Abstract
In the phytopathogenic fungus Ustilago maydis, pheromone-mediated cell fusion is a prerequisite for the generation of the infectious dikaryon. The pheromone signal elevates transcription of the pheromone genes and elicits formation of conjugation hyphae. Cyclic AMP and mitogen-activated protein kinase (MAPK) signaling are involved in this process. The MAPK cascade is presumed to be composed of Ubc4 (MAPK kinase kinase), Fuz7 (MAPK kinase), and Ubc3/Kpp2 (MAPK). We isolated the kpp4 gene and found it to be allelic to ubc4. Epistasis analyses with constitutively active alleles of kpp4 and fuz7 substantiate that Kpp4, Fuz7, and Kpp2/Ubc3 are components of the same module. Moreover, we demonstrate that Fuz7 activates Kpp2 and shows interactions in vitro. Signaling via this cascade regulates expression of pheromone-responsive genes, presumably through acting on the transcription factor Prf1. Interestingly, the same cascade is needed for conjugation tube formation, and this process does not involve Prf1. In addition, fuz7 as well as kpp4 deletion strains are nonpathogenic, while kpp2 deletion mutants are only attenuated in pathogenesis. Here we show that strains expressing the unphosphorylatable allele kpp2T182A/Y184F are severely affected in tumor induction and display defects in early infection-related differentiation.
The phytopathogenic fungus Ustilago maydis, the causal agent of corn smut disease, displays a complex life cycle, which is linked to distinct morphological changes (27). In its haploid form U. maydis divides by budding and is nonpathogenic. After fusion of two compatible haploid cells, the pathogenic dikaryon is formed, which grows filamentously. Compatibility is genetically regulated by two mating type loci. The biallelic a locus controls recognition and fusion, while the multiallelic b locus regulates filamentous growth and pathogenic development (5). To exert their regulatory function, the bE and bW homeodomain proteins encoded by the b locus have to dimerize, and a prerequisite for this is that they are derived from different alleles (20, 28). The a locus encodes pheromone precursor and receptor genes that allow recognition and fusion with nonself partners (9). Therefore, the generation of an infectious dikaryon is possible only if cells are compatible, i.e., if they differ at their a and b loci.
In response to the pheromone signal, conjugation tubes are formed and pheromone-responsive gene expression is elevated. Among the induced genes are the pheromone gene (mfa), the pheromone receptor gene (pra), and the b genes (54). Transcriptional activation as well as basal expression of these genes requires the high-mobility-group protein Prf1 (22). Prf1 activity is assumed to be controlled by cyclic AMP (cAMP) as well as by mitogen-activated protein kinase (MAPK) signaling. Adenylyl cyclase (Uac1) is activated through the Gα subunit of a heterotrimeric G protein (Gpa3) (29). This in turn leads to the activation of the protein kinase A (Adr1) by triggering dissociation from its regulatory subunit Ubc1 (18). When this signaling route is disturbed, pheromone-induced transcription of the a genes is blocked (29, 41), and such strains display filamentous growth that is independent of the b heterodimer (18, 21). Conversely, when this signaling route is activated, e.g., in strains either carrying constitutive alleles of gpa3 or lacking ubc1, strongly elevated expression of pheromone genes is observed (29, 41). Interestingly, these mutations do not lead to the induction of conjugation tubes. It has been hypothesized that the cAMP cascade acts on Prf1 (23, 29). Prf1 has also been postulated to act downstream of a MAPK module containing the MAPK Kpp2 (37). This inference stems from the observation that deletion of kpp2/ubc3 abolishes pheromone-dependent expression of the mfa genes as well as conjugation tube formation. Furthermore, deletion of fuz7, encoding a MAPK kinase (MAPKK), also results in defects in conjugation tube formation while still allowing pheromone-dependent gene expression (4, 41). On these grounds it has been difficult to place Fuz7 in the pheromone-signaling cascade. On the other hand, mutations in fuz7/ubc5 or ubc3/kpp2 were shown to suppress the filamentous phenotype of uac1 deletion mutants (35). The same screen also led to the isolation of ubc4, presumed to encode a MAPKK kinase (MAPKKK), and ubc2, encoding a protein with similarities to Ste50p of Saccharomyces cerevisiae and Ste4 of Schizosaccharomyces pombe (1, 36). All of these genes were placed in one cascade suppressing filamentous growth caused by low-cAMP conditions (1).
Here we provide genetic as well as biochemical evidence that Kpp2/Ubc3, Fuz7, and Kpp4/Ubc4 act in one cascade that is activated after pheromone perception. Our experiments show that the pathways leading to pheromone-dependent gene expression and conjugation tube formation separate downstream of Kpp2. In addition, the integrity of this MAPK module is also crucial for pathogenic development.
MATERIALS AND METHODS
Strains and growth conditions.
The Escherichia coli K-12 derivatives DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used for cloning purposes, and E. coli BL21(DE3)(pLysS) (Novagen) was used for protein expression. The U. maydis strains used in this study are listed in Table 1. Prior to transformation into U. maydis, plasmids were digested with DraI (pKpp4-1, pKpp4WT, pKpp4RA, pGFP-Kpp4WT, pGFP-Kpp4RA, pGE1, pKpp2WT, pKpp2AEF, pKpp2K50R, pKpp2WT-GFP, pKpp2AEF-GFP, and pKpp2K50R-GFP), SspI (p123Pcrg1:kpp4PS, p123Pcrg1:kpp4-2, p123Pcrg1:fuz7, and p123), or BsrGI (pOTEF:pra2). In all cases single homologous integration events into the respective loci were verified by Southern analysis. Single homologous integration in the ip locus was verified by PCR and Southern analysis as described previously (32).
TABLE 1.
U. maydis strains used in this study
| Strain | Reference | Plasmid transformed | Integration locus | Progenitor strain |
|---|---|---|---|---|
| FB1 (a1 b1) | 3 | |||
| FB2 (a2 b2) | 3 | |||
| SG200 (a1::mfa2 bW2bE1) | 8 | |||
| HA103 (a1 bW2bE1con) | 22 | |||
| FB1Δkpp2-1 | 37 | |||
| FB2Δkpp2-1 | 37 | |||
| FB1Δprf1 | 37 | |||
| FB2Δprf1 | 37 | |||
| FB1Δkpp4 | This study | pKpp4-1 | kpp4 | FB1 |
| FB2Δkpp4 | This study | pKpp4-1 | kpp4 | FB2 |
| SG200Δkpp4 | This study | pKpp4-1 | kpp4 | SG200 |
| HA103Δkpp4 | This study | pKpp4-1 | kpp4 | HA103 |
| FB1kpp4WT | This study | pKpp4WT | kpp4 | FB1 |
| FB2kpp4WT | This study | pKpp4WT | kpp4 | FB2 |
| FB1kpp4RA | This study | pKpp4RA | kpp4 | FB1 |
| FB2kpp4RA | This study | pKpp4RA | kpp4 | FB2 |
| SG200GFP-kpp4WT | This study | pGFP-Kpp4WT | kpp4 | SG200 |
| SG200GFP-kpp4RA | This study | pGFP-Kpp4RA | kpp4 | SG200 |
| FB1Δfuz7 | This study | pGE1 | fuz7 | FB1 |
| FB2Δfuz7 | This study | pGE1 | fuz7 | FB2 |
| SG200Δfuz7 | This study | pGE1 | fuz7 | SG200 |
| HA103Δfuz7 | This study | pGE1 | fuz7 | HA103 |
| FB1kpp2WT-GFP | This study | pKpp2WT-GFP | kpp2 | FB1 |
| FB2kpp2WT-GFP | This study | pKpp2WT-GFP | kpp2 | FB2 |
| FB1kpp2AEF-GFP | This study | pKpp2AEF-GFP | kpp2 | FB1 |
| FB2kpp2AEF-GFP | This study | pKpp2AEF-GFP | kpp2 | FB2 |
| FB1kpp2K50R-GFP | This study | pKpp2K50R-GFP | kpp2 | FB1 |
| FB2kpp2K50R-GFP | This study | pKpp2K50R-GFP | kpp2 | FB2 |
| FB1kpp2WT | This study | pKpp2WT | kpp2 | FB1 |
| FB2kpp2WT | This study | pKpp2WT | kpp2 | FB2 |
| SG200kpp2WT | This study | pKpp2WT | kpp2 | SG200 |
| FB1kpp2AEF | This study | pKpp2AEF | kpp2 | FB1 |
| FB2kpp2AEF | This study | pKpp2AEF | kpp2 | FB2 |
| SG200kpp2AEF | This study | pKpp2AEF | kpp2 | SG200 |
| FB1kpp2K50R | This study | pKpp2K50R | kpp2 | FB1 |
| FB2kpp2K50R | This study | pKpp2K50R | kpp2 | FB2 |
| FB1Pcrgl:kpp4PS | This study | p123Pcrgl:kpp4PS | ip | FB1 |
| FB1Pcrgl:kpp4-2 | This study | p123Pcrgl:kpp4-2 | ip | FB1 |
| FB1Δkpp2-1Pcrgl:kpp4-2 | This study | p123Pcrgl:kpp4-2 | ip | FB1Δkpp2-1 |
| FB1Δfuz7Pcrgl:kpp4-2 | This study | p123Pcrgl:kpp4-2 | ip | FB1Δfuz7 |
| FB1Δprf1Pcrgl:kpp4-2 | This study | p123Pcrgl:kpp4-2 | ip | FB1Δprf1 |
| FB1Δkpp6Pcrgl:kpp4-2 | This study | p123Pcrgl:kpp4-2 | ip | FB1Δkpp6 |
| FB1Pcrgl:fuz7DD | This study | p123Pcrgl:fuz7DD | ip | FB1 |
| FB1Δkpp2-1Pcrgl:fuz7DD | This study | p123Pcrgl:fuz7DD | ip | FB1Δkpp2-1 |
| FB1Δkpp4Pcrgl:fuz7DD | This study | p123Pcrgl:fuz7DD | ip | FB1Δkpp4 |
| FB1Δprf1Pcrgl:fuz7DD | This study | p123Pcrgl:fuz7DD | ip | FB1Δprf1 |
| FB2pra2con | This study | pOTEF:pra2 | ip | FB2 |
| FB2Δkpp2-1pra2con | This study | pOTEF:pra2 | ip | FB2Δkpp2-1 |
| FB2Δfuz7pra2con | This study | pOTEF:pra2 | ip | FB2Δfuz7 |
| FB2Δkpp4pra2con | This study | pOTEF:pra2 | ip | FB2Δkpp4 |
| FB2Δprf1pra2con | This study | pOTEF:pra2 | ip | FB2Δprf1 |
| FB1kpp2AEF/Pcrgl:fuz7DD | This study | p123Pcrgl:fuz7DD | ip | FB1kpp2AEF |
| FB1kpp2K50R/Pcrgl:fuz7DD | This study | p123Pcrgl:fuz7DD | ip | FB1kpp2K50R |
| FB1Pcrgl:fuz7DD/kpp2-GFP | This study | pKpp2WT-GFP | kpp2 | FB1Pcrgl:fuz7DD |
| FB1Pcrgl:fuz7DD/kpp2AEF-GFP | This study | pKpp2AEF-GFP | kpp2 | FB1Pcrgl:fuz7DD |
| FB1Pcrgl:fuz7DD/kpp2K50R-GFP | This study | pKpp2K50R-GFP | kpp2 | FB1Pcrgl:fuz7DD |
| SG200Potef:GFP | This study | p123 | ip | SG200 |
| SG200Δkpp4/Potef:GFP | This study | p123 | ip | SG200Δkpp4 |
| SG200kpp2AEF/Potef:GFP | This study | p123 | ip | SG200kpp2AEF |
U. maydis strains were grown at 28°C in liquid CM (25), YEPSL (0.4% yeast extract, 0.4% peptone, 2% sucrose), or potato dextrose (PD) (2.4% PD broth [Difco]) medium on a rotary shaker at 220 rpm or on solid PD agar. For induction of crg1 promoter activity, strains were grown in CM medium containing 1% glucose (CM-Glc) to an optical density at 600 nm (OD600) of 0.5, washed twice with water, and suspended in CM medium with 1% arabinose as a carbon source (CM-Ara).
