Abstract
Myc is a transcription factor frequently found deregulated in human cancer. The Myc-mediated cellular transformation process is associated with fast proliferative cells and inherent genomic instability, giving rise to malignant, invasive neoplasms with poor prognosis for survival. Transcription-independent functions of Myc include stimulation of replication. Excessive Myc expression stimulates a replication-associated DNA damage response that signals via the phosphoinositide-3-kinase (PI3K)-related protein kinases (PIKKs) ATM and ATR. These, in turn, activate the DNA damage transducers Chk1 and Chk2. Here, we show that Myc can stimulate Chek2 transcript indirectly in vitro as well as in B cells of λ-Myc transgenic mice or in the intestine of ApcMin mice. However, Chk2 is dispensable for Myc's ability to transform cells in vitro and for the survival of established lymphoma cells from λ-Myc transgenic mice. Chk2 deficiency induces polyploidy and slow growth, but the cells are viable and protected against DNA damage. Furthermore, inhibition of both Chk1/Chk2 with AZD7762 induces cell death and significantly delays disease progression of transplanted lymphoma cells in vivo. DNA damage recruits PARP family members to sites of DNA breaks that, in turn, facilitate the induction of DNA repair. Strikingly, combining Chk2 and PARP inhibition elicits a synergistic lethal response in the context of Myc overexpression. Our data indicates that only certain types of chemotherapy would give rise to a synergistic lethal response in combination with specific Chk2 inhibitors, which will be important if Chk2 inhibitors enter the clinic.
Key words: lymphoma, Myc, Chk1, Chk2, PARP, DNA damage, AZD-7762, ABT-888
Introduction
The MYC family of transcription factors, including c-Myc (hereafter Myc), L-Myc and N-Myc, are functionally redundant transcription factors known to be deregulated in a majority of human cancers. Myc regulates a vast number of genes,1 and cells respond by the reprogramming of major cellular functions, including cell cycle progression, cell growth and metabolism, all hallmarks of cancer progression and cellular transformation. Fortunately, major tumor suppressive mechanisms are used to protect the cell from deregulated oncogenes, such as Myc. Two of these, oncogene-induced apoptosis and senescence, need to be circumvented in order for tumor progression to occur.2,3
Tumor progression relies on a certain amount of genomic instability to accumulate mutations in key tumor suppressor genes, such as Tp53.4 Checkpoints controlling genomic stability include the DNA damage response and repair machinery.5 DNA insults leading to single- or double-strand DNA breaks activate distinct checkpoint programs that depend on the function of the phosphoinositide-3-kinase (PI3K)-related protein kinases (PIKKs), including ataxia-telangiectasia mutated (ATM), ATM and RAD3-related (ATR) and DNA-dependent protein kinase (DNA-PK).6 The signaling events triggered by these kinases induce cell cycle progression delay but also trigger DNA repair mechanisms, both ensuring the fidelity of the genome and protection against tranformation.5
Key signal transducers of the DNA damage response include the serine/threonine kinases Chk1 and Chk2. Upon DNA damage, ATM and ATR phosphorylate and activate Chk1 and Chk2,7–9 further conveying the DNA damage signal in the cell. Chk1 and Chk2 share substrate specificity but are not redundant kinases,10 and phosphorylation targets include Cdc25 family members that, upon inactivation, lead to cell cycle arrest. Another important phosphorylation target of these kinases is the p53 tumor suppressor.11,12 Stabilization of p53 ensures a prolonged G2 arrest, and the induction of DNA repair can also stimulate apoptosis depending on the extent of DNA damage and cell type.13 Targeted deletion of Chek1 has been shown to be embryonic lethal,14 whereas vertebrate cells can survive without Chk2 but show defective checkpoint signaling.15 Chk2 is an established tumor suppressor, and inactivation in humans lead to Li-Fraumeni-like syndrome16 and an increased risk of developing breast cancer.17,18
Myc has recently been shown to induce DNA damage via its role at the replication fork, where Myc stimulates replication fork firing.19 This transcription-independent function of Myc triggers a DNA damage signal that is relayed through the ATM-ATR-Chk1 axis. Here, we show that Myc regulates Chk2, but Myc-overexpressing cells are not dependent on Chk2 for their survival or transformation potential. Furthermore, Chk2 abrogation induces polyploidy and protects lymphoma cells from DNA damage. Using a dual Chk1/Chk2 inhibitor, we also reveal that, even though Chk2 abrogation induces polyploidy, which is, itself, a tumor-promoting condition, this therapeutic approach delays disease progression in vivo. Finally, we present data demonstrating that Chk2 deficiency synergizes with PARP inhibition.
Results
Myc regulates Chk2.
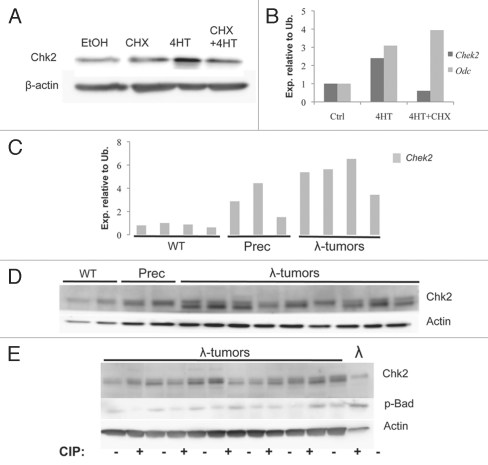
We have recently shown that Myc sensitizes cells to DNA damage.20,21 Following DNA damage, Myc can override several cell cycle checkpoints regulated by the PIKKs and downstream transducers Chk1 and Chk2 and further enforced by the p53 tumor suppressor, resulting in genomic destabilization and subsequent apoptosis.20 Since Myc deregulation has been shown to stimulate hyper-replication and DNA damage, we wanted to investigate the role and regulation of the DNA damage transducer Chk2 in a Myc-overexpressing context. To that end, we used NIH 3T3 fibroblasts and transduced these with a retrovirus engineered to express a fusion protein between c-Myc and the ligand-binding domain of the estrogen receptor (ER), the MycER protein.22 Addition of 4-hydroxytamoxifen (4-HT) to the cell culture media mediates the relocation of the MycER fusion protein from the cytoplasm to the cell nucleus, starting transcription of Myc target genes. Myc activation in these cells led to increased levels of Chk2 protein; this increase was not observed in cells pre-treated with the translation inhibitor cycloheximide (CHX, Fig. 1A). In order to investigate if Myc-mediated regulation of Chk2 was dependent on p53, we made mouse embryonic fibroblasts (MEFs) from E13.5 embryos from timed pregnancies between p53 heterozygous mice. Upon Myc activation, Chek2 transcript and protein was induced, but not when the cells were pre-treated with CHX. In contrast, Odc, a known Myc target gene,23 was regulated even in the presence of CHX, implying an indirect Chk2 regulation that requires de novo protein synthesis (Fig. 1B and data not shown).
