
Studies on ADRP-like NinaE mutations indicate that the UPR helps reduce degeneration of photoreceptor cells. In flies expressing Rh1P37H, the misfolded Rh1 becomes a substrate of VCP/ter94, a chaperone that extracts misfolded proteins for proteasomal degradation. A reduction of VCP/ter94 elevates the level of misfolded Rh1P37H, which triggers the UPR that ameliorates death of photoreceptors.87 Likewise, administration of inhibitors of the VCP/ter94 or proteasomes offers protection against photoreceptor degeneration. In contrast, suppression of the ER stress response by decreasing the xbp1 gene dosage accelerates degeneration.98
The beneficial effect of the UPR is also observed in ninaA photoreceptors. NINAA is a resident chaperone critical for folding and maturation of Rh1.51 Rh1 accumulates in the ER in the absence of NINAA, which triggers the UPR.99 Significantly, the UPR prevents cell death initiated by several apoptotic signals including RPR (reaper), p53 and death caspase-1.99 This protective effect appears to be contributed by the inhibition of caspases.99 Taken together, upregulation of the UPR triggered by ADRP-like Rh1 or a loss of NINAA prevents death of photoreceptors, while inhibition of the UPR accelerates.
Somewhat paradoxically, persistent activation of the UPR has been linked to cell death (reviewed in refs. 90, 91 and 100). This may be promoted by recruiting ASK1 (Apoptosis Signal-Regulating Kinase 1) via IRE1, which is critical for the activation of JNK (c-Jun N-terminal Kinase) and cell death programs.101 Indeed, expression of Rh1P37H in Drosophila results in the activation of JNK and p38, which initiate stress-induced cell death pathways. Importantly, Rh1P37H-dependent retinal degeneration is suppressed by the expression of p35, an anti-apoptotic protein that inhibits caspases.86
Cell Death Caused by Recessive nina Mutations
A loss of Rh1 also leads to retinal degeneration. How does a lack of Rh1 affect survival of photoreceptors? Rh1 serves dual roles as it not only acts as the light receptor that triggers rhythmic Ca2+ influx, but also supports the morphogenesis of rhabdomeres. Interestingly, transgenic expression of a dominant active Rac, the small Rho GTPase involved in the formation of the actin cytoskeleton, prevents retinal degeneration in ninaEI17 flies.102 Therefore defects in rhabdomere expansion can be overcome by the active elongation of the actin cytoskeleton. Retinal degeneration in ninaE also can be rescued by vitamin A deprivation, suggesting that accumulation of free retinal is toxic to photoreceptors.72 Consistently, both ninaB and ninaD flies that are incapable of generating 11-cis retinal, exhibit no overt degeneration phenotype.72 Free retinal may covalently modify both proteins and lipids. Indeed, a lipid-retinal adduct, N-retinylidene-N-retinyl ethanolamine, has been implicated in human retinal disease.103 It is likely that defective rhabdomere morphogenesis due to a lack of Rh1 rhodopsin results from accumulation of free actin and 11-cis retinal, which may initiate apoptotic pathways.
The class III genes are critical for either Ca2+ or phospholipid-dependent modulation of visual signaling.
The class III gene products include those mediating the distal events of the visual signaling, which can be subdivided into three groups (Table 1). The first group encodes proteins that modulate cytosolic Ca2+ levels including calx and cnx. CALX is the Na+/Ca2+ exchanger104 that exports Ca2+, and CNX50 controls the buffering of intracellular Ca2+, both of which allow re-establishment and maintenance of a low cytosolic Ca2+ level. A lack of CALX or CNX results in Ca2+ overload, and light-dependent retinal degeneration.50,104 CNX is also involved in Rh1 maturation (class II).
The second group encompasses genes that regulate the biosynthesis/recycling of PIP2 including those well known retinal degeneration genes such as rdgA (retinal degeneration A),29,105 and rdgB (retinal degeneration B).106,107 This group also includes genes required for functional NORPA activity, such as ceramide kinase.108 Mutations in the second group result in retinal degeneration because of either a lack of Ca2+-dependent feedback modulation or the uncontrolled Ca2+ entry.
The recycling of PIP2 is orchestrated by a series of enzymatic reactions catalyzed by the DAG kinase RDGA,109,110 CDS (CDP-DAG synthase),111 PIS (phosphatidylinositol synthase),111 RDGB (a phosphatidylinositol transfer protein),107,111 and SKTL (phosphatidylinositol-4-phosphate-5-kinase).112 Perturbation of these enzymes may reduce the level of PIP2 affecting both the visual signaling and morphology of photoreceptors. Both rdgB and cds display light-dependent degeneration, and the molecular mechanism involved in rdgB113 appears similar to that in norpA (see below), while rdgA exhibits light-independent retinal degeneration.110
RDGA is critical for the conversion of DAG into phosphatic acid (PA).105,114 A lack of RDGA thus extends the half-life of DAG leading to constitutive TRP channel activity,29 and degeneration of photoreceptors.105 Retinal degeneration caused by the persistence of DAG was also observed upon overexpression of pld (phospholipase D) in fly photoreceptors.115 PLD generates PA, which can be further converted into DAG by LAZA (lazaro), a lipid phosphate phosphatase.116,117 Similarly, persistent activation of TRP in dominant Trp alleles (e.g., Trp365) leads to light-independent retinal degeneration.118 In all cases, death of photoreceptors is triggered by excessive Ca2+ influx, which may result in the activation of Ca2+-regulated proteases, phospholipases and endonucleases leading to destruction of cellular contents, and apoptosis.119
The third group of the class III includes trp,120 inaD,109 inaC,46 and norpA,28 whose encoded proteins directly or indirectly control the activity of the TRP Ca2+ channel (Table 1). Loss-of-function mutations in this group fail to support rhythmic Ca2+ entry affecting Ca2+-dependent feedback regulation, which gives rise to light-dependent degeneration of photoreceptors. Degeneration in norpA has been especially well explored, serving as a model for the molecular events leading to retinal cell death.