Hygromycin B was purchased from Roche, nourseothricin (NAT) was purchased from the Hans-Knöll-Institute (Jena, Germany), and carboxin was purchased from Riedel de Haen (Seelze, Germany). All other chemicals were of analytical grade and were obtained from Sigma or Merck.
Isolation of the kpp4 gene.
Degenerate primers MEKK4 (GTITAYYTIGGNATGAAYGC) and MEKK6 (YTTYTTISWDATICCRAARTC) were used for amplification of U. maydis DNA. Reaction mixtures contained 10 mM Tris-HCl (pH 8.3), 3 mM MgCl2, 50 mM KCl, 50 pmol of primers, and 2 U of Taq polymerase. Amplification was achieved by 35 cycles of 1 min at 95°C, 1 min at 48°C, and 1 min at 72°C. For sequencing, PCR products of 420 bp were cloned into pCR2.1TOPO. The amplified kpp4 fragment was used to screen a genomic λEMBL3 library (45). From a hybridizing clone, kpp4 was subcloned as 5.2-kb HindIII and 7.4-kb BamHI fragments in pTZ19R, and the resulting plasmids were designated pKpp4H and pKpp4B, respectively. In addition, we cloned a 2.5-kb HindIII-BamHI fragment comprising the kpp4 gene into pSP72 to obtain pSP-kpp4H/B.
To isolate cDNA fragments of kpp4, we produced cDNA by using an oligo(dT)18 primer, Superscript II reverse transcriptase (Life Technologies), and RNA obtained from AB33 (12) as the template. For the subsequent PCR, the following primer combinations were used: kpp4-550 (CGACGCTTCAAGTCGTCC)-OPM45 (GCGTAGCCGGCCGACTG), OPM46 (CGAAGAGGCCAGATGCGAC)-OPM41 (AACCTTGCGTGCATCCTCAC), OMP40 (CAAAGCTCTCTCCGACAACG)-OPM21 (AGGGCCGTGTCGAGGCAG), OPM46 (CGAAGAGGCCAGATGCGAC)-kpp4rev (GCTTGACCGCCATCAGTAG), and kpp4+1490 (CGCGGATCCGCCTCTTCGCTGAACGC)-kpp4+974 (CCGGAATTCCGTCGATCGGTCCATGACC).
Plasmids and plasmid constructions.
Plasmids pTZ18R (Pharmacia), pTZ19R (Pharmacia), pSP72 (Promega), pSL1180 (Pharmacia), and pBS-SKII(+) (Stratagene) were used for cloning, subcloning, and sequencing of genomic fragments, and pCR2.1TOPO (Invitrogen) was used for cloning and sequencing of fragments generated by PCR. pGEX-2T (Pharmacia) was used for protein expression in E. coli. Primers were obtained from Sigma ARK. Sequence analysis of genomic sequences and fragments generated by PCR was performed with an automated sequencer (ABI 377) and standard bioinformatic tools.
pRU11 contains the crg1 promoter as a 3.5-kb NotI-NdeI fragment, and pSLHyg(−) contains a hygromycin resistance cassette as a NotI fragment (12). pCU3 is a pSP72 derivative harboring the tef1 promoter as a NotI-NdeI fragment (A. Brachmann and R. Kahmann, unpublished data). pNEBNat(+) and pSLNat(+) are pNEB193 (New England Biolabs) and pSL1180 (Pharmacia) derivatives, respectively, both containing a NAT resistance cassette as a NotI fragment (37; A. Brachmann and R. Kahmann, unpublished data). p123 is a pSP72 derivative containing the egfp gene (Clontech) fused to the otef promoter and nos terminator and a carboxin resistance cassette (55). pOTEF:pra2 is a p123 derivative. For construction of pOTEF:pra2, we isolated a 1.9-kb HindIII-NotI fragment encompassing the otef promoter and cDNA from pra2 as an ATG fusion from pJG10 (M. Feldbrugge, unpublished data) and ligated it into p123 digested with HindIII and NotI. The resulting plasmid provides for carboxin resistance and harbors the pra2 cDNA under the control of the otef promoter and nos terminator.
kpp4 plasmids.
In pkpp4-1 the kpp4 open reading frame (ORF) is deleted from bp +24 (XmaI site) to bp 74 after the stop codon (AvrII site). For construction, we ligated a 2.1-kb BamHI-XmaI fragment encompassing the 5′ region of kpp4 from pKpp2B, a 3-kb AgeI-SpeI fragment from pSLHyg(−) containing the hygromycin resistance cassette, and a 0.5-kb AvrII-EcoRI fragment from pKpp4B containing the 3′ region into pTZ19R opened with BamHI and EcoRI.
Plasmid pKpp4WT is a pTZ19R derivative containing the 0.8-kb 5′ region of kpp4 as a EcoRI-NotI fragment generated by PCR, the hygromycin resistance cassette as a 2.9-kb NotI-NotI fragment from pSLHyg(−), and the tef1 promoter derived from pCU3 as a NotI-NdeI fragment fused to a 2.7-kb fragment encompassing kpp4. At position 1 of the kpp4 ORF, an NdeI site was introduced by using the annealed oligonucleotides kpp4Linker-I (TATGAGTGCTGCAACACCTACCAGC) and kpp4Linker-II (CCGGGCTGGTAGGTGTTGCAGCACTCA).
pKpp4RA is identical to pKpp4WT except for the K481E mutation, which was generated by PCR with primers kpp4RAIII (GCCTCTTCGCAGCGCTATGC), kpp4RAV (GCCGCTAGGCGGCTTGCCGAATTCTTTGAGCACGCGCGCCATGAC), and kpp4RAIV (CCACAGGCATGCGCTCACC).
pGFP-Kpp4WT is a pKpp4WT derivative in which the tef promoter was replaced by a 1.6-kb NotI-NdeI fragment encompassing sgfp under the control of the otef promoter (47). This results in a translational fusion of the green fluorescent protein (GFP) gene to the kpp4 ORF.
pGFP-Kpp4RA is identical to pGFP-Kpp4WT except for the K481E mutation.
Plasmid p123Pcrg1:kpp4PS is a p123 derivative in which the otef promoter and GFP gene were replaced by the crg1 promoter (3.5-kb NotI-NdeI fragment from pRU11) fused to kpp4P681S including the 0.55-kb 3′ region. To introduce the P681S mutation, we performed a PCR with primers kpp4PS (AAGATCCGCAACTTCTTCGGCCAGCGATCGCCCTCAGAACTCATC) and kpp4+2239 (GGTGACCATCCATGGAACC).
p123Pcrg1:kpp4-2 is a p123 derivative in which the otef promoter and GFP gene were replaced by the crg1 promoter (3.5-kb NotI-NdeI fragment from pRU11) fused to kpp4-2 including the 0.55-kb 3′ region. To generate the kpp4-2 allele, we ligated a 0.5-kb NheI-PvuII fragment from pKpp4B and a 4.75-kb PvuI (blunted)-HindIII fragment from pKpp4H into pBS-SKII(+) cut with XbaI-HindIII.
fuz7 plasmids.
pHA42 is a pSP72 derivative that contains a 3.3-kb SphI genomic fragment encompassing the fuz7 gene obtained from pFuz7 (41).
pGE1 is a pHA42 derivative in which a 0.9-kb NaeI-NsiI fragment encompassing bp +146 to +1057 of the fuz7 ORF was replaced by a NAT resistance cassette derived as a 1.5-kb StuI-PstI fragment from pSLNat(+).
p123Pcrg1:fuz7DD is a p123 derivative in which the otef promoter and GFP gene were replaced by the crg1 promoter (3.5-kb NotI-NdeI fragment from pRU11) fused to fuz7DD including the 0.2-kb 3′ region. To introduce the S259D and T263D mutations, we performed a PCR with primers fuz7DD (ACATGTAGGTACTTGTACCAACAAAGTCGTCTGCGATATCGTTGATGAGC) and fuz7+1NdeI (CATATGCTTTCGTCCGGTGCG).
pGEX-Fuz7 is derivative of pGEX-2T containing an NcoI-MfeI fragment encoding a His6-tagged version of fuz7 derived from pET-Fuz7 (P. Muller and R. Kahmann, unpublished data).
kpp2 plasmids.
p123kpp2 is a p123 derivative in which the GFP gene was replaced by a 1-kb NcoI-NotI fragment that codes for Kpp2 and was generated by PCR with primers kpp2A (CATGCCATGGCACATGCCCACGGACAGC) and kpp2B (ATTTGCGGCCGCAAGATCAACGCATGATCTC).
pKpp2WT is a pTZ19R derivative that contains a 1.3-kb 5′ region of kpp2 derived as a HindIII-BglII fragment from pKpp2H, a 0.6-kb BglII-NotI fragment encoding the 3′ part of kpp2 from p123kpp2, the mfa2 terminator as a 0.4-kb NotI-BamHI fragment, a NAT resistance cassette obtained as a 1.5-kb MfeI-BamHI fragment from pNEBNat(+), and a 1.1-kb EcoRI-XhoI fragment from pKpp2H representing the 3′ region of kpp2.