Figure 1.
Myc upregulates Chk2 mRNA and protein levels. (A) Protein gel blot analysis of NIH 3T3 fibroblasts infected with MSCV-MycER-IRES-puro retrovirus. The nuclear translocation of MycER was induced by 4-HT for 24 h. Whole-cell lysates were harvested and analyzed using antibodies directed against the indicated proteins. (B) qRT-PCR analysis of Chek2 and Odc transcript levels in p53-knockout mEFs infected with MSCV-MycER-IRES-GFP retrovirus. 4-HT was added to the cells, and transcript expression was measured 24 h later, in the presence or absence of 1 µg/ml cycloheximide (CHX) in the growth media. (C) qRT-PCR analysis of Chek2 transcript levels in (B) cells from WT and γ-Myc mice as well as tumors developed in the γ-Myc transgenic animals. (D) Protein gel blot analysis of Chk2 protein levels in < 6-week-old wild-type (WT) and pre-cancerous γ-Myc mice compared with palpable lymphomas harvested from sick animals. (E) Lymphomas from sick γ-Myc mice were either treated with FastAP™ alkaline phosphatase or mock-treated (heat-inactivated alkaline phosphatase) and then analyzed by protein gel blot.
To assess if Chk2 is a Myc-regulated gene in vivo, we investigated the expression of Chk2 in γ-Myc transgenic mice, where the human MYC gene is expressed under the control of the Immunoglobulin γ enhancer to recapitulate the translocation occurring in a subset of Burkitt lymphoma. Splenic B cells from either precancerous γ-Myc transgenic mice or wild-type C57BL/6 littermates were magnetically sorted using IgM-specific antibodies. These cells and palpable lymphomas harvested from sick γ-Myc animals were then used to make protein lysates and RNA for protein gel blot and qRT-PCR analysis. Precancerous cells and all lymphomas exhibited high levels of Chek2 transcript as compared with wild-type control cells (Fig. 1C). However, analysis of Chk2 protein levels in the tumors revealed that these were comparable to wild-type and precancerous controls with the exception that a second band also was detectable (Fig. 1D). It is conceivable that this form represents an alternatively phosphorylated form of Chk2. Chk2 dimerization and auto-phosphorylation is required for Chk2 activity,24 and has previously been shown to give rise to such a band shift on SDS page.25 In order to investigate if this form was phosphorylated, we treated lysates of lymphomas from the γ-Myc mouse with FastAP™ Alkaline phosphatase (AP) and compared these to untreated lysates from the same tumor. Intriguingly, this treatment did not affect the band suspected to be the phosphorylated form of Chk2 but did reduce phosphorylation of the anti-apoptotic Bcl-2 family member Bad (Fig. 1E). Moreover, a cell line established from a tumor of a γ-Myc mouse did not display the lower of the detected bands, suggesting that this alternate form of Chk2 is an effect of in vivo tumor progression.
Myc is deregulated in most human cancers due to indirect activation by upstream pathways. Most colon cancer carries a mutation in the APC gene, giving rise to excessive Wnt/β-catenin signaling and downstream c-Myc activation.26 We wanted to investigate if tumors arising in this setting regulate Chk2. In order to answer this question, we screened ApcMin mice that carry a mutation in the adenomatus polyposis coli (APC) gene. These mice develop spontaneous adenomas in the colon and small intestine at around 120 d of age.27 Comparing normal tissue with palpable adenomas of the small intestine, we detected an upregulation of Chek2 transcript that also correlated with Myc expression (Fig. S1).
Chk2 is dispensable for Myc induced colony formation.
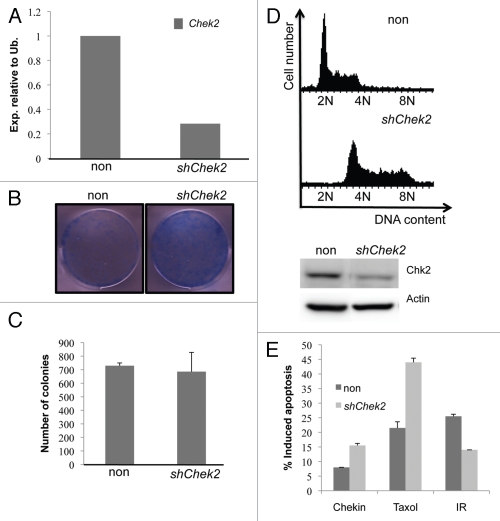
Chk2 is, as shown above, regulated by Myc in vitro and in vivo, suggesting that it could be important for Myc-mediated transformation. In order to investigate this, we genetically depleted Chek2 mRNA using shRNA in Myc-overexpressing NIH 3T3 fibroblasts (Fig. 2A). Clonogenic survival assays over 10 days showed that removal of Chek2 did not compromise the ability of Myc to colonize these plates (Fig. 2B), nor did it affect Myc's ability to transform cells in soft agar (Fig. 2C). Interestingly, however, the Chek2 deficient fibroblasts appeared distorted in morphology (data not shown). Many of these were larger than control-infected cells, and immunofluorescence analysis of mitotic cells using antibodies against tubulin demonstrated a higher percentage of Chk2-deficient cells stuck in mitosis (Fig. S2). These data suggests a dependency of these cells on Chk2 to properly execute mitosis.