Retinal Degeneration in Flies that Lack NORPA
In Drosophila photoreceptors, NORPA breaks down PIP2 to generate the second messenger for gating the TRP Ca2+ channel. The rise of cytoplasmic Ca2+ orchestrates feedback regulation,121 which is achieved in part by activating both CaMKII and RDGC. CaMKII regulates the phosphorylation of Arr2,43,122 while RDGC catalyzes the dephosphorylation of Rh1,41,42,123 both contributing to the inactivation of activated Rh1. Activated Rh1 may become phosphorylated by rhodopsin kinase,124 and Rh1 interaction with arrestins (Arr1 or Arr2) greatly diminishes its ability to turn on Gq.
In norpA photoreceptors, both RDGC and CaMKII fail to become activated resulting in hyperphosphorylated Rh1 yet unphosphorylated Arr2, both of which promote the formation of Rh1/Arr2 complexes leading to light-dependent retinal degeneration.113 Similarly, rdgC photoreceptors have an elevated level of hyperphosphorylated Rh1, which also results in photoreceptor degeneration.41,56 Retinal degeneration in rdgC and norpA appear to share similar mechanisms as degeneration depends on both Rh1 and Arr2.41,56,113 Significantly, the interaction between Rh1 and Arr2 appears to be more persistent, as Arr2 is not readily released from the membrane when Rh1 becomes inactivated.113 Based on these findings, it has been proposed that formation of a stable Rh1/Arr2 complex, and its internalization triggers retinal cell death.56,113,125
Internalization of the Rh1/Arr2 is the Initial Event Leading to Degeneration of Photoreceptor Cells
The mechanism that regulates the internalization of activated Rh1 appears similar to that of a prototypical GPCR in response to agonists, a phenomenon termed homologous desensitization. Here, activation of GPCR results in its phosphorylation, and phosphorylated GPCR recruits β-arrestin. Binding of β-Arrestin downregulates GPCR activity, and also promotes GPCR endocytosis thus reducing the receptor level in the plasma membrane. Internalized GPCR, depending on its affinity with β-arrestin, is sorted in the endocytic compartment for either recycling/reinsertion into the plasma membrane, or degradation and subsequent removal.126
The internalization of Rh1/Arr2 that initiates retinal degeneration may employ the clathrin-dependent endocytic pathway. Indeed, when dynamin is rendered inactive (as in shibire mutants), the degeneration phenotype in either norpA or rdgC is eliminated.56,113 The interaction with the endocytic machinery may be regulated by phosphorylation of Arr2 as phosphorylated Arr2 fails to associate with purified clathrin cage in vitro.56 However, internalization of the Rh1/Arr2 complex may occur via direct binding of Arr2 to the AP-2 adaptor complex, as a reduced AP-2 interaction in flies expressing modified Arr2 suppresses photoreceptor death in norpA.127 Significantly, a lack of AP-2 interaction also results in retinal degeneration, indicating that endocytosis of Rh1 is required for the maintenance of photoreceptors.127
Endocytosed Rh1 is often destined for degradation, and is trafficked to the lysosome. This trafficking may become a rate-limiting step during massive internalization of Rh1 resulting in the accumulation of activated Rh1 in the Rab7-positive late endosome.58 The endosome to lysosomal trafficking is regulated partly by the so-called ‘granule group’ genes including car (carnation),128,129 lt (light),130 and cm (carmine). Importantly, mutations in one of the ‘granule group’ genes lead to light-dependent degeneration, which appears to result from defective trafficking of internalized Rh1.58 Indeed, by reducing the Rh1 level the degeneration phenotype of these trafficking mutants is greatly suppressed.58 These findings strongly support the notion that defects in trafficking and degradation of photoactivated Rh1 are detrimental to R1–R6 photoreceptors.
It is likely that the endosomal retention of activated Rh1 initiates a cell death program. However, it is also possible that endocytic accumulation of Rh1 interferes with the uptake and/or trafficking of secreted proteins such as growth factors involved in photoreceptor survival. Interestingly, the endocytic turnover of activated Rh1 requires ceramidase secreted from neighboring cells.131 Ceramidase is involved in the breakdown of ceramide to generate sphingosine; both sphingosine and ceramide are bioactive lipids that mediate diverse signaling including cell proliferation, differentiation and apoptosis.132 Significantly, photoreceptor cells that are devoid of ceramidase undergo light-dependent degeneration, while overexpression rescues retinal degeneration of norpA flies.133
Role of Programmed Cell Death
The degeneration phenotype observed in norpA is consistent with inappropriate initiation of programmed cell death (PCD). To date, PCD can be subdivided into two common forms, apoptotic cell death (type I) and autophagic cell death (type II),134 based on the underlying mechanisms. Type I PCD is characterized by the activation of caspases (cysteine-dependent aspartate-directed proteases).135 In healthy cells, caspases (e.g., death caspase-1) are kept in an inactive state by the inhibitor of apoptosis (IAP, e.g., DIAP1), which inactivates caspases through ubiquitination and proteasomal degradation of pro-caspases.136 In response to cell death signals, activation of RHG (RPR, HID and GRIM) results in proteosomal degradation of DIAP1, leading to increased levels of active caspases that initiate apoptosis (reviewed in refs. 137 and 138).
Type I PCD may be involved in the degeneration of norpA photoreceptors. However, genetically removing or reducing the cell death inducers from photoreceptors including RHG fails to prevent death of norpA photoreceptors.139 Moreover, overexpression of DIAP1 is not able to suppress degeneration, nor is the expression of a dominant negative caspase (Dronc). Thus, retinal degeneration observed in norpA is not attributed to the caspase-dependent PCD, unlike that of NinaE mutants expressing ADRP-like Rh1 rhodopsin. The cell death of these dominant NinaE mutants can be abrogated by the expression of p35, a caspase inhibitor.86,140 While the molecular events leading to apoptotic cell death are known, the mechanisms underlying type II autophagic cell death are less understood.
Morphologically, type II PCD is characterized by the presence of excess vacuoles or autophagosomes, which are formed during autophagy.141 Autophagy is defined as a lysosome-dependent degradation process, which is a protective and self-renewal mechanism that allows cells to degrade damaged proteins and organelles, as well as to recycle obsolete cellular constituents (reviewed in ref. 142). Autophagy requires the participation of many atg's (autophagy related genes).142 Several of the ATG proteins interact and collaborate to form the ‘core machinery’ to promote autophagosome assembly. Subsequently, most of the core machinery is retrieved, and autophagosomes then fuse with lysosomes for initiating degradation of the encapsulated contents by lysosomal hydrolases (reviewed in ref. 142).