pKpp2AEF is identical to pKpp2WT except for the mutations T182A and Y184F. These were introduced by PCRs with primers kpp2C (CCATCGTGTGGCAACGAATTCGGCCATGAAACCC), kpp2D (GGAGCTCTCCGATGACCAC), and kpp2B (see above).
pKpp2K50R is identical to pKpp2WT except for the K50R mutation introduced by PCRs with primers K50RI (CTCGTGTCGCCATCCGGAAGATCACCCCATTCGATCAC), K50RII (TGACGCGATGCATGTCGG), and K50RIII (CAAAAGACGCGTCGCTGC).
pKpp2WT-GFP is a pKpp2WT derivative containing a kpp2-gfp fusion. The GFP gene was isolated as 0.7-kb NcoI-NotI fragment from p123. This fragment was ligated to a 2.1-kb HindIII-NcoI fragment of kpp2 in which an NcoI site was introduced at codon 351 by PCR with primers kpp2+379 (TATCAAACACTGCGTGGCTTG) and kpp2C′NcoI (CCATGGTCTCGTTATAAATCAACCTCTTG).
pKpp2AEF-GFP and pKpp2K50R-GFP were constructed by replacing the 2.7-kb BstXI fragments of pKpp2AEF and pKpp2K50R, respectively, with a 3.4-kb BstXI fragment encompassing the kpp2-gfp fusion derived from pKpp2WT-GFP.
pGEX-Kpp2 is a pGEX-2T derivative containing a 1.3-kb NcoI-NotI fragment derived from p123kpp2.
pGEX-Kpp2K50R is a pGEX-Kpp2 derivative in which a 0.5-kb NcoI-BglII fragment was replaced by a 0.5-kb NcoI-BglII fragment harboring the K50R mutation.
DNA and RNA procedures.
Standard molecular techniques were used (43). Transformation of U. maydis was performed as published previously (45). U. maydis DNA was isolated as described previously (24). RNA from strains grown in liquid culture was prepared as described previously (29). The following probes were used for Northern analyses: a 0.67-kb EcoRV fragment and a 1.3-kb EcoRI-EcoRV fragment from pSP4.2EcoRV (9) for mfa1 and pra1, respectively; a 0.4-kb SpeI-PstI fragment from pTZa2XhoI3.5 (9) for mfa2; a 2.6-kb PvuII fragment from pbW2-Nde-bE1 (12) for bE and bW; and a 1.2-kb NdeI-MluI fragment from p123Pcrg1:kpp4-2 and a 1.4-kb NdeI-SphI fragment from p123Pcrg1:fuz7DD for kpp4-2 and fuz7, respectively. Radioactive labeling was performed with the NEBlot kit (New England Biolabs). A 5′-end-labeled oligonucleotide complementary to the U. maydis 18S rRNA (10) was hybridized as a loading control in Northern analyses. A PhosphorImager (Storm 840; Molecular Dynamics) and the program ImageQuant (Molecular Dynamics) were used for visualization and quantification of radioactive signals.
Mating, pheromone stimulation, and pathogenicity assays.
To test for mating, compatible strains were cospotted on charcoal-containing PD plates (25), and the plates were sealed with Parafilm and incubated at 24°C for 48 h. For pheromone stimulation, strains were grown in CM-Glc to an OD600 of 0.6. Synthetic pheromone (49) dissolved in dimethyl sulfoxide (DMSO) was added to a final concentration of 2.5 μg/ml, and cells were harvested for microscopic observations and RNA preparations after 5 h of incubation in a 15-ml plastic tube on a tissue culture roller at 28°C. Quantification was done with photomicrographs by manual counting.
Plant infections of the corn variety Early Golden Bantam (Olds Seeds, Madison, Wis.) were performed as described previously (37). For coinoculations of SG200 and derivatives, cells were mixed in equal amounts prior to infection. Fungal structures on the plant surface were visualized by Chlorazole Black E and by Calcofluor staining as described previously (11).
U. maydis cell lysates, glutathione S-transferase (GST) pulldown, and kinase assay.
U. maydis protein extracts were prepared with a French press (gauge pressure of 1,000 lb/in2) and cleared by centrifugation (4°C, 30 min, 33,300 rpm [Sorvall TH-660 rotor]).
GST-Kpp2-, GST-Kpp2K50R-, GST-Fuz7-, or GST-expressing BL21(DE3)(pLysS) cells were grown in dYT containing 1% glucose, ampicillin (100 μg/ml), and chloramphenicol (34 μg/ml) at 37°C. At an OD600 of ∼0.5, cell suspensions were cooled, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and incubation was continued for 16 h at 16°C. Cells were harvested, washed once in buffer A (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 2.5 mM EDTA, 2.5 mM EGTA, 1% Triton X-100, 1 mM dithiothreitol [DTT]), and resuspended in 1 ml of buffer A containing complete protease inhibitor cocktail (catalog no. 1873580; Roche). Cells were freeze-thawed, and after DNase treatment, insoluble components were removed by centrifugation (4°C, 30 min, 28,000 × g). The supernatant was incubated with 50 μl of glutathione-Sepharose beads (catalog no. 17075601; Amersham Bioscience) for 1 h at 4°C. The beads were washed once with buffer A and five times with 1 ml of buffer B (50 mM Tris-HCl [pH 7.5], 125 mM NaCl, 2.5 mM EDTA, 2.5 mM EGTA, 0.1% Triton X-100, 1 mM DTT).
To assay the kinase activity of GST-Kpp2 and GST-Kpp2K50R, beads were washed once with 1 ml of kinase buffer (KB) (20 mM HEPES [pH 7.4], 15 mM MgCl2, 5 mM EGTA, 1 mM DTT) and split into two portions. One was subjected to kinase assay (see below); the other was resuspended in 50 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and after 5 min of boiling, GST-Kpp2 levels were assayed by SDS-10% PAGE and Coomassie blue staining.
For GST pulldown, beads with GST-Fuz7 or GST alone were washed once with GST pulldown buffer (GPB) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 5 mM EDTA, 1 mM DTT) and incubated for 1 h at 4°C with cleared portions of U. maydis cell extracts (1 mg of protein) prepared in GPB containing complete protease inhibitor cocktail (catalog no. 1873580; Roche). After five washes with 1 ml of GPB, the beads were resuspended in 50 μl of 2× SDS-PAGE sample buffer and boiled for 5 min at 95°C. After centrifugation, 10 μl was loaded on two SDS-10% polyacrylamide gels to assay GST or GST-Fuz7 levels by Coomassie blue staining and to assay GFP-Kpp2 levels by Western analysis.
To obtain extracts from pheromone-treated cells, U. maydis strains were grown in CM-Glc to an OD600 of ∼0.8 with rotary shaking at 28°C. Cultures were transferred to 50-ml plastic tubes, pheromone was added to a final concentration of 2.5 μg/ml, and cultures were incubated on a tissue culture roller at 28°C. For induction of fuz7DD, strains were grown in CM-Glc to an OD600 of ∼0.8, washed with water twice, and suspended in CM-Ara. After the cultures were harvested, cells were washed once in immunoprecipitation (IP) buffer (25 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 15 mM EGTA, 75 mM NaCl, 0.1% Tween 20, 1 mM DTT) and resuspended in ice-cold IP buffer containing cocktails of protease and phosphatases inhibitors (catalog no. 1873580 [Roche] and catalog no. P-2850 and P-5726 [Sigma]). U. maydis protein extracts were prepared with a French press and cleared by centrifugation.
GFP-tagged Kpp2 was immunoprecipitated by adding 0.5 μg of rabbit anti-GFP polyclonal antibody (catalog no. 3999100; Biocat, Heidelberg, Germany), immobilized to Dynabeads protein G (catalog no. 100.03; Dynal) by cross-linking, to portions (1 mg) of cleared cell extracts and mixing for 1 h at 4°C. Precipitated beads were washed once with 1 ml of IP buffer, five times with 1 ml of IP wash buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 5 mM EGTA, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT), and once with 1 ml of KB; all buffers contained protease and phosphatase inhibitors. The washed beads were split in two portions to assay kinase activity and precipitated Kpp2-GFP levels.
To determine Kpp2-GFP levels, beads were suspended in 20 μl of SDS-PAGE sample buffer and boiled for 5 min. Ten microliters was separated on an SDS-10% polyacrylamide gel, followed by semidry transfer to a Hybond-P membrane (catalog no. RPN303F; Amersham Bioscience).
GFP-tagged Kpp2 or Kpp4 derivatives were detected by using mouse anti-GFP monoclonal antibody (clones 7.1 and 13.1) (catalog no. 1814460; Roche) at a 1:5,000 dilution, and α-tubulin (α-Tub) was detected with anti-α-Tub monoclonal antibody (Oncogene) at a 1:5,000 dilution. To detect phosphorylated Kpp2, the polyclonal phosphoepitope pTEpY-specific antibody 9101 from New England Biolabs was used at a 1:1,000 dilution. As the secondary antibody we used horseradish peroxidase-conjugated goat anti-mouse antibody (diluted 1:10,000) (catalog no. W4021; Promega) or goat anti-rabbit antibody (diluted 1:10,000) (catalog no. 1706515; Bio-Rad) followed by detection with ECL+ (catalog no. RPN2132; Amersham Bioscience).
To assay kinase activity, residual supernatant was removed and beads were resuspended in 20 μl of room temperature kinase assay buffer (KB containing 1 mg of myelin basic protein (MBP) (catalog no. 13228-010; Gibco) per ml, 50 mM Na-β-glycerol phosphate, 5 mM NaVO3, 50 μM ATP, 5 mM MgCl2, and 0.2 μM [γ-32P]ATP [6,000Ci/mmol]) and incubated for 20 min at 28°C. Reactions were stopped by adding 20 μl of 2× SDS-PAGE sample buffer and boiling for 5 min. Phosphorylation was analyzed by SDS-15% PAGE, and dried gels were exposed to a PhosphorImager (Molecular Dynamics).
Microscopic observation.
For microscopic observation, we used a Zeiss Axiophot microscope with differential interference contrast optics. Calcofluor fluorescence was observed with a standard DAPI (4′,6′-diamidino-2-phenylindole) filter set. GFP fluorescence was detected with a specific filter set (BP 470/20, FT 493, BP 505-530; Zeiss, Jena, Germany). Pictures were taken with a charge-coupled device camera (catalog no. C4742-95; Hamamatsu, Herrsching, Germany). Image processing was done with Image Pro (Media Cybernetics), Adobe Photoshop 6.0, and Canvas 6.0 (Deneba Systems).