Figure 2.
Chk2 is dispensable for Myc-induced colony formation, and Chk2 deficiency protects lymphoma cells against DNA damage. (A) qRT-PCR analysis of Chek2 transcript levels NIH-3T3 fibroblasts infected with MSCV-Myc-IRES-GFP retroviruses and shRNA against Chek2 or a non-target shRNA. (B) Clonogenic survival assay of 104 NIH-3T3 fibroblasts of cells in (A). The cells were grown to confluence during the course of 10 d and then stained with Commasie blue. (C) The cells constructed in (A) were also put in soft agar and grown for two weeks. Colonies were counted using bright-field microscopy on three separate plates, here presented as an average of these. (D) A mouse lymphoma cell line from the γ-Myc transgenic mouse was infected with shRNA against Chek2 or control vector (protein gel blot inset). After a few passages, the cells were harvested, stained with propidium iodine (PI) and analyzed by flow cytometry to generate DNA histograms. (E) The Chk2-deficient cells were treated for 48 h with 1 µM Chekin, 50 nM Taxol or 5 Gy of gamma irradiation (IR). Apoptosis was scored by analysis of the sub-G1 population of PI stained cells using flow cytometry.
Recently, Chk2-dependent BRCA1 phosphorylation was implicated as an important regulator of chromosomal instability (CIN).28 BRCA1 localizes to mitotic centrosomes29 and is required for proper spindle assembly,30 thus Chk2 deficiency results in a failure to properly align duplicated chromosomes, leading to lagging chromosomes and increased genomic instability. Interestingly, when we introduced shRNA against Chek2 in a mouse lymphoma cell line derived from the γ-Myc transgenic mouse, these cells became severely polyploid within a few passages (Figs. 2D and S3A). Even though the cells tolerated this genomic instability, their generation time was severely affected compared with control infected cells (Fig. S3B). Genomic instability has been proposed to be an emerging hallmark of cancer that drives tumor progression.31 Because of this, we went on to transplant the Chk2-deficient polyploid lymphoma cells into recipient animals and monitored these for visible signs of disease. The cells lacking Chk2 expression had a significantly slower disease progression than control-infected cells (p = 0.0295) (Fig. S3C), in line with the slower growth phenotype observed in vitro. When sick, mouse tumor material was snap frozen and prepared for protein gel blot analysis. Interestingly, tumors did not retain Chk2 knockdown (protein gel blot inset Fig. S2C) but remained polyploid (data not shown), suggesting that a selection against cells with low Chk2 expression had occurred in vivo. Furthermore, the tumors that emerged also retained the band shift observed in the γ-Myc mice tumors; this band was not present in the parental cell line injected (Fig. 2D). Importantly, moribund mice transplanted with Chk2-deficient cells did not exhibit a different or more invasive tumor spectra then control animals (data not shown). Thus, the slower growth rate of the Chk2-deficient cells was dominant in vivo, and the polyploidization induced by Chk2 removal did not negatively affect disease progression.
Chk2 is an important cell cycle regulator in response to DNA damage, affecting both the S-phase32 and G2-phase checkpoints.33 Chk2-targeted therapy is currently being pursued in order to augment the effect of DNA damage-related therapy.34 In light of this, we wanted to investigate the potential behind Chk2 abrogation in combination with DNA damage in a Myc-overexpressing setting. We applied a lethal dose (5 Gy) of irradiation (IR) to the above-generated Chk2-deficient lymphoma cells and scored for apoptotic cells following propidium iodine (PI) staining and flow cytometry (FACS) analysis. Strikingly, the Chk2-deficient cells did not respond as potently as control cells (Fig. 2E). We also treated the same cells with the microtubule-stabilizing drug Taxol or the novel Chk1 inhibitor Chekin.62 Interestingly, these drugs generated a more potent response in the cells lacking Chk2 expression (Fig. 2E). Collectively, these data suggest that Chk2-targeted therapy could be useful when combined with some but not all chemotherapies.
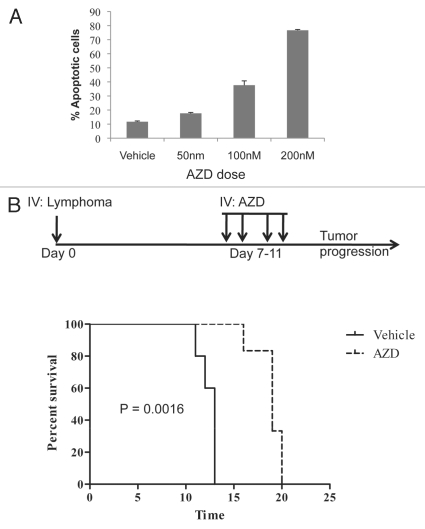
The dual Chk1/Chk2 inhibitor AZD7762 delays disease onset of transplanted lymphoma cells in vivo.