Role of Autophagy in Retinal Degeneration
Autophagy is an evolutionarily conserved mechanism for maintaining cellular homeostasis, and for multicellular development, and it can be upregulated by intracellular (e.g., ER stress) or extracellular stresses (e.g., pathogen infection, growth factor deprivation) (reviewed in ref. 143). Defective autophagy has been implicated in several pathological conditions including cancer, microbial infection and neurodegeneration.144 Importantly, the activity of some ATG proteins is negatively regulated by TOR (target of rapamycin):145 Inhibition of TOR promotes autophagy, whereas hyperactivation of TOR suppresses.
Inhibition of autophagy results in light-dependent photoreceptor death146 while induction greatly reduces retinal degeneration in norpA flies. Thus, autophagy appears to have a protective role towards photoreceptor death caused by endosomal accumulation of activated Rh1. Consistently, a reduction of essential components including atg7/atg8, Psd (phosphatidylserine decarboxylase) and Ept (CDP-ethanolamine:diacylglycerol ethanolamine phosphotransferase) results in retinal degeneration,147 which may be contributed by the accumulation of activated Rh1 in Rab7-positive late endosomes. Importantly, overexpression of Rab7 prevents death of photoreceptors in these atg's knockdown flies.147 These results support the notion that induction of autophagy protects photoreceptor cells from degeneration possibly by promoting autophagic degradation of activated Rh1 in late endosomes. Although autophagy has been shown to relieve the ER stress, it fails to suppress retinal degeneration caused by the ER accumulation of misfolded Rh1 in ADRP-type NinaE mutants.146
A Summary of Diverse Degeneration Mechanisms
Many mutations lead to retinal degeneration. In general, the downstream events leading to death of photoreceptor cells fall into five distinct mechanisms contributed by (1) defective rhabdomere formation (class I), (2) a reduced Rh1 level [ninaE (class II)], (3) misfolded Rh1 [NinaE (LOF), ninaA, cnx (class II)], (4) trafficking defects associated with internalization of photoactivated Rh1 [some of class III; NinaE (GOF) (class II)], and (5) the Ca2+-mediated excitatoxicity due to overactive TRP [Trp(GOF), rdgA (class III); arr2, NinaE(GOF) (class II)] (Table 1). It is important to note that some mutations may initiate multiple mechanisms to trigger cell death. For example, expression of constitutively active Rh1 (NinaE) promotes massive endocytosis of activated Rh1, and also orchestrates the Ca2+-dependent excitatoxicity due to un-regulated Ca2+ influx.
During the terminal differentiation of photoreceptors, elaboration of rhabdomeres provides the specialized compartment for housing the visual signaling proteins. Misformed rhabdomeres fail to support the subcellular localization, affecting both the visual signaling and maintenance of photoreceptors. The most abundant membrane protein in the rhabdomere is Rh1, which is critical for rhabdomere assembly and elongation. Loss-of-function mutations affecting Rh1 biosynthesis usually lead to photoreceptor degeneration. Photoreceptor death may be caused by defective rhabdomere biogenesis and by the accumulation of cytotoxic 11-cis retinal.
Newly translated Rh1 may form oligomeric complexes to assist its folding and maturation. Mutations in Rh1 or proteins involved in its post-translational maturation lead to accumulation of Rh1 in the ER resulting in degeneration of photoreceptors. In NinaE heterozygotes, co-expression of misfolded with wild-type Rh1 has more debilitating consequences as it causes overproliferation of the ER cisternae, and a more severe degeneration phenotype than in NinaE homozygotes.
Mutants affecting the intermediate steps of the visual signaling cascade may lead to accumulation of photoactivated Rh1, due to a lack of the Ca2+-dependent feedback regulation. Activated Rh1 forms a stable complex with Arr2 resulting in its rapid internalization. Internalization of massive Rh1 may overwhelm the endocytic pathway, and persistence of activated Rh1 in the endosome may activate pro-apoptotic signaling leading to the demise of photoreceptors.
Unregulated visual signaling also contributes to the pathogenesis of retinal degeneration, which could be due to either gain-of-function mutations in key signaling proteins involved in the activation of visual cascade (e.g., Rh1 rhodopsin or TRP) or loss-of-function mutations involved in either negative regulatory mechanisms (e.g., arr2), or inactivation of the second messenger (e.g., rdgA). In all cases, the activity of the downstream TRP Ca2+ channel fails to become properly regulated leading to constitutive Ca2+ entry and the Ca2+-mediated excitatoxicity.
Conclusions
Based on the proximal events that initiate death of photoreceptors, here I divide most of the known retinal degeneration mutations into three classes. It appears that defects associated with trafficking of Rh1 are the major cause of retinal dystrophy (Fig. 3). This may be due to the fact that Rh1 is the most abundant membrane protein in photoreceptors (65% estimated by freeze fracture EM analysis148). In ADRP-type NinaE mutants, misfolded Rh1 hinders the maturation of the wild-type Rh1 by forming oligomeric Rh1 complexes. The ER accumulation of Rh1 complexes orchestrates the UPR by activating the transcription of several proteins to promote re-folding and degradation of misfolded proteins. Conceivably, the UPR is protective as its induction suppresses photoreceptor death. Persistent ER stress caused by the accumulation of misfolded Rh1, however, negatively impacts the survival of the R1–R6 photoreceptors. The links between the UPR and programmed cell death remain to be explored.
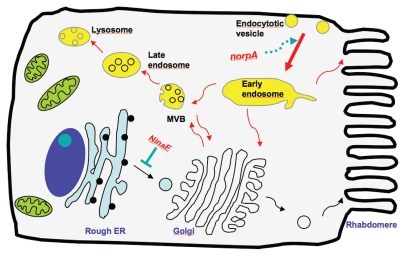
Figure 3.