Nucleotide sequence accession number.
The U. maydis kpp4 gene has been assigned GenBank accession number AF542505.
RESULTS
Isolation of kpp4, encoding a MAPKKK homologue.
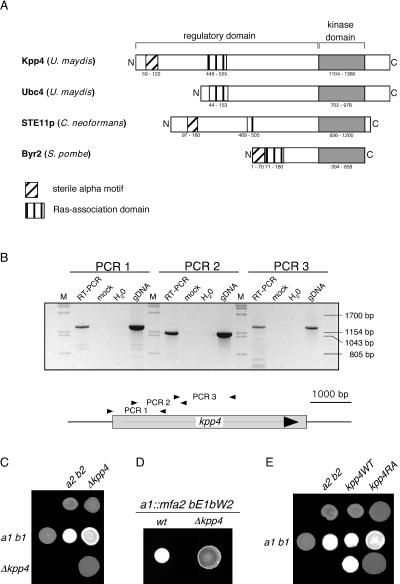
In a PCR approach using degenerate primers designed according to conserved sequences of two MAPKKK genes, STE11 of S. cerevisiae and byr2 of S. pombe, we isolated the gene kpp4 of U. maydis. After sequencing a corresponding genomic clone, kpp4 turned out to be 95% identical to ubc4 on the nucleotide level. kpp4 is predicted to encode a polypeptide of 1,567 amino acids, while an ORF coding for 1,166 amino acids had been assigned to ubc4 (Fig. 1A) (1). The polypeptides are identical after the N-terminal 441 and 39 amino acids, respectively (except for a P794S substitution in Kpp4). Reverse transcription-PCR analysis of kpp4 revealed the absence of introns and placed the 5′ end of the mRNA upstream of position −100 (Fig. 1B and data not shown). This reinforces the assertion that kpp4 codes for a protein of 1,567 amino acids. Therefore, it is likely that the start codon of the ubc4 ORF was wrongly assigned due to sequencing errors. Inspection of the N-terminal region of Kpp4 with ISREC (http://hits.isb-sib.ch/cgi-bin/hits_motifscan) identified a sterile-alpha motif (SAM) domain which is not present in Ubc4 but can be found in other fungal MAPKKKs, such as Ste11p of Cryptococcus neoformans and Byr2 of S. pombe (Fig. 1A). This domain is thought to mediate both homo- and hetero-oligomerization and plays a role in signaling (39, 44). In addition, a Ras association (RA) domain, which is known to interact with small G proteins (40), could be identified between positions 446 and 555 (Fig. 1A). Despite the substantial differences in the N-terminal domain, we nevertheless consider kpp4 and ubc4 to represent the same gene.
FIG. 1.
Deletion of kpp4 affects mating and filamentous growth. (A) Schematic representation of Kpp4 domain structure. Domains were identified with ISREC, and domain annotations are PS50105 (SAM domain), PS50200 (RA domain), and PS500011 (kinase domain). Accession numbers are AAN63948 for Kpp4 (1,567 amino acids), AAF86841 for Ubc4 (1,166 amino acids), AAG30205 for Ste11p of C. neoformans (1,230 amino acids), and P28829 for Bry2 of S. pombe (689 amino acids). Interestingly, at bp +4438 of the kpp4 ORF we found hexanucleotide repeats (GCTGCG) which encode an alanine stretch and are present in six copies in FB1 (a1 b1) and FB2 (a2 b2) but only in five copies in RK32 (a1 b3). (B) Reverse transcription-PCR analysis of kpp4. RNA isolated from AB33 was reverse transcribed and then subjected to three different PCRs, i.e., PCR 1 with primers kpp4-550 and OPM45, PCR 2 with primers OPM46 and OPM41, and PCR 3 with primers OPM40 and OPM21, amplifying the regions between positions −99 and +1172, +547 and +1695, and +1566 and +2872 of the kpp4 ORF, respectively. As controls, we performed reactions without reverse transcriptase (mock) or with water or genomic DNA (gDNA) as the template. Lanes M, molecular size markers. (C) The strains indicated on the top were spotted alone and in combinations with the strains indicated on the left on charcoal-containing PD plates. Dikaryotic filaments appear as white fuzziness. (D) The strains indicated the on top were spotted on charcoal-containing PD plates. SG200 developed filaments characterized by white fuzziness, while SG200Δkpp4 deletion strains are severely affected in filamentation. wt, wild type. (E) The strains indicated on the top were spotted alone (top row) and in combinations with the wild type (middle row) on charcoal-containing PD plates. In the bottom row compatible combinations of either kpp4WT (left) or kpp4RA (right) strains were cospotted.
Deletion of kpp4 attenuates mating, impairs conjugation tube formation, and abolishes pathogenic development.
The similarity of Ubc4 to Byr2 of S. pombe and Ste11p of S. cerevisiae led Andrews et al. to propose a function of ubc4 during the mating process besides suppressing filamentous growth of a uac1 deletion mutant (1). To address this question, we have generated deletion mutations of kpp4 in compatible haploid strains FB1 (a1 b1) and FB2 (a2 b2) as well as in the haploid solopathogenic strain SG200 (a1::mfa2 bE1bW2).
In plate mating assays, successful fusion of compatible strains results in the formation of the filamentous dikaryon, which appears as white fuzziness on charcoal plates (Fig. 1C). In this assay we observed no significant reduction in dikaryon formation of kpp4 deletion strains crossed with compatible wild-type strains (Fig. 1C). This illustrates that deletion of kpp4 does not lead to sterility. However, a mixture of two compatible Δkpp4 strains failed to develop fuzzy filaments (Fig. 1C). To investigate the mating process in more detail, FB1 and FB1Δkpp4 were stimulated with synthetic a2 pheromone. While the wild-type strain formed conjugation tubes, the kpp4 deletion strain showed no response (Table 2). This demonstrates that the MAPKKK Kpp4 is essential for this pheromone-specific change in morphology, as was shown for fuz7 and kpp2/ubc3, coding for a MAPKK and a MAPK, respectively (Table 2) (4, 35, 37). Moreover, deletion of kpp4 in SG200 (a1::mfa2 bE1bW2), which grows filamentously because of active a and b loci, resulted in strongly attenuated filament formation (Fig. 1D), as described for SG200Δkpp2-1 and for diploid Δfuz7/Δfuz7 strains (4, 37). This illustrates that kpp4 affects postfusion processes such as filament formation.
TABLE 2.
Conjugation tube formationa
| Strain | No. of cells | % Conjugation tubes |
|---|---|---|
| FB1 | 506 | 80 |
| FB1Δkpp4 | 503 | 0 |
| FB1kpp4WT | 340 | 96 |
| FB1kpp4RA | 318 | 19 |
| FB1Δfuz7 | 508 | 0 |
| FB1Δkpp2-1 | 546 | 0 |
| FB1Δprf1 | 550 | 0 |
| FB1kpp2AEF | 532 | 0 |
| FB1kpp2K50R | 541 | 0 |
Cells were exposed to synthetic a2 pheromone, and conjugation tube formation was determined by microscopic observation.
To assay the function of kpp4 during pathogenic development, corn plants were infected with mixtures of compatible Δkpp4 mutants or with wild-type strains as a control. We observed tumors in 90% of plants infected with wild-type strains, while kpp4 deletion strains failed to induce tumors (155 plants were tested) (Table 3). To exclude the possibility that this outcome results from cell fusion defects of Δkpp4 strains, we also performed plant infections with the haploid solopathogenic strain SG200 (a1::mfa2 bE1bW2), which induces tumors without prior fusion (Table 3). Its derivative SG200Δkpp4 was unable to induce disease symptoms (Table 3), demonstrating an essential function of kpp4 during pathogenic development.
TABLE 3.
Plant infection assays
| Inoculum | No. of:
|
% Tumor formationb | |
|---|---|---|---|
| Infected plantsa | Plants with tumors | ||
| FB1 × FB2 | 40 | 36 | 90 |
| FB1Δkpp4 × FB2Δkpp4 | 155 | 0 | 0 |
| SG200 | 40 | 30 | 75 |
| SG200Δkpp4 | 117 | 0 | 0 |
| FB1kpp4WT × FB2kpp4WT | 44 | 38 | 86 |
| FB1kpp4RA × FB2kpp4RA | 37 | 15 | 40 |
| HA103 | 135 | 82 | 60 |
| HA103Δkpp4 | 156 | 0 | 0 |
| FB1 × FB2 | 18 | 14 | 93 |
| FB1Δfuz7 × FB2Δfuz7 | 61 | 0 | 0 |
| SG200Δfuz7 | 42 | 0 | 0 |
| HA103 | 38 | 32 | 84 |
| HA103Δfuz7 | 75 | 0 | 0 |
| FB1 × FB2 | 40 | 35 | 87 |
| SG200 | 64 | 58 | 91 |
| FB1kpp2-GFP × FB2kpp2-GFP | 77 | 60 | 78 |
| FB1Δkpp2-1 × FB2Δkpp2-1 | 77 | 16 | 20 |
| FB1kpp2WT × FB2kpp2WT | 40 | 38 | 95 |
| SG200kpp2WT | 40 | 37 | 80 |
| FB1kpp2AEF × FB2 | 36 | 18 | 50 |
| FB1kpp2AEF × FB2kpp2AEF | 76 | 2c | 3 |
| SG200kpp2AEF | 115 | 0 | 0 |
All infections were performed twice with two independently generated mutants.
Percentage of plants that developed at least one tumor on a stem or leaf.
Tumors observed were found on leaves only and did not extend 2 mm in diameter. This is significantly different from infections with wild-type strains, where tumors develop on all green parts of the plant and reach diameters of up to 50 mm.
The RA domain of Kpp4 is important for function.