Several dual Chk1/Chk2 inhibitors, including UCN-01, PF-00477736 and AZD7762, are currently in clinical trials.34 In order to model the effect of dual Chk1/Chk2 inhibition, we obtained AZD7762 (AZD), which has been shown to potentiate the effect of DNA damage in xenograft studies.35 Treatment with increasingly higher doses of AZD over the course of 48 h correlated with an increased apoptotic response in mouse lymphoma cells with close to 80% apoptotic cells scored at a concentration of 200 nM AZD (Fig. 3A). To evaluate the effect of AZD in vivo, we generated a transplantable lymphoma model by infecting bone marrow-derived B cells from p53-knockout mice with an MSCV-Myc-IRESGFP virus. Mice transplanted with these cells develop aggressive B-cell lymphomas.62 The lymphomas were injected into recipient C57BL/6 animals and divided into two groups receiving injections for four days of either vehicle (n = 5) or 25 mg/kg/qd AZD (n = 6) (Fig. 3B, dosage schedule). The mice were then observed for signs of disease. Strikingly, AZD-treated animals had a significantly slower disease progression (Median survival; AZD: 13 d vs. Vehicle: 19 d, p = 0.0016) (Fig. 3B, survival curve). These data are consistent with the Chek2 RNAi results.
Figure 3.
Chk1/Chk2 inhibition induces apoptosis in mouse lymphoma cells and delays disease onset in vivo. (A) Lymphoma cells from the γ-Myc transgenic mouse were treated with increasingly higher doses of AZD7762 (AZD) during the course of 48 h as indicated. Apoptosis was scored by analysis of the sub-G1 population of PI-stained cells using flow cytometry. (B) Recipient mice were transplanted with B-cell lymphoma cells and, after a week, treated (25 mg/kg/qd) for 4 d with IV injections of AZD (dose schedule insert). The mice were then monitored for palpable lymphomas. AZD-treated animals had a median survival time of 19 d compared with 13 d for vehicle-treated animals (p = 0.0016).
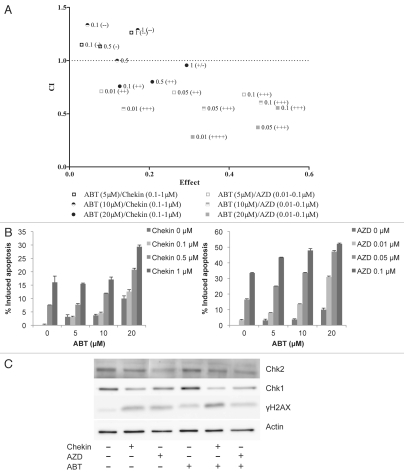
Dual PARP and Chk2 inhibition elicits a synergistic response in mouse lymphoma cells.
In response to cellular stress by, for exmaple, reactive oxygen species (ROS), PARP family members modulate cellular response by physical interaction or poly-(ADP-ribosyl)ation of partner proteins. PARP family members have been implicated in genome maintenance functions like DNA repair, chromatin remodulation and transcription.36 However, PARP-1 activation is also implicated in a number of age-related diseases due to its role as a transcriptional co-factor to NFκB and inflammationpromoting abilities.37 Inhibition of PARP has beneficial implications for certain age-related diseases, but also results in accumulation of single-stranded DNA breaks (SSBs) that, when encountered by a replication fork, get converted into double-stranded DNA breaks (DSBs).38 Homologs recombination (HR) is the preferred DNA repair pathway for such lesions and is activated via ATM-dependent phosphorylation cascades. Specifically, ATM activation leads to DNA end resection controlled by the MRN-BRCA1 complex.39,40 The exact functions of BRCA1 in the induction of HR is unclear, but DNA end resection leads to the formation of 3′ ssDNA and RPA recruitment, followed by BRCA2-mediated RAD51 filament formation, in turn stimulating HR. The use of PARP inhibitors has been shown to cause synergistic lethality in the context of BRCA1 and BRCA2 deficiency where HR is inactive.38,41 Since both Chk1 and Chk2 stimulate HR,42–44 we wanted to evaluate the use of combinatorial Chk1/Chk2- and PARP inhibition in our model system. In order to do so, we used the PARP inhibitor ABT-888 (ABT).45 Combination therapy of ABT with either AZD (inhibits both Chk1 and Chk2) or Chekin (inhibits Chk1)62 in a mouse lymphoma cell line produced a synergistic effect in both treatment regimes when using the median effect analysis by Chou and Talalay,46 as assessed by PI staining and flow cytometry analysis (Fig. 4A). However, all doses of AZD evaluated produced a synergistic effect when combined with ABT (CI < 1, Range 0.282–0.711), whereas Chekin treatment only slightly synergized with the highest dose of ABT (CL < 1, Range 0.759–0.956) (Fig. 4A and Table 1). The increase in apoptosis was moderate in Chekin- and ABT-treated samples but produced a robust enhancement of apoptosis with increasing doses of ABT in combination with AZD (Fig. 4B). In order to check target specificity, we treated lymphoma cells with select doses of Chekin and AZD in combination with ABT (Fig. 4C). Chk1 stability is affected when activity is inhibited and DNA damage is applied,47 and, predictably, Chekin potently reduced Chk1 protein levels whereas AZD did so to a lesser degree. Chekin and AZD, as well as combinations with ABT, also induced an increased DNA damage as scored by phosphorylated histone H2AX (γH2AX, Fig. 4C). Our data suggests that Chk2 appears to be dominant when compared with Chk1 in determining sensitivity to combinatorial PARP inhibition in our model system.
Figure 4.
Dual Chk2 and PARP inhibition elicits a synergistic lethal response. (A) Lymphoma cells from the γ-Myc transgenic mouse were treated with combinations of either AZD and ABT-888 (ABT) or Chekin and ABT and assessed for apoptosis by analysis of the sub-G1 population of PI-stained cells using flow cytometry. Synergistic response was calculated using the median effect analysis of the CalcuSyn software (Biosoft).46 Depicted are also drug doses of Chekin, ABT and AZD used (µM). [Combination Index (CI) values of < 1 indicate synergy; effect describes the fraction of apoptotic cells at the various drug combinations]. (B) Apoptotic response in samples described in (A). AZD and ABT combinatorial treatment leads to an increase in apoptosis with increasing concentrations of the drugs as indicated. (C) Mouse lymphoma cells were treated for 48 h with either Chekin (1 µM) or AZD (0.1 µM) alone or in combinations with ABT (10 µM) and then harvested for protein gel blot analysis.