Retinal degeneration contributed by defective trafficking of Rh1 in NinaE and norpA photoreceptors. Shown is a diagram depicting trafficking of newly translated Rh1 rhodopsin (black arrows) and of internalized photoactivated Rh1 rhodopsin (red arrows) in photoreceptors. Rh1 opsin is translated, and undergoes glycosylation and maturation in the rough ER. Mature opsin is conjugated with 11-cis 3-hydroxy-retinal to become rhodopsin, which is transported through the Golgi for its delivery to the rhabdomere. In NinaE heterozygotes, folding deficient Rh1 may form oligomeric complexes with wild-type Rh1 affecting the transport of wild-type Rh1, and resulting in degeneration of photoreceptors. In norpA photoreceptors, photoactivated Rh1 becomes rapidly internalized. However, the endosome to lysosome trafficking of endocytosed Rh1 becomes a rate-limiting step leading to accumulation of activated Rh1 in the late endosome, which leads to death of photoreceptor cells. MVB, multivesicular body.
While photoactivated Rh1 may be taken up and targeted for degradation, the sorting at the late endosome may become a ratelimiting step to cope with the massive internalization of Rh1. The endosomal accumulation of Rh1 may trigger retinal degeneration. Significantly, induction of autophagy facilitates removal and degradation of internalized Rh1, and protects retinal degeneration of norpA flies. In both norpA and NinaE photoreceptor cells, the trafficking defects associated with activated or misfolded Rh1 can be corrected by reducing the expression of Rh1 via dietary vitamin A restriction. Vitamin A deprivation is also beneficial for mutants affecting rhabdomere morphogenesis (class I), as a low level of Rh1 alleviates the demand of the compromised rhabdomere. Moreover, in flies containing a reduced level of Rh1, vitamin A restriction limits the cytotoxicity of free 11-cis retinal.
Retinal degeneration in Drosophila occurs via diverse mechanisms, some of which also are involved in that of humans. Our knowledge in the fly will provide a better understanding into the molecular basis of prevalent eye diseases such as ADRP. The distinct mechanistic insights uncovered in Drosophila may bring new avenues towards rationale therapeutic interventions for treating human retinal dystrophy. For example, administration of the UPR activators may alleviate death of photoreceptors in ADRP patients that express misfolded rod rhodopsin. Likewise, drugs that induce autophagy through the inhibition of mammalian target of rapamycin, mTOR, may slow down the progression of degeneration contributed by mutant rod rhodopsin that displays abnormal endocytosis. The promise of these pharmacological agents is also enhanced by the accessibility of the eye for drug application: drugs can be formulated as eye drops for topical application, thus minimizing their systemic toxicity.
Acknowledgments
I thank members of the laboratory for critical reading of the manuscript. This work is supported by a grant from NIH.
Abbreviations
- ADRP
autosomal dominant retinitis pigmentosa
- Arr2
arrestin 2
- Arr1
arrestin 1
- atg
autophagy related gene
- Baz
bazooka
- CalX
Na+/Ca2+ exchanger
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- cGMP
cyclic GMP
- CNX
calnexin
- DAG
diacylglycerol
- dpp
deep pseudopupil
- GPCR
G-protein coupled receptor
- nina
neither-inactivation-nor-afterpotential
- norpA
no-receptor-potential A
- IAP
inhibitor of apoptosis
- ina
inactivation-no-afterpotential
- IP3
inositol trisphosphate
- PA
phosphatic acid
- PATJ
PALS1-associated tight junction protein
- PCD
programmed cell death
- PDZ
PSD95, discs-large, ZO-1
- PKC
protein kinase C
- PIP2
phosphatidylinositol-4,5-bisphosphate
- PIP3
phosphatidylinositol-3,4,5-trisphosphate
- PLD
phospholipase D
- rdgA
retinal degeneration A
- rdgB
retinal degeneration B
- rdgC
retinal degeneration C
- TOR
target of rapamycin
- TRP
transient receptor potential
- TRPL
trp-like
- UPR
unfolded protein response
References
- 1.Franceschini N. Pupil and psueopupil in the compound eye of Drosophila. In: Wehner R, editor. Information Processing in the Visual Systems of Arthropods. Berlin: Springer-Verlag; 1972. pp. 75–82. [Google Scholar]
- 2.Knust E. Photoreceptor morphogenesis and retinal degeneration: lessons from Drosophila. Curr Opin Neurobiol. 2007;17:541–547. doi: 10.1016/j.conb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 4.Elliott DA, Brand AH. The GAL4 system: a versatile system for the expression of genes. Methods Mol Biol. 2008;420:79–95. doi: 10.1007/978-1-59745-583-1_5. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW. Gene silencing by double-stranded RNA. Curr Opin Cell Biol. 2001;13:244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 6.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelhof AC, Hardy RW. WASp is required for the correct temporal morphogenesis of rhabdomere microvilli. J Cell Biol. 2004;164:417–426. doi: 10.1083/jcb.200307048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt SG, Feiler R, Kirschfeld K, Zuker CS. Spectral tuning of rhodopsin and metarhodopsin in vivo. Neuron. 1993;11:29–39. doi: 10.1016/0896-6273(93)90268-v. [DOI] [PubMed] [Google Scholar]
- 9.Lin SW, Sakmar TP. Colour tuning mechanisms of visual pigments. Novartis Found Symp. 1999;224:124–135. doi: 10.1002/9780470515693.ch8. [DOI] [PubMed] [Google Scholar]
- 10.O'Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 11.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 12.Montell C, Jones K, Zuker C, Rubin G. A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J Neurosci. 1987;7:1558–1566. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker CS, Montell C, Jones K, Laverty T, Rubin GM. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules. J Neurosci. 1987;7:1550–1557. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 15.Huber A, Schulz S, Bentrop J, Groell C, Wolfrum U, Paulsen R. Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells. FEBS Lett. 1997;406:6–10. doi: 10.1016/s0014-5793(97)00210-x. [DOI] [PubMed] [Google Scholar]