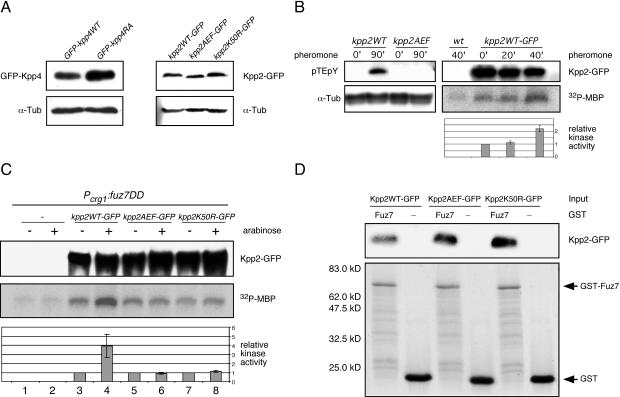
To elucidate the role of the RA domain (Fig. 1A) in Kpp4 function, we introduced a K481E mutation into kpp4 and replaced the endogenous allele in FB1 and FB2 with Ptef1:kpp4K481E as well as with Ptef1:kpp4WT as a control. Both alleles were expressed from the constitutive tef1 promoter (47). The corresponding mutation in the RA domain of S. pombe Byr2p was shown to abolish Ras1 binding (52). In a plate mating assay, strains carrying Ptef1:kpp4WT (FB1kpp4WT and FB2kpp4WT) were indistinguishable from wild-type strains (Fig. 1E), indicating that expression of kpp4 from the tef1 promoter does not interfere with function. In addition, Western analysis with protein extracts from strains expressing either GFP-Kpp4 or GFP-Kpp4RA fusion proteins under the control of the otef promoter (47) revealed that the K481E substitution does not influence Kpp4 protein stability (see Fig. 5A, left panel).
FIG. 5.
Pheromone as well as fuz7DD activates Kpp2 kinase activity. (A) Expression levels of kpp4 and kpp2 alleles constructed in this study. Left panel, Western analysis of protein extracts from SG200GFP-kpp4WT and SG200GFP-kpp4RA grown in CM-Glc by using anti-GFP antibody. In the lower panel the Western blot was stained with anti-α-Tub antibody as a loading control. Right panel, protein extracts from FB1kpp2WT-GFP, FB1kpp2AEF-GFP, and FB1kpp2K50R-GFP grown in CM-Glc were analyzed with anti-GFP antibody. In the lower panel the Western blot was stained with anti-α-Tub antibody as loading control. (B) Left panel, before and after pheromone stimulation, phosphorylation of Kpp2 was monitored with anti-pTEpY antibody. Extracts were prepared from FB1kpp2WT and FB1kpp2AEF 90 min after stimulation with synthetic a2 pheromone (90′) or after 90 min of DMSO treatment (0′). The upper panel shows a Western blot with anti-pTEpY antibody, and the lower panel shows a Western blot with anti-α-Tub antibody as a loading control. Right panel, Kpp2 kinase activity in FB2 (wild type [wt]) and FB2kpp2WT-GFP was assayed by MBP phosphorylation at 0, 20, and 40 min after pheromone addition. The upper panel shows precipitated Kpp2WT-GFP detected with anti-GFP antibody. The middle panel shows MBP phosphorylation, and the lower panel summarizes relative kinase activity measured by quantification of incorporated phosphate in three independent experiments. Error bars indicate standard deviations. (C) FB1 and the FB1-derived strains indicated on the top were shifted to arabinose-containing medium. Extracts were prepared prior to (−) and 90 min after (+) the shift. The upper panel shows precipitated Kpp2-GFP derivatives detected with anti-GFP antibody. The middle panel illustrates MBP phosphorylation, and the lower panel depicts relative kinase activity measured by quantification of incorporated phosphate in three independent experiments. (D) To demonstrate Kpp2 interaction with Fuz7 in vitro, protein extracts were prepared from strains expressing either kpp2-GFP, kpp2AEF-GFP, or kpp2K50R. These extracts were incubated with either GST-Fuz7 (Fuz7) or GST (−) bound to glutathione-Sepharose. The upper panel shows precipitated Kpp2-GFP (67 kDa) detected with anti-GFP antibody, and the lower panel illustrates GST fusion proteins (GST-Fuz7 [76 kDa] and GST [27 kDa]) bound to glutathione-Sepharose as detected by Coomassie blue staining. Experiments were performed twice with similar results.
In contrast to the kpp4WT strains, mixtures of compatible Ptef1:kpp4K481E strains (FB1kpp4RA and FB2kpp4RA) did not develop dikaryotic filaments, while the combination of FB2kpp4RA with the compatible wild-type strain FB1 displayed only a slight reduction in the formation of dikaryotic hyphae (Fig. 1E). The kpp4K481E strains thus resemble kpp4 deletion strains. To investigate whether conjugation tube formation is also affected by kpp4K481E, we stimulated FB1kpp4WT and FB1kpp4RA with synthetic a2 pheromone. Only 59 out of 318 FB1kpp4RA cells (19%) responded to stimulation, whereas in the control, 96% of the cells developed tubes (Table 2). This result indicates that a functional RA domain in Kpp4 is necessary for an efficient response to pheromone.
To assay the role of the RA domain during pathogenic growth, we infected corn plants with mixtures of FB1kpp4RA and FB2kpp4RA strains, both carrying the kpp4K481E mutant allele. While in control experiments with mixtures of FB1kpp4WT and FB2kpp4WT, 86% of infected plants showed tumor formation, compatible kpp4K481E strains induced tumors in only 40% of the infected plants (Table 3). Thus, kpp4K481E strains are reduced in pathogenicity but differ from kpp4 deletion mutants, which are completely impaired in pathogenic development. This suggests that a functional RA domain in Kpp4 is required for full virulence only.
kpp4, fuz7, and kpp2 act in one cascade.
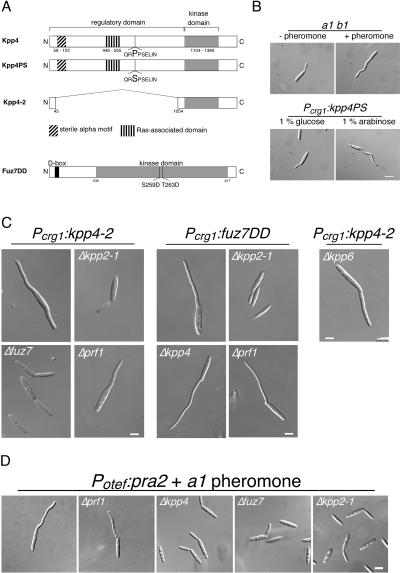
To analyze whether Kpp4 acts in one module with the known MAPKK Fuz7 and the MAPK Kpp2 we carried out genetic epitasis analyses. To this end we constructed constitutively active alleles of kpp4 by introducing mutations that were shown to confer constitutive activity to STE11 of S. cerevisiae (15, 48). kpp4PS carries a P681S substitution, and in kpp4-2 the coding region for amino acids 45 to 1055 of the presumed regulatory domain was deleted (Fig. 2A). To generate a constitutively active allele of fuz7 (fuz7DD), we introduced two point mutations resulting in S259D and T263D substitutions, which likely mimic an activated kinase. These alleles were placed under the control of the crg1 promoter, which is repressed by glucose and induced by arabinose (10), and were introduced in single copy into the ip locus of FB1 (32). Under repressing conditions, strains harboring kpp4PS, kpp4-2, or fuz7DD were morphologically indistinguishable from wild-type strains (Fig. 2B and data not shown). However, 4 h after transfer to arabinose-containing medium, cells expressing either kpp4PS, kpp4-2, or fuz7DD developed irregular filaments at one or both poles of the cell (Fig. 2B, lower panel, and C). These filamentous structures appeared to be curved and resembled conjugation tubes (Fig. 2B, upper panel). Moreover, in cells expressing kpp4PS, kpp4-2, or fuz7DD, we could detect only one nucleus, which localized to the growing filament (not shown). These results show that kpp4PS, kpp4-2, and fuz7DD induce the formation of conjugation tube-like structures.
FIG. 2.
Constitutively active alleles of kpp4 and fuz7 induce conjugation tube-like structures. (A) Schematic representation of the kpp4 and fuz7 alleles used in this study. Kpp4PS harbors a P681S substitution in a conserved region, the so-called catalytic binding domain of STE11p-like kinases. In Kpp4-2 the regulatory domain is deleted. In Fuz7DD (accession number Q99078, 435 amino acids) two amino acid substitutions, S259D and T263D, were introduced in the conserved phosphorylation motif of MAPKK as described for, e.g., MEK1 (26). The D box (amino acids 13 to 21), described to be essential for MAPKK-MAPK interaction, is located in the N terminus of Fuz7 (6). (B) Expression of constitutively active kpp4PS induces structures that resemble conjugation tubes. FB1 cells were treated with a2 pheromone (upper right panel), and FB1Pcrg1:kpp4PS was incubated with arabinose for 4 h (lower right panel). All pictures were taken with the same magnification. Bar, 10 μm. (C) Kpp4, Fuz7, and Kpp2 act in one cascade. FB1 derivatives harboring either kpp4-2 or fuz7DD are indicated on top and with the indicated gene deletions were used. Cell morphology was scored at 5 h after growth in arabinose-containing medium. All pictures were taken with the same magnification. Bar, 5 μm. (D) Overexpression of the pheromone receptor rescues conjugation tube formation in prf1 deletion strains. FB2 and all FB2-derived strains expressed pra2 constitutively, carried the indicated gene deletions, and were stimulated with a1 pheromone. All pictures were taken with the same magnification. Bar, 5 μm.
As shown in Fig. 2C, the MAPK cascade components fuz7 and kpp2 are essential for the morphological transition induced by kpp4-2. The ability of fuz7DD to trigger tube formation required kpp2 but was independent of kpp4 (Fig. 2C). These results demonstrate that fuz7 acts downstream of kpp4 and upstream of kpp2. Unexpectedly, deletion of prf1 did not abolish the morphological transition induced by either kpp4-2 or fuz7DD (Fig. 2C).
Prf1 is dispensable for conjugation tube formation.
We wondered whether a genetically activated MAPK cascade is equivalent to a pheromone stimulus, since deletion of prf1 impairs conjugation tube formation (Table 2) but appeared to be dispensable for the morphological transition triggered by an activated cascade. Since prf1 is required for the basal transcription of the a and b genes (22), prf1 deletion mutants lack the pheromone receptor and are blind to pheromone. Therefore, we overexpressed the pheromone receptor pra2 in FB2Δprf1 by introducing the gene under the control of the constitutive strong otef promoter in single copy into the ip locus. Upon stimulation with synthetic a1 pheromone, this strain (FB2pra2Δprf1) formed conjugation tubes, while the progenitor strain FB2Δprf1 did not react (Fig. 2D and Table 2). This shows that the inability of Δprf1 strains to develop conjugation tubes is due to insufficient expression of the receptor gene. In contrast, overexpression of pra2 in kpp4, fuz7, or kpp2 deletion strains did not rescue impaired conjugation tube formation, indicating that signaling via this cascade is crucial for this morphological response (Fig. 2D).
kpp4-2 and fuz7DD induce pheromone gene expression.