Table 1.
Statistical analysis of Chekin/ABT and AZD/ABT treated mouse lymphoma cells.
| ABT (µM) | Chekin (µM) | Fa | CI | Description |
| 5 | 0.1 | 0.0305 | 1.149 | Slight antagonism (−) |
| 5 | 0.5 | 0.077 | 1.133 | Slight antagonism (−) |
| 5 | 1 | 0.155 | 1.263 | Moderate antagonism (−−) |
| 10 | 0.1 | 0.045 | 1.335 | Moderate antagonism (−−) |
| 10 | 0.5 | 0.1195 | 1.000 | Additive |
| 10 | 1 | 0.171 | 1.289 | Moderate antagonism (−−) |
| 20 | 0.1 | 0.126 | 0.759 | Moderate synergism (++) |
| 20 | 0.5 | 0.2075 | 0.801 | Moderate synergism (++) |
| 20 | 1 | 0.2935 | 0.956 | Near additive (+/−) |
| ABT (µM) | AZD (µM) | Fa | CI | Description |
| 5 | 0.01 | 0.08 | 0.711 | Moderate synergism (++) |
| 5 | 0.05 | 0.262 | 0.701 | Moderate synergism (++) |
| 5 | 0.1 | 0.437 | 0.681 | Synergism (+++) |
| 10 | 0.01 | 0.1355 | 0.547 | Synergism (+++) |
| 10 | 0.05 | 0.3365 | 0.550 | Synergism (+++) |
| 10 | 0.1 | 0.4795 | 0.606 | Synergism (+++) |
| 20 | 0.01 | 0.3085 | 0.282 | Strong synergism (++++) |
| 20 | 0.05 | 0.472 | 0.372 | Synergism (+++) |
| 20 | 0.1 | 0.5215 | 0.555 | Synergism (+++) |
Mathematical analysis of synergy was calculated using CalcuSyn software (Biosoft) and has been described in reference 46. Indicated are the various drug combination doses used and the synergistic response elicited as assessed by apoptotic analysis cells of the sub-G1 population of PI stained cells using flow cytometry. (Fa = Fraction affected, CI = Combinatorial Index; <1 indicates synergy).
Discussion
The Myc family of transcription factors are deregulated in a majority of human cancers,3 making the pathways regulated by Myc, and Myc itself, attractive targets for chemotherapy. The challenge lies with the identification of target proteins in Myc-overexpressing tumors that govern key signaling hubs essential for tumor maintenance. Targeting proteins in the Myc transcriptome has been shown by us to be a valid approach for treatment of disease, both as chemoprevention and in treatment of solid tumors.48–50 Here, we show that the checkpoint kinase Chk2 is indirectly regulated at the RNA level by Myc in vitro and in vivo. Even though Chk1 and Chk2 share substrate specificity, they are not redundant kinases. Chek1-knockout mice are embryonically lethal,14 and mutations or silencing of this kinase are seldom found in human cancer.51,52 Chek2, on the other hand, is not essential for embryonic survival15 but is an established tumor suppressor, where Chk2 deficiency predisposes to several types of human cancer.53,54
Over 90 splice variants of CHEK2 have been reported in human breast cancer cell lines.55 The function of all of these remains to be elucidated, but at least a subset seems to interfere with wild-type Chk2 function,56 which, in turn, promotes tumor progression due to the role of Chk2 as a tumor suppressor. In several γ-Myc lymphomas, we detect the expression of another form of Chk2 that does not appear to be derived from a phosphorylation event. This could, therefore, be an alternatively spliced form of Chek2 mRNA. In our model system, the same size of protein is observed in all tumors. The splice variants observed in reference 55, on the other hand, appear to be randomly selected for because of the observed complexity in the Chek2 splice forms. This suggests that specific regulation occurs in γ-Myc lymphomas in vivo, which is not seen in in vitro growth conditions. It would appear highly unlikely that the alternatively expressed form of Chk2 would exert any kind of DN effects on wt Chk2, since in our lymphoma model, Chk2 deficiency results in slower cell growth in vitro and in vivo. A previous report has shown splice variants of Chk2 without DN effects on wt Chk2 and also with specific cellular localization,57 which provocatively would exert a positive influence on genomic stability in our model system. The mechanism of Myc-dependent Chk2 regulation observed herein remains elusive, but it is not unlikely that Chk2 is regulated due to Myc's ability to induce S-phase progression and/or DNA damage.19
Our data suggests that Chk2 is dispensable for Myc-overexpressing NIH 3T3 fibroblasts' ability to survive and form colonies in in vitro transformation assays. Interestingly, removing Chek2 using shRNA in lymphoma cells from γ-Myc mice induces polyploidy and growth retardation, both in vitro and in vivo. This is in line with a previous study showing a connection between Chk2 and proper chromosomal segregation, where Chk2 deficiency induces aneuploidy in HCT 116 colon cancer cells.28 Clearly, Chk2 is dispensable for Myc-overexpressing cancer cells to survive, and the induced polyploidy could even benefit tumor progression long-term, as genomic instability has been proposed as an emerging hallmark that drives multistep tumor progression.31
Targeting the Chk1 and Chk2 kinases in combination with various DNA damage agents are currently being pursued as a means of producing better clinical outcome in the treatment of various human cancers.34 In our lymphoma cells, Chk2 deficiency resulted in radioprotection. Most likely this was an effect of the severe growth retardation seen in these cells. Considering that the experiments were run over short time points (48 h), and because the apoptotic effect of DNA damage correlates to genomic instability acquired with the number of cells doublings, it is possible that, over a longer time, the effect would be equivalent, independent of Chk2 status. However, Carlessi et al. also show that Chk2 inhibition in combination with radiotherapy results in protection.58 This, along with the fact that Chk2 deficiency induces polyploidy, which, in itself, could drive more aggressive clonal outgrowth, highlights the need for more studies before Chk2-specific inhibitors are introduced into the clinic. Our data also implies that the enhanced effect of DNA damage-related therapies in combination with dual Chk1/Chk2 inhibitors like AZD7762 is the result of Chk1 inhibition,35 but could also be cell context-dependent, since both radioprotection and radiosensitization have been reported in Chk2-deficient settings.58,59 Interestingly though, Chk2 deficiency resulted in sensitization to Chk1 inhibition and Taxol treatment. These data suggest that the mitotic defects observed in these cells renders them more sensitive to further genomic destabilization by drugs that affect the mitotic checkpoint. Taxol causes a mitotic defect by stabilization of microtubules, whereas Chk1 not only share ssubstrate specificity with Chk2, but has also been implicated in mechanisms of proper chromosome segregation in unperturbed cells.60