- 16.Cowman AF, Zuker CS, Rubin GM. An opsin gene expressed in only one photoreceptor cell type of the Drosophila eye. Cell. 1986;44:705–710. doi: 10.1016/0092-8674(86)90836-6. [DOI] [PubMed] [Google Scholar]
- 17.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, et al. International Union of Pharmacology. XLVI. G protein-coupled receptor list. Pharmacol Rev. 2005;57:279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 18.Hong Y, Ackerman L, Jan LY, Jan YN. Distinct roles of Bazooka and Stardust in the specification of Drosophila photoreceptor membrane architecture. Proc Natl Acad Sci USA. 2003;100:12712–12717. doi: 10.1073/pnas.2135347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- 21.Stocker H, Andjelkovic M, Oldham S, Laffargue M, Wymann MP, Hemmings BA, Hafen E. Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science. 2002;295:2088–2091. doi: 10.1126/science.1068094. [DOI] [PubMed] [Google Scholar]
- 22.Pinal N, Goberdhan DC, Collinson L, Fujita Y, Cox IM, Wilson C, Pichaud F. Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr Biol. 2006;16:140–149. doi: 10.1016/j.cub.2005.11.068. [DOI] [PubMed] [Google Scholar]
- 23.Berger S, Bulgakova NA, Grawe F, Johnson K, Knust E. Unraveling the genetic complexity of Drosophila stardust during photoreceptor morphogenesis and prevention of light-induced degeneration. Genetics. 2007;176:2189–2200. doi: 10.1534/genetics.107.071449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- 25.Richard M, Muschalik N, Grawe F, Ozuyaman S, Knust E. A role for the extracellular domain of Crumbs in morphogenesis of Drosophila photoreceptor cells. Eur J Cell Biol. 2009;88:765–777. doi: 10.1016/j.ejcb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Zuker CS. The biology of vision of Drosophila. Proc Natl Acad Sci USA. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott K, Becker A, Sun Y, Hardy R, Zuker C. Gqalpha protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron. 1995;15:919–927. doi: 10.1016/0896-6273(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 28.Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, et al. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 29.Raghu P, Usher K, Jonas S, Chyb S, Polyanovsky A, Hardie RC. Constitutive activity of the light-sensitive channels TRP and TRPL in the Drosophila diacylglycerol kinase mutant, rdgA. Neuron. 2000;26:169–179. doi: 10.1016/s0896-6273(00)81147-2. [DOI] [PubMed] [Google Scholar]
- 30.Leung HT, Tseng-Crank J, Kim E, Mahapatra C, Shino S, Zhou Y, et al. DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron. 2008;58:884–896. doi: 10.1016/j.neuron.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghu P, Colley NJ, Webel R, James T, Hasan G, Danin M, et al. Normal phototransduction in Drosophila photoreceptors lacking an InsP(3) receptor gene. Mol Cell Neurosci. 2000;15:429–445. doi: 10.1006/mcne.2000.0846. [DOI] [PubMed] [Google Scholar]
- 32.Pak WL, Leung HT. Genetic approaches to visual transduction in Drosophila melanogaster. Receptors Channels. 2003;9:149–167. [PubMed] [Google Scholar]
- 33.Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 34.Shieh BH, Zhu MY. Regulation of the TRP Ca2+ channel by INAD in Drosophila photoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 35.Adamski FM, Zhu MY, Bahiraei F, Shieh BH. Interaction of eye protein kinase C and INAD in Drosophila. Localization of binding domains and electrophysiological characterization of a loss of association in transgenic flies. J Biol Chem. 1998;273:17713–17719. doi: 10.1074/jbc.273.28.17713. [DOI] [PubMed] [Google Scholar]
- 36.Shieh BH, Zhu MY, Lee JK, Kelly IM, Bahiraei F. Association of INAD with NORPA is essential for controlled activation and deactivation of Drosophila phototransduction in vivo. Proc Natl Acad Sci USA. 1997;94:12682–12687. doi: 10.1073/pnas.94.23.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, et al. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 38.van Huizen R, Miller K, Chen DM, Li Y, Lai ZC, Raab RW, et al. Two distantly positioned PDZ domains mediate multivalent INAD-phospholipase C interactions essential for G protein-coupled signaling. EMBO J. 1998;17:2285–2297. doi: 10.1093/emboj/17.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu XZ, Choudhury A, Li X, Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fain GL, Hardie R, Laughlin SB. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010;20:114–124. doi: 10.1016/j.cub.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vinos J, Jalink K, Hardy RW, Britt SG, Zuker CS. A G protein-coupled receptor phosphatase required for rhodopsin function. Science. 1997;277:687–690. doi: 10.1126/science.277.5326.687. [DOI] [PubMed] [Google Scholar]
- 42.Steele FR, Washburn T, Rieger R, O'Tousa JE. Drosophila retinal degeneration C (rdgC) encodes a novel serine/threonine protein phosphatase. Cell. 1992;69:669–676. doi: 10.1016/0092-8674(92)90230-a. [DOI] [PubMed] [Google Scholar]
- 43.Kahn ES, Matsumoto H. Calcium/calmodulin-dependent kinase II phosphorylates Drosophila visual arrestin. J Neurochem. 1997;68:169–175. doi: 10.1046/j.1471-4159.1997.68010169.x. [DOI] [PubMed] [Google Scholar]
- 44.Alloway PG, Dolph PJ. A role for the light-dependent phosphorylation of visual arrestin. Proc Natl Acad Sci USA. 1999;96:6072–6077. doi: 10.1073/pnas.96.11.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolph PJ, Ranganathan R, Colley NJ, Hardy RW, Socolich M, Zuker CS. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science. 1993;260:1910–1916. doi: 10.1126/science.8316831. [DOI] [PubMed] [Google Scholar]
- 46.Smith DP, Ranganathan R, Hardy RW, Marx J, Tsuchida T, Zuker CS. Photoreceptor deactivation and retinal degeneration mediated by a photoreceptor-specific protein kinase C. Science. 1991;254:1478–1484. doi: 10.1126/science.1962207. [DOI] [PubMed] [Google Scholar]
- 47.Popescu DC, Ham AJ, Shieh BH. Scaffolding protein INAD regulates deactivation of vision by promoting phosphorylation of transient receptor potential by eye protein kinase C in Drosophila. J Neurosci. 2006;26:8570–8577. doi: 10.1523/JNEUROSCI.1478-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardie RC, Peretz A, Suss-Toby E, Rom-Glas A, Bishop SA, Selinger Z, Minke B. Protein kinase C is required for light adaptation in Drosophila photoreceptors. Nature. 1993;363:634–637. doi: 10.1038/363634a0. [DOI] [PubMed] [Google Scholar]