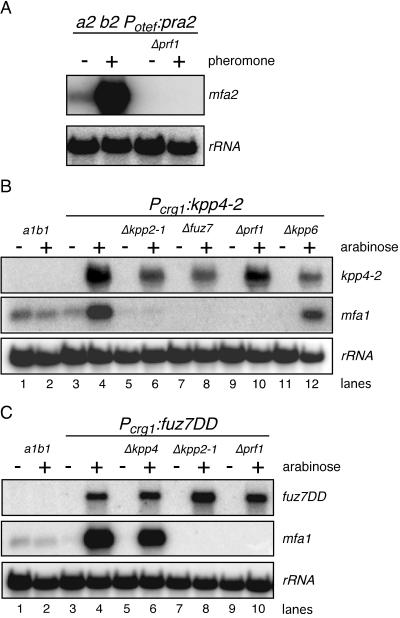
In previous experiments, conjugation tube formation and pheromone-dependent gene expression always appeared to be linked. The finding that conjugation tubes can be formed in the absence of Prf1 led us to reinvestigate the prf1 requirement for mfa1 gene induction. We performed Northern analysis with RNA isolated from FB2pra2 as well as FB2pra2Δprf1 before and 5 h after pheromone stimulation and observed that prf1 was essential for pheromone-induced mfa2 gene expression (Fig. 3A).
FIG. 3.
Constitutively active kpp4-2 and fuz7DD trigger pheromone gene expression. (A) Overexpression of pra2 does not rescue pheromone gene expression in prf1 deletion strains. The strains indicated on the top were treated for 5 h with synthetic a1 pheromone dissolved in DMSO (+) or with the same volume of DMSO (−). RNA was isolated, and 10 μg of total RNA was loaded per lane. The blot was probed with mfa2 and with rRNA as loading control. (B and C) Activation of the MAPK cascade elevates pheromone gene expression. FB1 and the FB1-derived strains indicated on the top were grown with glucose (−) or arabinose (+) as a carbon source. RNA was isolated, 15 μg of total RNA were loaded per lane and the same filters were hybridized in succession with the probes indicated on the right.
Next we analyzed pheromone gene transcription in strains harboring either kpp4-2 or fuz7DD before and after induction of these alleles. Expression of either kpp4-2 or fuz7DD elevated pheromone gene transcription (Fig. 3B and C). Furthermore, mfa1 induction by kpp4-2 required the downstream components fuz7 and kpp2, while the response to fuz7DD was independent of kpp4 but dependent on kpp2 (Fig. 3B and C). These results demonstrate that components of the cascade which regulate the morphological response to pheromone are also required to observe pheromone-dependent gene expression. Prf1 was essential for mfa1 gene expression induced by either kpp4-2 or fuz7DD (Fig. 3B and C), while it was not needed for the morphological transition. Thus, signaling through this MAPK cascade appears to branch downstream of Kpp2.
MAPK cascade mutants are affected in pheromone-responsive gene expression.
To investigate pheromone-responsive gene expression in Δkpp4 strains, we used synthetic a2 pheromone. As shown in Fig. 4A, pheromone stimulation of a wild-type strain led to induced expression of the a and b genes (pra, mfa and bE, bW), as was shown before with compatible strains (54). Analysis of the mutant strains revealed that in Δkpp4 mutants pheromone-induced b gene expression was completely abolished, while the induction of mfa1 and pra1 was not affected (Fig. 4A, lanes 3 and 4). A comparable response was observed in Δfuz7 strains (Fig. 4A, lanes 5 and 6). However, in Δkpp2-1 mutants, pheromone induction of mfa1, pra1, and b was severely attenuated (37) (Fig. 4A, lanes 7 and 8). This indicates that upon pheromone stimulation the entire MAPK module is required for b gene expression. In contrast, induction of genes in the a locus requires Kpp2, but the upstream components, Kpp4 and Fuz7, are not needed.
FIG. 4.
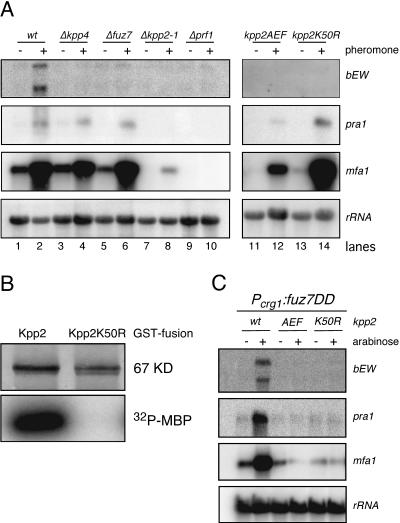
Pheromone signaling via the Kpp4/Fuz7/Kpp2 cascade affects b gene expression. (A) FB1 and the FB1-derived strains indicated on the top were treated for 5 h with synthetic a2 pheromone dissolved in DMSO (+) or with DMSO (−). RNA was isolated, and 10 μg of total RNA was loaded per lane. Blots were hybridized with the probes indicated on the right. wt, wild type. (B) kpp2K50R encodes a kinase-dead Kpp2 protein. The GST fusion proteins indicated at the top were purified from E. coli and subjected to kinase assay with MBP as the substrate. The upper panel shows Coomassie blue staining, and the lower panel depicts the incorporated radioactive phosphate in MBP. (C) fuz7DD elevates b gene expression. FB1 and the FB1-derived strains indicated on the top were grown with glucose (−) or arabinose (+) as a carbon source. RNA was isolated, and 15 μg of total RNA was loaded per lane. The filter was hybridized in succession with the probes indicated on the right.
We wondered whether this regulation of the a locus genes might rely on Kpp2 kinase activity. To test this, we generated two different kpp2 alleles, kpp2AEF and kpp2K50R. Kpp2AEF contains two amino acid substitutions in the conserved phosphate acceptor site (T182A and Y184F) and is therefore an unphosphorylatable MAPK. The kpp2K50R allele should encode a phosphorylatable but kinase-dead mutant protein due to a defect in ATP binding capacity. We assayed the kinase activity of Kpp2 and Kpp2K50R by mixing the respective proteins purified as GST fusions from E. coli with [γ-32P]ATP and MBP. These reactions were analyzed by SDS-PAGE for the presence of radioactively labeled MBP. As shown in Fig. 4B, GST-Kpp2 efficiently phosphorylated MBP, while the mutant GST-Kpp2K50R did not show kinase activity. Thus, kpp2K50R encodes a kinase-dead protein.
Next we constructed strains carrying kpp2AEF and kpp2K50R as well as strains expressing GFP-tagged Kpp2, Kpp2AEF, or Kpp2K50R by replacing the endogenous kpp2 allele in wild-type strains with kpp2AEF, kpp2K50R, kpp2WT-GFP, kpp2AEF-GFP, or kpp2K50R-GFP (see Materials and Methods). Western analysis revealed that kpp2WT-GFP, kpp2AEF-GFP, and kpp2K50R-GFP are expressed to comparable levels (Fig. 5A, right panel). Moreover, the resulting kpp2WT-GFP strains showed no significant defect in mating or conjugation tube formation (not shown) or in pathogenic development (Table 3), demonstrating that the GFP moiety does not interfere with the function of Kpp2 in vivo.
Upon pheromone stimulation, FB1kpp2AEF and FB1kpp2K50R failed to form conjugation tubes, indicating that phosphorylation as well as kinase activity of Kpp2 is essential for the morphological response (Table 2). When assayed for pheromone-induced gene expression, neither mutant strain showed b gene expression (Fig. 4A, lanes 11 to 14). However, the mutants differed with respect to mfa1 induction: while pheromone-responsive mfa1 expression was attenuated in FB1kpp2AEF (Fig. 4A, lanes 11 and 12), FB1kpp2K50R displayed a reduced basal level of mfa1 transcripts, while pheromone-induced mfa1 expression was comparable to the levels seen in FB1 (Fig. 4A, lanes 13 and 14). These findings suggest that the involvement of Kpp2 in the regulation of a locus gene expression is independent of its catalytic activity.
To elucidate whether Kpp2 kinase activity is required for fuz7DD-induced pheromone gene expression, we introduced Pcrg1:fuz7DD in FB1kpp2AEF and FB1kpp2K50R. After expression of fuz7DD, both strains exhibited budding growth (not shown) and did not show elevated expression of a and b locus genes (Fig. 4C). These results demonstrate that Fuz7DD can trigger expression of the a and b locus genes, presumably by increasing Kpp2 kinase activity.
Taken together, our findings illustrate that genetic activation of the MAPK module elevates transcription of genes in the a locus as well as in the b locus. After pheromone stimulation, this cascade is required only for pheromone-responsive b gene expression and is dispensable for the induction of the genes located in the a locus.
Pheromone stimulation increases Kpp2 kinase activity.
MAPK activation results from phosphorylation on the threonine and the tyrosine residues in the TXY activation loop. With antibody raised against the phosphoepitope pTEpY of mammalian ERK1 and ERK2, we examined pheromone-stimulated phosphorylation of Kpp2. When protein extracts from pheromone-treated kpp2WT strains were used, the anti-pTEpY antibody reacted specifically with a protein of the size expected for Kpp2 (41 kDa) (Fig. 5B, left panel), while no reaction was observed with proteins extracted from a strain expressing the unphosphorylatable derivative Kpp2AEF. This result suggested that Kpp2 is phosphorylated and activated upon pheromone stimulation. To assay the latter biochemically, the catalytic activity of Kpp2 was monitored in an immunoprecipitation-kinase assay. Protein extracts from FB2kpp2WT-GFP were prepared before and after treatment with pheromone. Kpp2WT-GFP was immunoprecipitated with anti-GFP antibody, and its ability to phosphorylate MBP was determined. In three independent experiments we observed a twofold induction of Kpp2 kinase activity after 40 min of stimulation (Fig. 5B). This indicates that Kpp2 becomes activated after pheromone stimulation.
Kpp2 is activated after expression of fuz7DD and interacts with Fuz7.
To verify the activation of Kpp2 by constitutively active fuz7DD, we replaced the endogenous kpp2 gene in FB1Pcrg1:fuz7DD with either kpp2WT-GFP, kpp2AEF-GFP, or kpp2K50R-GFP and performed immunoprecipitation kinase assays after 90 min of growth in arabinose-containing medium. As shown in Fig. 5C, the kinase activity of Kpp2WT-GFP increased fourfold after expression of fuz7DD (compare lanes 3 and 4). As expected, expression of fuz7DD did not enhance the kinase activity of Kpp2AEF-GFP and Kpp2K50R (Fig. 5C, lanes 5 to 8). However, in all assays we detected more phosphorylated MBP than in control experiments in which protein extracts from FB1Pcrg1:fuz7DD lacking Kpp2-GFP were used (Fig. 5C, compare lanes 1 and 2 with lanes 5 to 8). This might indicate that another kinase, whose activity was not altered by Fuz7DD, coimmunoprecipitated with Kpp2-GFP.