The established role of Chk2 as a tumor suppressor, as well as the consequences of Chk2 abrogation discussed above, puts Chk2-targeted therapy in question. However, pursuit of synergistic pharmacological interactions could establish a use for specific Chk2 inhibitors in the clinic. The use of PARP inhibitors in anticancer therapy shows potential in combination with genotoxic insult that would normally be repaired through base excision repair,61 but also exhibits synthetic lethality with HR deficient tumor cells.38,41 Both Chk1 and Chk2 have previously been implicated as important for the induction of HR following DSBs.42–44 Intriguingly, our data demonstrate that, in the context of Myc overexpression, Chk2 inhibition appears to be the determining factor in combinatorial synergistic lethality with PARP inhibition. However, we cannot exclude the possibility that both Chk1 and Chk2 are important for regulation of HR in our model system, and that the effect seen with the dual Chk1/Chk2 inhibitor AZD reflects this fact. Anderson et al. recently published a synergistic lethal response in human cancer cells to dual PARP and Chk2 inhibition using a new novel Chk2 inhibitor with minimal specificity for Chk1.25 These data together demonstrate a possible therapeutic application for specific Chk2 inhibitors.
Collectively, our data show that the usage of specific Chk2-targeted therapy needs to be selective in a clinical setting. Not only could Chk2 abrogation cause more aggressive tumor outgrowth due to the polyploidy observed herein and reference 28, but it could also protect against certain types of chemotherapeutic approaches. On the other hand, our data also demonstrates that PARP inhibition holds promise as an anticancer strategy in tumors with inherent or induced Chk2 deficiency.
Materials and Methods
Materials.
Primary antibodies were obtained from Santa Cruz (Myc, p19), Sigma (β-actin and Tubulin) and Cell Signaling (Chk2, p-Bad). Horseradish peroxidise-conjugated antibodies against mouse and rabbit antibodies were from GE Healthcare Life Sciences. Secondary antibody anti-mouse DyLight 488 was purchased from Immunkemi F&D AB. The Chk1 inhibitor “Chekin” was synthesized by Abbott Laboratories and is described elsewhere.62 AZD7762 and ABT-888 were obtained from Axon Medchem. FastAPTM Alkaline phosphatase was purchased from Fermentas.
Cell culture.
293T human kidney cells and NIH 3T3 fibroblasts were purchased from ATCC and cultured in Dulbecco's modified Eagle medium with 10% fetal calf serum (FCS), 2 mM L-glutamine, 1 mM sodium pyruvate and antibiotics. Mouse lymphoma cell lines established from tumors arising in the λ-Myc transgenic mice were cultured at a density of 105 cell/ml in RPMI1640 medium with 5% FCS, 2 mM L-glutamine, 50 µM β-mercaptoethanol, 0.1875% sodium bicarbonate and antibiotics. Mouse embryo fibroblasts (MEFs) were generated from E13.5-E15 embryos from timed mating between p53 heterozygous males and females according to previous methodology.
Viral infections.
Retroviruses were made by calcium phosphate-mediated co-transfection of 293T cells with MSCV-IRES-puro (with or without MycER) together with ecotropic helper plasmids expressing gag, pol and env. Twenty-four h post-transfection supernatants from the cells were harvested three times every eight hours, filtered and used to infect p53−/− MEFs in the presence of 8 µg/ml polybrene. Cells infected with MSCV-IRES-puro-based retroviruses were selected in the presence of 6 µg puromycin.
Lentiviral infections were made by calcium phosphate-mediated co-transfection of 293T cells with packaging plasmids pCMV-dR8.2 dvpr and pHCMV-Eco (Addgene) using five different MISSION shRNA constructs (Sigma) directed against Chek2. Twenty-four h post-transfection, the different supernatants were harvested three times every eight hours, filtered and then used to infect target cells. Mouse lymphoma cells were infected by two rounds of spinoculation (20 min at 50x g) 24 h apart in the presence of 2 µg/ml polybrene. Mouse fibroblasts were infected by culturing the cells in the presence of viral particles and 8 ug/ml of polybrene. The cells were selected by culturing them in the presence of 2–6 µg/ml puromycin.
Cell cycle and apoptosis analyses.
For cellular staining with propidium iodine (PI), mouse B cells were collected by centrifugation together with its original culture supernatant. The cells were resuspended in 0.5 ml Vindelovs reagent (10.0 mM Tris, 10 mM NaCl, 75 µM propidium iodine, 0.1% Igepal and 700 units/liter RNase adjusted to pH 8.0). The PI-stained cells were kept in the dark at 4°C for 30–60 min and then analyzed with a FACScalibur flow cytometer (BD Biosciences) using the FL3 channel in a linear scale. Apoptosis was determined using DNA histograms on PI-stained cells (as above) and was based on the number of cells that carried less than diploid DNA content (subG1) in a logarithmic FL2 channel.