- 49.Ranganathan R, Harris GL, Stevens CF, Zuker CS. A Drosophila mutant defective in extracellular calcium-dependent photoreceptor deactivation and rapid desensitization. Nature. 1991;354:230–232. doi: 10.1038/354230a0. [DOI] [PubMed] [Google Scholar]
- 50.Rosenbaum EE, Hardie RC, Colley NJ. Calnexin is essential for rhodopsin maturation, Ca2+ regulation and photoreceptor cell survival. Neuron. 2006;49:229–241. doi: 10.1016/j.neuron.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker EK, Colley NJ, Zuker CS. The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shetty KM, Kurada P, O'Tousa JE. Rab6 regulation of rhodopsin transport in Drosophila. J Biol Chem. 1998;273:20425–20430. doi: 10.1074/jbc.273.32.20425. [DOI] [PubMed] [Google Scholar]
- 53.Satoh AK, O'Tousa JE, Ozaki K, Ready DF. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development. 2005;132:1487–1497. doi: 10.1242/dev.01704. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto H, Isono K, Pye Q, Pak WL. Gene encoding cytoskeletal proteins in Drosophila rhabdomeres. Proc Natl Acad Sci USA. 1987;84:985–989. doi: 10.1073/pnas.84.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satoh AK, Ready DF. Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Curr Biol. 2005;15:1722–1733. doi: 10.1016/j.cub.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 56.Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, Ranganathan R. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 57.Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, et al. A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chinchore Y, Mitra A, Dolph PJ. Accumulation of rhodopsin in late endosomes triggers photoreceptor cell degeneration. PLoS Genet. 2009;5:1000377. doi: 10.1371/journal.pgen.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webel R, Menon I, O'Tousa JE, Colley NJ. Role of asparagine-linked oligosaccharides in rhodopsin maturation and association with its molecular chaperone, NinaA. J Biol Chem. 2000;275:24752–24759. doi: 10.1074/jbc.M002668200. [DOI] [PubMed] [Google Scholar]
- 60.Katanosaka K, Tokunaga F, Kawamura S, Ozaki K. N-linked glycosylation of Drosophila rhodopsin occurs exclusively in the amino-terminal domain and functions in rhodopsin maturation. FEBS Lett. 1998;424:149–154. doi: 10.1016/s0014-5793(98)00160-4. [DOI] [PubMed] [Google Scholar]
- 61.O'Tousa JE. Requirement of N-linked glycosylation site in Drosophila rhodopsin. Vis Neurosci. 1992;8:385–390. doi: 10.1017/s0952523800004910. [DOI] [PubMed] [Google Scholar]
- 62.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 63.Schneuwly S, Shortridge RD, Larrivee DC, Ono T, Ozaki M, Pak WL. Drosophila ninaA gene encodes an eye-specific cyclophilin (cyclosporine A binding protein) Proc Natl Acad Sci USA. 1989;86:5390–5394. doi: 10.1073/pnas.86.14.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shieh BH, Stamnes MA, Seavello S, Harris GL, Zuker CS. The ninaA gene required for visual transduction in Drosophila encodes a homologue of cyclosporin A-binding protein. Nature. 1989;338:67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- 65.Colley NJ, Baker EK, Stamnes MA, Zuker CS. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- 66.von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K. Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation in vivo. Proc Natl Acad Sci USA. 2001;98:1130–1135. doi: 10.1073/pnas.031576398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiefer C, Sumser E, Wernet MF, Von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc Natl Acad Sci USA. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voolstra O, Kiefer C, Hoehne M, Welsch R, Vogt K, von Lintig J. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry. 2006;45:13429–13437. doi: 10.1021/bi060701u. [DOI] [PubMed] [Google Scholar]
- 69.Ahmad ST, Joyce MV, Boggess B, O'Tousa JE. The role of Drosophila ninaG oxidoreductase in visual pigment chromophore biogenesis. J Biol Chem. 2006;281:9205–9209. doi: 10.1074/jbc.M510293200. [DOI] [PubMed] [Google Scholar]
- 70.Wang T, Jiao Y, Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozaki K, Nagatani H, Ozaki M, Tokunaga F. Maturation of major Drosophila rhodopsin, ninaE, requires chromophore 3-hydroxyretinal. Neuron. 1993;10:1113–1119. doi: 10.1016/0896-6273(93)90059-z. [DOI] [PubMed] [Google Scholar]
- 72.Voolstra O, Oberhauser V, Sumser E, Meyer NE, Maguire ME, Huber A, von Lintig J. NinaB is essential for Drosophila vision but induces retinal degeneration in opsin-deficient photoreceptors. J Biol Chem. 2010;285:2130–2139. doi: 10.1074/jbc.M109.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kock I, Bulgakova NA, Knust E, Sinning I, Panneels V. Targeting of Drosophila rhodopsin requires helix 8 but not the distal C-terminus. PLoS One. 2009;4:6101. doi: 10.1371/journal.pone.0006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahmad ST, Natochin M, Artemyev NO, O'Tousa JE. The Drosophila rhodopsin cytoplasmic tail domain is required for maintenance of rhabdomere structure. Faseb J. 2007;21:449–455. doi: 10.1096/fj.06-6530com. [DOI] [PubMed] [Google Scholar]
- 75.Leonard DS, Bowman VD, Ready DF, Pak WL. Degeneration of photoreceptors in rhodopsin mutants of Drosophila. J Neurobiol. 1992;23:605–626. doi: 10.1002/neu.480230602. [DOI] [PubMed] [Google Scholar]
- 76.Kumar JP, Ready DF. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- 77.Iakhine R, Chorna-Ornan I, Zars T, Elia N, Cheng Y, Selinger Z, et al. Novel dominant rhodopsin mutation triggers two mechanisms of retinal degeneration and photoreceptor desensitization. J Neurosci. 2004;24:2516–2526. doi: 10.1523/JNEUROSCI.5426-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurada P, Tonini TD, Serikaku MA, Piccini JP, O'Tousa JE. Rhodopsin maturation antagonized by dominant rhodopsin mutants. Vis Neurosci. 1998;15:693–700. doi: 10.1017/s0952523898154093. [DOI] [PubMed] [Google Scholar]