Interactions of MAPKs and regulatory proteins, e.g., MAPKKsor specific phosphatases, are known to be mediated by a so-called docking site (D box), which is defined by the consensus sequence (R/K)2-(x)2-6-L/I-x-L/I (19). Since such a D box is located in the N terminus of Fuz7 (Fig. 2A) (6), we tested the interaction of Fuz7 and Kpp2 with a GST pulldown assay. For this purpose, bacterially expressed GST and GST-Fuz7 fusion proteins were immobilized on glutathione-Sepharose beads and incubated with protein extracts prepared from either FB2kpp2WT-GFP, FB2kpp2AEF-GFP, or FB2kpp2K50R-GFP. After extensive washing, the beads were subjected to SDS-PAGE to examine for the presence of GST-Fuz7 and Kpp2 fusion proteins by Coomassie blue staining (Fig. 5D, lower panel) and by immunoblotting with anti-GFP antibody (Fig. 5D, upper panel), respectively. The results showed that Kpp2WT-GFP, Kpp2AEF-GFP, and Kpp2K50R-GFP interacted with GST-Fuz7 but not with GST alone (Fig. 5D).
Integrity of the MAPK module is essential for pathogenic development.
As described above, deletion of kpp4 abolishes pathogenic development. In this respect Δkpp4 strains behave like fuz7 deletion strains, which in our hands are nonpathogenic (Table 3). Furthermore, we did not observe disease symptoms in corn plants infected with SG200Δfuz7, which is consistent with the described lack of tumor formation in diploid Δfuz7/Δfuz7 strains (Table 3) (4). These results make it likely that Fuz7 and Kpp4 act in one cascade also during pathogenic development. Since Δkpp4 and Δfuz7 strains are impaired in b gene expression, we analyzed whether the observed loss of tumor formation is simply due to insufficient b gene expression. For this purpose, we introduced the deletion alleles into the haploid solopathogenic strain HA103 (a1 bcon), which expresses a bE1-bW2 heterodimer from constitutive promoters (22). HA103 as well as the resulting strains HA103Δkpp4 and HA103Δfuz7 exhibited filamentous growth, indicating functionality of the bE1-bW2 heterodimer (not shown). However, plants infected with HA103Δkpp4 or HA103Δfuz7 showed no disease symptoms, while the progenitor strain HA103 was able to induce plant tumors efficiently (Table 3). This indicates that kpp4 and fuz7 are crucial for pathogenic development and exert their function in parallel or downstream of the b heterodimer.
If Kpp2 acts downstream of Fuz7 and Kpp4 during pathogenic development as it does during mating, kpp2 deletions strains should be unable to cause disease. However, Δkpp2-1 deletion strains are only reduced in pathogenicity (35, 37). This finding could reflect genetic redundancy on the level of the MAPK, as was observed in the yeast pheromone pathway. To analyze this possibility, we assayed the pathogenicities of strains carrying either the inactive mutant allele kpp2AEF or wild-type kpp2 as control. Interestingly, only 3% of plants infected with mixtures of FB1kpp2AEF and FB2kpp2AEF showed small leaf tumors, while combinations of compatible kpp2WT (FB1kpp2WT × FB2kpp2WT) strains were able to induce tumors in 95% of the infected plants (Table 3). Since SG200kpp2AEF was also nonpathogenic, the pathogenicity defect seen in mixtures of compatible kpp2AEF mutant strains cannot simply be a result of inefficient cell fusion (Table 3). In addition, mixtures of FB1kpp2AEF and FB2 induced tumor development in 50% of the infected plants, which makes it unlikely that kpp2AEF is a dominant-negative allele. Thus, the unphosphorylatable allele of kpp2 (kpp2AEF) blocks disease development much more efficiently than the deletion of kpp2.
To address the question of which stage during pathogenesis is affected in Δkpp4, Δfuz7, or kpp2AEF mutants, we stained fungal material on the plant surface with Calcofluor and Chlorazole Black E. The latter stain has recently been adapted for the visualization of infection structures of U. maydis (11). On leaves prepared from plants infected with either SG200 or HA103, we observed vigorous filaments and formation of appressoria (not shown). In contrast, after infections with either SG200Δkpp4, SG200kpp2AEF, HA103Δkpp4, or HA103Δfuz7, only a small proportion of the inoculum developed filaments, and none of these formed appressoria (not shown). As we could not exclude the possibility that we might have overlooked some appressoria, we directly examined development of appressoria by coinfecting SG200 with either SG200Δkpp4 or SG200kpp2AEF (SG200 and derivatives cannot fuse and therefore allow for the simultaneous analysis of different strains). In order to distinguish between appressoria developed by SG200 and the mutant derivatives, we used combinations, in which one of the strains expressed GFP from the strong otef promoter. While in control infections with SG200 and SG200Potef:GFP approximately 50% of Calcofluor-stained appressoria showed green fluorescence, in combinations of SG200Potef:GFP and either SG200Δkpp4 or SG200kpp2AEF all stained infection structures derived from SG200Potef:GFP (Table 4). When plants were infected with the reciprocal combinations (SG200 with either SG200Δkpp4/Potef:GFP or SG200kpp2AEF/Potef:GFP), none of the appressoria found showed GFP fluorescence (Table 4). These results show that integrity of the MAPK module is important for filament formation as well as for development of appressorial structures on the plant surface.
TABLE 4.
Formation of appressoria
| Inoculum | No. of appressoria counteda | No. of appressoria showing green fluorescence |
|---|---|---|
| SG200 + SG200Potef:GFP | 32 | 17 |
| SG200Δkpp4 + SG200Potef:GFP | 37 | 37 |
| SG200 + SG200Δkpp4/Potef:GFP | 40 | 0 |
| SG200kpp2AEF + SG200Potef:GFP | 35 | 35 |
| SG200 + SG200kpp2AEF/Potef:GFP | 31 | 0 |
Appressoria were counted on at least 10 different plants.
DISCUSSION
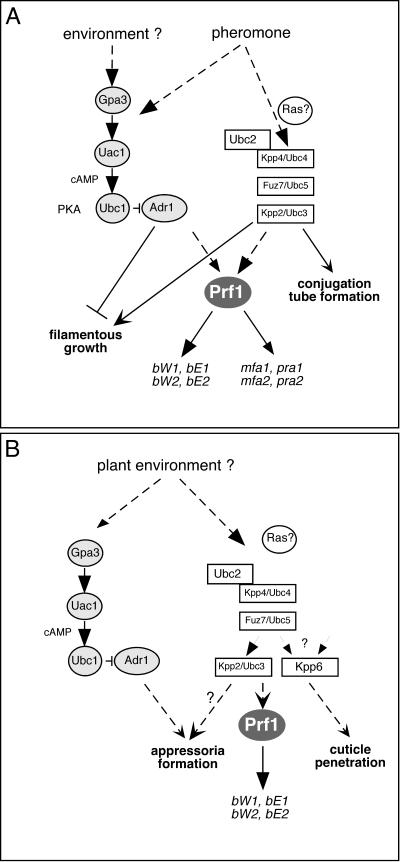
In this study we have demonstrated genetically as well as biochemically that ubc4, fuz7, and ubc3/kpp2, which encode components of a MAPK module, act in one cascade during mating and pathogenic development. This extends previous studies which demonstrated that the same three components are essential for the filamentous phenotype of mutants lacking adenylyl cyclase (1, 34, 35). The full characterization of the MAPKKK gene kpp4 shows that this gene is identical to ubc4. The deduced N terminus of Kpp4 contains a conserved protein-protein interaction domain termed the SAM domain (not present in Ubc4), which is a hallmark of other fungal MAPKKKs. In addition to the SAM domain (amino acids 59 to 122), Kpp4 harbors a putative RA domain (amino acids 446 to 555). These domains are also present in the MAPKKK Byr2 of S. pombe and provide for a direct interaction with Ste4 and Ras1, respectively (52). Recently, two U. maydis genes, ras2 and ubc2, displaying homology to Ras1 and Ste4 of S. pombe, respectively, were identified. The respective deletion strains are impaired in conjugation tube formation and pathogenic growth, phenotypes that are also associated with kpp4 deletion (30, 36). In this report we have shown that strains expressing kpp4K481E are attenuated in mating, conjugation tube formation, and pathogenic development. The kpp4K481E allele carries a mutation analogous to Byr2FBR that prevents interaction with Ras1 and results in strongly reduced conjugation of S. pombe cells (52). Therefore, we consider it likely that the regulation of Kpp4 is similar to Byr2 regulation in S. pombe and involves interactions of Kpp4 with Ras2 via the RA domain and with Ubc2 via the respective SAM domains (Fig. 6).
FIG. 6.
Proposed signaling processes during saprophytic growth and mating (A) as well as during the early steps of infection (B). The broken arrows indicate missing components or putative signaling events. See text for a detailed discussion.
Regulation of mating by the Kpp4/Fuz7/Kpp2 MAPK module.
Our data show that Kpp4, Fuz7, and Kpp2 act in one module during mating. Structures resembling conjugation tubes are induced by a constitutively active allele of kpp4 (kpp4-2), and this is dependent on the downstream components Fuz7 and Kpp2. The same morphological switch is triggered by a constitutively active allele of fuz7 (fuz7DD), and here it requires kpp2 but not kpp4. These genetic experiments are supported by biochemical data. The kinase activity of Kpp2 is increased after expression of fuz7DD, and this enhanced activity requires the conserved phosphate-acceptor sites (TEY) in Kpp2. In addition, Fuz7 interacts with Kpp2 in vitro as shown by a GST pulldown assay. On these grounds, we assume that Fuz7 activates Kpp2 in vivo (Fig. 6A).