Protein gel blot analysis.
Cell pellets or tumors crushed in liquid nitrogen were lysed essentially as described before.20 The debris was removed by centrifugation, and the protein concentrations were determined using Bio-Rad's protein determination reagent. 30–50 µg proteins per lane were separated on SDS-PAGE gels and subsequently transferred to nitrocellulose membranes (Schleicher-Schuell). Membranes were stained with Ponceau S red dye to verify equal loading. All subsequent steps were performed in TBS-Tween (10 mM TRIS-HCl, pH 7.6, 150 mM NaCl and 0.05% Tween-20) either containing 5% milk (blocking and antibody incubations), or 5% BSA (phospho-specific antibody incubations). Antibody binding was visualized by enhanced chemiluminescence using the SuperSignal West Dura or Pico reagents from Pierce. For FastAP™ Alkaline phosphatase (AP) treatment, crushed tumor pieces were either lysed in a buffer containing phosphatase inhibitors or in a lysis buffer without inhibitors. They were then either mock treated (Heat inactivated AP) or treated with AP (0.5 U/µg), respectively, for 1 h at 37°C. The reaction was stopped by heat inactivation at 75°C and by supplement of 10 mM of sodium orthovanadate to the lysis buffer. The samples were then separated on a SDS page gel and transferred to nitrocellulose membranes.
Immunoflourescence.
Briefly, cells were fixed in MeOH at −20°C for 1 h and then blocked in phosphatase buffered saline containing 10% FCS and 0.1% Saponin. Samples were then incubated for 16 h at 4°C with tubulin antibodies. Secondary anti-mouse Dylight-488 staining was performed during 1 h at 37°C. Cells were counterstained with PI and mounted for microscopy analysis using a standard cytospin protocol.
RNA preparation and analysis by quantitative reverse transcription PCR (qRT-PCR).
RNA from cultured cells was isolated using NucleoSpin RNA II (Macherey-Nagel). cDNA synthesis was performed on 1 µg RNA using an iScript first strand synthesis kit (Bio-Rad). qRT-PCR was performed using the KAPA SYBR FAST qPCR Kit (Kapabiosystems); cDNA and primers directed against Odc, Chek2, Myc and Ubiquitin were run on an IQ real-time PCR machine (Bio-Rad). Relative mRNA levels were calculated using the ΔΔCT method.
Mouse experiments.
All animal experiments were performed in accordance with the Regional Animal Ethic Committee Approval #A6-08 or #A18-08. The p53-knockout mice and ApcMin, both on a C57BL/6 background, were obtained from the Jackson lab. The γ-Myc mice were a kind gift from Dr. Georg Bornkamm (GSF). All transgenic mice were observed daily for signs of disease. All moribund mice were immediately sacrificed. When tumor-bearing mice were sacrificed, tumors and lymphoid organs were collected for analyses or tissue banking. Tumors were either snap frozen down as pieces and/or dispersed into single-cell suspensions by scalpels and cell strainers.
For the lymphoma transplant assay, recipient C57BL/6 mice were injected via intravenous injection of 500,000 cells carrying either an shRNA against Chek2 or a non-targeting vector and then monitored for tumor progression. When palpable lymphoma was observed, the mice were sacrificed, and tumor material was snap frozen for protein gel blot analysis.
To develop a p53-deficient Myc-driven in vivo model, we magnetically sorted bone marrow-derived B cells by labeling them with an anti-B220-R-PE antibody and anti-PE magnetic microbeads, followed by loading on a MACS column (Miltenyi). The purified B cells were cultured overnight in RPMI1640 medium with 10% FCS, 2 mM L-glutamine, 50 µM β-mercaptoethanol, 0.1875% sodium bicarbonate and antibiotics in the presence of MSCV-Myc-IRES-GFP retrovirus, produced as described above, and 4 µg/ml polybrene. Infected cells were injected into C57BL/6 mice, and tumor development was monitored and frozen down in medium containing 10% DMSO for banking.62 For drug experiments, cells were thawed, and 150,000 cells were intravenously injected per mouse. After one week, AZD7762 (25 mg/kg, n = 6) or vehicle (β-hydroxypropylcyclodextrin 10%, n = 5) was injected once daily via intravenous injection, for four days after which tumor development was observed.
Statistical analysis.
Statistical analyses of mouse survival curves were performed using a Log Rank (Mantel Cox) Test in GraphPad Prism (GraphPad Software) and only p-values < 0.05 were considered statistically significant. The error bars shown in experiments represent the mean of triplicates ± standard deviation (SD) as calculated by the STDEVA function in Excel. For drug synergy calculations, we used the median effect analysis by Chou and Talalay46 in the CalcuSyn software from Biosoft.
Acknowledgments
We thank the personnel at Umeå Transgene Core Facility for animal care. This work was supported by the Swedish Cancer Society, the Association of International Cancer Research, The Swedish Research Council, the Kempe foundation, Norrland's/Lion's Cancer foundation and Umeå University (career grant).