- 79.Sung CH, Davenport CM, Hennessey JC, Maumenee IH, Jacobson SG, Heckenlively JR, et al. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dryja TP, McEvoy JA, McGee TL, Berson EL. Novel rhodopsin mutations Gly114Val and Gln184Pro in dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:3124–3127. [PubMed] [Google Scholar]
- 81.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol Med. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 82.Noorwez SM, Ostrov DA, McDowell JH, Krebs MP, Kaushal S. A high-throughput screening method for small-molecule pharmacologic chaperones of misfolded rhodopsin. Invest Ophthalmol Vis Sci. 2008;49:3224–3230. doi: 10.1167/iovs.07-1539. [DOI] [PubMed] [Google Scholar]
- 83.Illing ME, Rajan RS, Bence NF, Kopito RR. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277:34150–34160. doi: 10.1074/jbc.M204955200. [DOI] [PubMed] [Google Scholar]
- 84.Colley NJ, Cassill JA, Baker EK, Zuker CS. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc Natl Acad Sci USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurada P, O'Tousa JE. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 86.Galy A, Roux MJ, Sahel JA, Leveillard T, Giangrande A. Rhodopsin maturation defects induce photoreceptor death by apoptosis: a fly model for RhodopsinPro23His human retinitis pigmentosa. Hum Mol Genet. 2005;14:2547–2557. doi: 10.1093/hmg/ddi258. [DOI] [PubMed] [Google Scholar]
- 87.Griciuc A, Aron L, Roux MJ, Klein R, Giangrande A, Ueffing M. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 2010;6:1001075. doi: 10.1371/journal.pgen.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dryja TP, McGee TL, Hahn LB, Cowley GS, Olsson JE, Reichel E, et al. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 89.Robinson PR, Cohen GB, Zhukovsky EA, Oprian DD. Constitutively active mutants of rhodopsin. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 90.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 91.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 93.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 94.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 95.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Urano F, Bertolotti A, Ron D. IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci. 2000;113:3697–3702. doi: 10.1242/jcs.113.21.3697. [DOI] [PubMed] [Google Scholar]
- 97.Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mendes CS, Levet C, Chatelain G, Dourlen P, Fouillet A, Dichtel-Danjoy ML, et al. ER stress protects from retinal degeneration. EMBO J. 2009;28:1296–1307. doi: 10.1038/emboj.2009.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang HY, Ready DF. Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated Rac1. Science. 2000;290:1978–1980. doi: 10.1126/science.290.5498.1978. [DOI] [PubMed] [Google Scholar]
- 103.Sparrow JR, Boulton M. RPE lipofuscin and its role in retinal pathobiology. Exp Eye Res. 2005;80:595–606. doi: 10.1016/j.exer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 104.Wang T, Xu H, Oberwinkler J, Gu Y, Hardie RC, Montell C. Light activation, adaptation and cell survival functions of the Na+/Ca2+ exchanger CalX. Neuron. 2005;45:367–378. doi: 10.1016/j.neuron.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 105.Masai I, Suzuki E, Yoon CS, Kohyama A, Hotta Y. Immunolocalization of Drosophila eye-specific diacylgylcerol kinase, rdgA, which is essential for the maintenance of the photoreceptor. J Neurobiol. 1997;32:695–706. doi: 10.1002/(sici)1097-4695(19970620)32:7<695::aid-neu5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 106.Vihtelic TS, Hyde DR, O'Tousa JE. Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics. 1991;127:761–768. doi: 10.1093/genetics/127.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stark WS, Sapp R. Retinal degeneration and photoreceptor maintenance in Drosophila: rdgB and its interaction with other mutants. Prog Clin Biol Res. 1989;314:467–489. [PubMed] [Google Scholar]
- 108.Dasgupta U, Bamba T, Chiantia S, Karim P, Tayoun AN, Yonamine I, et al. Ceramide kinase regulates phospholipase C and phosphatidylinositol-4,5-bisphosphate in phototransduction. Proc Natl Acad Sci USA. 2009;106:20063–20068. doi: 10.1073/pnas.0911028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Georgiev P, Garcia-Murillas I, Ulahannan D, Hardie RC, Raghu P. Functional INAD complexes are required to mediate degeneration in photoreceptors of the Drosophila rdgA mutant. J Cell Sci. 2005;118:1373–1384. doi: 10.1242/jcs.01712. [DOI] [PubMed] [Google Scholar]
- 110.Johnson MA, Frayer KL, Stark WS. Characterization of rdgA: mutants with retinal degeneration in Drosophila. J Insect Physiol. 1982;28:233–242. [Google Scholar]
- 111.Wang T, Montell C. A phosphoinositide synthase required for a sustained light response. J Neurosci. 2006;26:12816–12825. doi: 10.1523/JNEUROSCI.3673-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hassan BA, Prokopenko SN, Breuer S, Zhang B, Paululat A, Bellen HJ. skittles, a Drosophila phosphatidylinositol-4-phosphate 5-kinase, is required for cell viability, germline development and bristle morphology, but not for neurotransmitter release. Genetics. 1998;150:1527–1537. doi: 10.1093/genetics/150.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Alloway PG, Howard L, Dolph PJ. The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28:129–138. doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 114.Inoue H, Yoshioka T, Hotta Y. Diacylglycerol kinase defect in a Drosophila retinal degeneration mutant rdgA. J Biol Chem. 1989;264:5996–6000. [PubMed] [Google Scholar]