Abbreviations
- ABT
ABT-888
- ATM
ataxia-telangiectasia mutated
- ATR
ATM and RAD3 related
- AP
alkaline phosphatase
- APC
adenomatus polyposis coli
- AZD
AZD7762
- CHX
cycloheximide
- CIN
chromosomal instability
- DNA-PK
DNAdependent protein kinase
- DN
dominant negative
- DSB
double-stranded DNA break
- ER
estrogen receptor
- FACS
flow cytometry
- FCS
fetal calf serum
- HR
homologous recombination
- IR
γ-irradiation
- MEF
mouse embryonic fibroblast
- PI
propidium iodide
- PIKK
phosphoinositide-3-kinase (PI3K)-related protein kinase
- SSB
single-stranded DNA break
- 4-HT
4-hydroxy tamoxifen
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Hydbring P, Larsson LG. Cdk2: a key regulator of the senescence control function of Myc. Aging (Albany NY) 2010;2:244–250. doi: 10.18632/aging.100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22:9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 5.Seviour EG, Lin SY. The DNA damage response: Balancing the scale between cancer and ageing. Aging (Albany NY) 2010;2:900–907. doi: 10.18632/aging.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 7.Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/S1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 11.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 12.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 13.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 14.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 15.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 17.Cipollini G, Tommasi S, Paradiso A, Aretini P, Bonatti F, Brunetti I, et al. Genetic alterations in hereditary breast cancer. Ann Oncol. 2004;15:7–13. doi: 10.1093/annonc/mdh651. [DOI] [PubMed] [Google Scholar]
- 18.Kwak EL, Kim S, Zhang J, Cardiff RD, Schmidt EV, Haber DA. Mammary tumorigenesis following transgenic expression of a dominant negative CHK2 mutant. Cancer Res. 2006;66:1923–1928. doi: 10.1158/0008-5472.CAN-05-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 20.Höglund A, Nilsson LM, Forshell LP, Maclean KH, Nilsson JA. Myc sensitizes p53-deficient cancer cells to the DNA-damaging effects of the DNA methyltransferase inhibitor decitabine. Blood. 2009;113:4281–4288. doi: 10.1182/blood-2008-10-183475. [DOI] [PubMed] [Google Scholar]
- 21.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol. 2003;23:7256–7270. doi: 10.1128/MCB.23.20.7256-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem. 2002;277:19389–19395. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- 25.Anderson VE, Walton MI, Eve PD, Boxall KJ, Antoni L, Caldwell JJ, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Res. 2011;71:463–472. doi: 10.1158/0008-5472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-Myc as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 27.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 28.Stolz A, Ertych N, Kienitz A, Vogel C, Schneider V, Fritz B, et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol. 2010;12:492–499. doi: 10.1038/ncb2051. [DOI] [PubMed] [Google Scholar]
- 29.Hsu LC, White RL. BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci USA. 1998;95:12983–12988. doi: 10.1073/pnas.95.22.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 33.Brown AL, Lee CH, Schwarz JK, Mitiku N, Piwnica-Worms H, Chung JH. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashwell S, Zabludoff S. DNA damage detection and repair pathways—recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14:4032–4037. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- 35.Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 36.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 37.Altmeyer M, Hottiger MO. Poly(ADP-ribose) polymerase 1 at the crossroad of metabolic stress and inflammation in aging. Aging (Albany NY) 2009;1:458–469. doi: 10.18632/aging.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 39.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 40.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 42.Sørensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-18.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC, et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006;66:1401–1408. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 45.Donawho CK, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 46.Chou TC. Theoretical basis, experimental design and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 47.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forshell TP, Rimpi S, Nilsson JA. Chemoprevention of B-cell lymphomas by inhibition of the Myc target spermidine synthase. Cancer Prev Res (Phila) 2010;3:140–147. doi: 10.1158/1940-6207.CAPR-09-0166. [DOI] [PubMed] [Google Scholar]
- 49.den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, et al. Aurora kinases A and B are upregulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116:1498–1505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forshell LP, Li Y, Forshell TP, Rudelius M, Nilsson L, Keller U, et al. The direct Myc target Pim3 cooperates with other Pim kinases in supporting viability of Myc-induced B-cell lymphomas. Oncotarget. 2011;2:448–460. doi: 10.18632/oncotarget.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertoni F, Codegoni AM, Furlan D, Tibiletti MG, Capella C, Broggini M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer. 1999;26:176–180. doi: 10.1002/(SICI)1098-2264(199910)26:2<176::AID-GCC11>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 53.Williams LH, Choong D, Johnson SA, Campbell IG. Genetic and epigenetic analysis of CHEK2 in sporadic breast, colon and ovarian cancers. Clin Cancer Res. 2006;12:6967–6972. doi: 10.1158/1078-0432.CCR-06-1770. [DOI] [PubMed] [Google Scholar]
- 54.Zhang P, Wang J, Gao W, Yuan BZ, Rogers J, Reed E. CHK2 kinase expression is downregulated due to promoter methylation in non-small cell lung cancer. Mol Cancer. 2004;3:14. doi: 10.1186/1476-4598-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staalesen V, Falck J, Geisler S, Bartkova J, Borresen-Dale AL, Lukas J, et al. Alternative splicing and mutation status of CHEK2 in stage III breast cancer. Oncogene. 2004;23:8535–8544. doi: 10.1038/sj.onc.1207928. [DOI] [PubMed] [Google Scholar]
- 56.Berge EO, Staalesen V, Straume AH, Lillehaug JR, Lonning PE. Chk2 splice variants express a dominant-negative effect on the wild-type Chk2 kinase activity. Biochim Biophys Acta. 2010;1803:386–395. doi: 10.1016/j.bbamcr.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Golan A, Pick E, Tsvetkov L, Nadler Y, Kluger H, Stern DF. Centrosomal Chk2 in DNA damage responses and cell cycle progression. Cell Cycle. 2010;9:2647–2656. doi: 10.4161/cc.9.13.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6:935–944. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- 59.Yu Q, Rose JH, Zhang H, Pommier Y. Antisense inhibition of Chk2/hCds1 expression attenuates DNA damage-induced S and G2 checkpoints and enhances apoptotic activity in HEK-293 cells. FEBS Lett. 2001;505:7–12. doi: 10.1016/S0014-5793(01)02756-9. [DOI] [PubMed] [Google Scholar]
- 60.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.dev-cel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, et al. Chemopotentiation of temozolomide, irinotecan and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol Cancer Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 62.Höglund A, Nilsson L, Muralidharan SV, Hasvold LA, Merta P, Rudelius M, et al. Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-11-1198. In press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.