- 115.LaLonde MM, Janssens H, Rosenbaum E, Choi SY, Gergen JP, Colley NJ, et al. Regulation of phototransduction responsiveness and retinal degeneration by a phospholipase D-generated signaling lipid. J Cell Biol. 2005;169:471–479. doi: 10.1083/jcb.200502122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garcia-Murillas I, Pettitt T, Macdonald E, Okkenhaug H, Georgiev P, Trivedi D, et al. lazaro encodes a lipid phosphate phosphohydrolase that regulates phosphatidylinositol turnover during Drosophila phototransduction. Neuron. 2006;49:533–546. doi: 10.1016/j.neuron.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 117.Kwon Y, Montell C. Dependence on the Lazaro phosphatidic acid phosphatase for the maximum light response. Curr Biol. 2006;16:723–729. doi: 10.1016/j.cub.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 118.Yoon J, Ben-Ami HC, Hong YS, Park S, Strong LL, Bowman J, et al. Novel mechanism of massive photoreceptor degeneration caused by mutations in the trp gene of Drosophila. J Neurosci. 2000;20:649–659. doi: 10.1523/JNEUROSCI.20-02-00649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arundine M, Tymianski M. Molecular mechanisms of calcium-dependent neurodegeneration in excitotoxicity. Cell Calcium. 2003;34:325–337. doi: 10.1016/s0143-4160(03)00141-6. [DOI] [PubMed] [Google Scholar]
- 120.Wang T, Jiao Y, Montell C. Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. J Cell Biol. 2005;171:685–694. doi: 10.1083/jcb.200508030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.O'Tousa JE. Ca2+ regulation of Drosophila phototransduction. Adv Exp Med Biol. 2002;514:493–505. [PubMed] [Google Scholar]
- 122.Matsumoto H, Kurien BT, Takagi Y, Kahn ES, Kinumi T, Komori N, et al. Phosrestin I undergoes the earliest light-induced phosphorylation by a calcium/calmodulin-dependent protein kinase in Drosophila photoreceptors. Neuron. 1994;12:997–1010. doi: 10.1016/0896-6273(94)90309-3. [DOI] [PubMed] [Google Scholar]
- 123.Byk T, Bar-Yaacov M, Doza YN, Minke B, Selinger Z. Regulatory arrestin cycle secures the fidelity and maintenance of the fly photoreceptor cell. Proc Natl Acad Sci USA. 1993;90:1907–1911. doi: 10.1073/pnas.90.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee SJ, Xu H, Montell C. Rhodopsin kinase activity modulates the amplitude of the visual response in Drosophila. Proc Natl Acad Sci USA. 2004;101:11874–11879. doi: 10.1073/pnas.0402205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Orem NR, Dolph PJ. Loss of the phospholipase C gene product induces massive endocytosis of rhodopsin and arrestin in Drosophila photoreceptors. Vision Res. 2002;42:497–505. doi: 10.1016/s0042-6989(01)00229-2. [DOI] [PubMed] [Google Scholar]
- 126.Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu Rev Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- 127.Orem NR, Xia L, Dolph PJ. An essential role for endocytosis of rhodopsin through interaction of visual arrestin with the AP-2 adaptor. J Cell Sci. 2006;119:3141–3148. doi: 10.1242/jcs.03052. [DOI] [PubMed] [Google Scholar]
- 128.Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- 129.Sriram V, Krishnan KS, Mayor S. Deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J Cell Biol. 2003;161:593–607. doi: 10.1083/jcb.200210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Warner TS, Sinclair DA, Fitzpatrick KA, Singh M, Devlin RH, Honda BM. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome. 1998;41:236–243. [PubMed] [Google Scholar]
- 131.Acharya JK, Dasgupta U, Rawat SS, Yuan C, Sanxaridis PD, Yonamine I, et al. Cell-nonautonomous function of ceramidase in photoreceptor homeostasis. Neuron. 2008;57:69–79. doi: 10.1016/j.neuron.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ishii I, Fukushima N, Ye X, Chun J. Lysophospholipid receptors: signaling and biology. Annu Rev Biochem. 2004;73:321–354. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 133.Acharya U, Patel S, Koundakjian E, Nagashima K, Han X, Acharya JK. Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science. 2003;299:1740–1743. doi: 10.1126/science.1080549. [DOI] [PubMed] [Google Scholar]
- 134.Bursch W, Ellinger A, Gerner C, Frohwein U, Schulte-Hermann R. Programmed cell death (PCD). Apoptosis, autophagic PCD or others? Ann NY Acad Sci. 2000;926:1–12. doi: 10.1111/j.1749-6632.2000.tb05594.x. [DOI] [PubMed] [Google Scholar]
- 135.Nicholson DW. Caspase, structure proteolytic substrates and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 136.Yang YL, Li XM. The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000;10:169–177. doi: 10.1038/sj.cr.7290046. [DOI] [PubMed] [Google Scholar]
- 137.Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- 138.Conradt B. Genetic control of programmed cell death during animal development. Annu Rev Genet. 2009;43:493–523. doi: 10.1146/annurev.genet.42.110807.091533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hsu CD, Whaley MA, Frazer K, Miller DA, Mitchell KA, Adams SM, O'Tousa JE. Limited role of developmental programmed cell death pathways in Drosophila norpA retinal degeneration. J Neurosci. 2004;24:500–507. doi: 10.1523/JNEUROSCI.3328-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davidson FF, Steller H. Blocking apoptosis prevents blindness in Drosophila retinal degeneration mutants. Nature. 1998;391:587–591. doi: 10.1038/35385. [DOI] [PubMed] [Google Scholar]
- 141.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 143.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Komatsu M, Kominami E, Tanaka K. Autophagy and neurodegeneration. Autophagy. 2006;2:315–317. doi: 10.4161/auto.2974. [DOI] [PubMed] [Google Scholar]
- 145.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 146.Wang T, Lao U, Edgar BA. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J Cell Biol. 2009;186:703–711. doi: 10.1083/jcb.200904090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Midorikawa R, Yamamoto-Hino M, Awano W, Hinohara Y, Suzuki E, Ueda R, Goto S. Autophagy-dependent rhodopsin degradation prevents retinal degeneration in Drosophila. J Neurosci. 2010;30:10703–10719. doi: 10.1523/JNEUROSCI.2061-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schinz RH, Lo MV, Larrivee DC, Pak WL. Freeze-fracture study of the Drosophila photoreceptor membrane: mutations affecting membrane particle density. J Cell Biol. 1982;93:961–967. doi: 10.1083/jcb.93.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ni L, Guo P, Reddig K, Mitra M, Li HS. Mutation of a TADR protein leads to rhodopsin and Gq-dependent retinal degeneration in Drosophila. J Neurosci. 2008;28:13478–13487. doi: 10.1523/JNEUROSCI.2122-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]