Several observations suggest that the Kpp4/Fuz7/Kpp2 module is directly involved in transmitting the pheromone signal (Fig. 6A). Disruption of kpp4 attenuates mating and impairs conjugation tube formation, phenotypes that were described before for fuz7 and kpp2 deletion strains (4, 35, 37). In none of these deletion strains does overexpression of the pheromone receptor rescue conjugation tube formation, indicating that all three components are required for transmitting the signal. Activation of the cascade by either kpp4-2 or fuz7DD induces conjugation tube-like structures, and concomitantly a and b gene expression is increased. Finally, upon pheromone stimulation, Kpp2 was phosphorylated and its kinase activity was shown to increase twofold. In comparable experiments with S. cerevisiae, the MAPK activity of Kss1p as well as Fus3p was increased five- to sixfold (13, 42). It is presently not clear why the increase of Kpp2 activity in U. maydis does not reach such levels. One possibility might be the lower solubility or activity of the lipopeptide pheromone applied to U. maydis cells compared to the peptide pheromone used in the respective yeast experiments.
In line with the assertion that the Kpp4/Fuz7/Kpp2 module transmits the pheromone signal, we detected elevated levels of b gene expression after expression of fuz7DD, and this induction required the kinase activity of Kpp2. In the absence of kpp4, fuz7, or kpp2, pheromone-induced b gene expression was prevented. However, with respect to pheromone-responsive a gene expression the picture is different: mfa1 induction is not affected in kpp4 and fuz7 deletion strains (41) or in strains expressing a kinase-dead allele of kpp2 (kpp2K50R), while it is strongly reduced in Δkpp2-1 mutants (37). This indicates that Kpp2 influences a gene expression via a kinase-independent mechanism. Precedence for kinase-independent functions of MAPK proteins is found in S. cerevisiae, where the MAPK Kss1p regulates invasive growth in a phosphorylation-dependent manner. Unphosphorylatable Kss1p-[AEF] but not kinase-dead Kss1p-[K42R] is able to prevent invasive growth of a FUS3 KSS1 double-deletion strain (7). Phosphorylated Kss1p is a positive regulator of the transcription factor Ste12p, while unphosphorylated Kss1p inhibits Ste12p function by direct interaction (7, 13, 42). In contrast to this situation in S. cerevisiae, unphosphorylatable Kpp2AEF in U. maydis is not able to exert the kinase-independent function, since strains expressing Kpp2AEF are reduced in pheromone-dependent a gene transcription. This suggests that the mechanism of kinase-independent function of Kpp2 in U. maydis differs from that of Kss1p. Preliminary results suggest that this function of Kpp2 may affect the transcription factor Prf1, which is absolutely required for pheromone-responsive gene expression. In Δkpp2-1 as well as kpp2AEF strains, prf1 transcription is affected and overexpression of prf1 rescues the attenuated pheromone gene expression (P. Muller and M. Feldbrugge, unpublished data). It is conceivable that Kpp2 and Prf1 interact, leading to a stabilization of Prf1 protein, and Kpp2AEF might no longer interact with Prf1, while stabilization could still be carried out by the kinase-dead Kpp2 protein.
In contrast to the situations in S. cerevisiae, S. pombe, C. neoformans, and Candida albicans (2, 16, 33), the pheromone MAPK cascade is not essential for cell fusion in U. maydis. This indicates that at least one additional pathway must participate. One likely candidate is the cAMP signaling cascade composed of the Gα subunit Gpa3, adenylyl cyclase Uac1, and cAMP-dependent protein kinase A (Fig. 6A). Disruption of the cAMP cascade affects cell fusion and abolishes pheromone-induced a gene expression (18, 41). On the other hand, an activated cAMP cascade results in elevated pheromone gene expression that depends on prf1 (23, 29, 41). Thus, we consider it likely that the pheromone signal is somehow transmitted by the cAMP pathway as well as the MAPK module to Prf1 which in turn promotes pheromone-regulated a and b gene expression (Fig. 6A).
In S. cerevisiae the pheromone signal is transmitted exclusively via a MAPK module and the pheromone-responsive MAPK Fus3p regulates the transcription factor STE12p, presumably by the phosphorylation-dependent inactivation of the Dig1/2 proteins, which inhibit the transcription factor STE12p (17, 46, 51). Once activated, Ste12p triggers a transcriptional program that is necessary for the mating response. Another Fus3p substrate is Far1p, which subsequently inhibits cyclin-Cdc28p kinase activity, resulting in cell cycle arrest, and establishes a site of polarized growth toward the mating partner (14, 38, 53). STE12 is absolutely required for projection formation, even when the pheromone MAPK cascade is activated by a dominant-active allele of STE11 (15). However, conjugation tube formation in U. maydis does not require the transcription factor prf1. This was evident only when the pheromone receptor was constitutively expressed or when the MAPK cascade was genetically activated by expressing either kpp4-2 or fuz7DD. Thus, pheromone signal transmission bifurcates downstream of Kpp2 (Fig. 6A). One branch must control the activation of Prf1 and subsequent induction of the genes located in the a and b loci. The other branch induces a morphological reprogramming, which might involve a second transcription factor and/or proteins regulating the cell cycle and the cytoskeleton.
Regulation of pathogenic development by the Kpp4/Fuz7/Kpp2 MAPK module.
In this study we have demonstrated that kpp4 and fuz7 are essential for pathogenesis even under conditions in which cell fusion and pheromone stimulation are not required. Thus, it is likely that Kpp4 and Fuz7 transmit additional signals during pathogenic development (Fig. 6B) (4). The defect in tumor formation caused by deletion of either kpp4 or fuz7 cannot be rescued by overexpression of b. Thus, signaling via Kpp4 and Fuz7 appears to regulate pathogenicity in parallel to b (Fig. 6B). Further analysis revealed that kpp4 and fuz7 are necessary for filamentous growth on the plant surface and development of appressoria even under conditions where cell fusion was bypassed. While kpp4 and fuz7 are absolutely required for pathogenicity, deletion of kpp2 only moderately reduced tumor development (35, 37). This is most likely due to genetic redundancy at the level of the MAPK. U. maydis encodes a Kpp2-related MAPK, Kpp6, which has no obvious role during mating, and kpp2 kpp6 double-deletion strains are nonpathogenic (11). Since strains expressing an inactive allele of kpp2 (kpp2AEF) display a stronger phenotype than the Δkpp2-1 mutants, we were able to elucidate the function of kpp2 during the infection process. Such strains behaved like Δkpp4 and Δfuz7 mutants and were unable to develop appressoria. Given these results, Kpp2 can be placed in one cascade with Kpp4 and Fuz7 during appressorial formation.
In a variety of phytopathogenic fungi MAPKs were shown to control appressoria differentiation, e.g., Pmk1 in Magnaporthe grisea, Cmk1 in Colletotrichum lagenarium, and Chk1 in Cochliobolus heterostrophus (31, 50, 56). Hence, signaling processes that regulate the infection process appear to be conserved in phytopathogenic fungi. U. maydis is so far unique among phytopathogenic fungi in having two Pmk1-like MAPKs, Kpp2 and Kpp6. These kinases control discrete steps of the infection-related development, with kpp2 being needed for formation of appressoria and kpp6 being needed for appressorial function (Fig. 6B) (11) Thus, the early infection processes in U. maydis is controlled by a stepwise activation of two highly homologous MAPKs.
Acknowledgments
We thank J. Görl for construction of pGEX-Kpp2 and pGEX-Fuz7 and Manuel Tönnis and Horst Kessler for providing synthetic pheromone. We thank Jan Schirawski and Anja Volz-Peters for their critical comments on the manuscript.
This work was supported by the DFG through grant SFB369.
REFERENCES
- 1.Andrews, D. L., J. D. Egan, M. E. Mayorga, and S. E. Gold. 2000. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 13:781-786. [DOI] [PubMed] [Google Scholar]
- 2.Banuett, F. 1998. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62:249-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banuett, F., and I. Herskowitz. 1989. Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. [DOI] [PubMed] [Google Scholar]
- 5.Banuett, F., and I. Herskowitz. 1994. Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and b loci. Exp. Mycol. 18:247-266. [Google Scholar]
- 6.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell, L., J. G. Cook, D. Voora, D. M. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolker, M., S. Genin, C. Lehmler, and R. Kahmann. 1995. Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73:320-325. [Google Scholar]
- 9.Bolker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 10.Bottin, A., J. Kamper, and R. Kahmann. 1996. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253:342-352. [DOI] [PubMed] [Google Scholar]
- 11.Brachmann, A., J. Schirawski, P. Muller, and R. Kahmann. 2003. An unusual MAP kinase is required for efficient penetration of the plant surface by U. maydis. EMBO J. 22:2199-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brachmann, A., G. Weinzierl, J. Kamper, and R. Kahmann. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047-1063. [DOI] [PubMed] [Google Scholar]
- 13.Breitkreutz, A., L. Boucher, and M. Tyers. 2001. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11:1266-1271. [DOI] [PubMed] [Google Scholar]
- 14.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 15.Cairns, B. R., S. W. Ramer, and R. D. Kornberg. 1992. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 6:1305-1318. [DOI] [PubMed] [Google Scholar]
- 16.Clarke, D. L., G. L. Woodlee, C. M. McClelland, T. S. Seymour, and B. L. Wickes. 2001. The Cryptococcus neoformans STE11alpha gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol. Microbiol. 40:200-213. [DOI] [PubMed] [Google Scholar]
- 17.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 18.Durrenberger, F., K. Wong, and J. W. Kronstad. 1998. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc. Natl. Acad. Sci. USA 95:5684-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enslen, H., and R. J. Davis. 2001. Regulation of MAP kinases by docking domains. Biol. Cell 93:5-14. [DOI] [PubMed] [Google Scholar]
- 20.Gillissen, B., J. Bergemann, C. Sandmann, B. Schroeer, M. Bolker, and R. Kahmann. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647-657. [DOI] [PubMed] [Google Scholar]
- 21.Gold, S., G. Duncan, K. Barrett, and J. Kronstad. 1994. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8:2805-2816. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann, H. A., R. Kahmann, and M. Bolker. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 23.Hartmann, H. A., J. Kruger, F. Lottspeich, and R. Kahmann. 1999. Environmental signals controlling sexual development of the corn Smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell 11:1293-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E. coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 25.Holliday, R. 1974. Ustilago maydis, p. 575-595. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Press, New York, N.Y.
- 26.Huang, W., and R. L. Erikson. 1994. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc. Natl. Acad. Sci. USA 91:8960-8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahmann, R., S. G., C. Basse, M. Feldbrügge, and J. Kämper. 2000. Ustilago maydis, the causative agent of corn smut disease, p. 347-371. In J. W. Kronstad (ed.), Fungal pathology. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 28.Kamper, J., M. Reichmann, T. Romeis, M. Bolker, and R. Kahmann. 1995. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73-83. [DOI] [PubMed] [Google Scholar]
- 29.Kruger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 30.Lee, N., and J. W. Kronstad. 2002. ras2 controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryot. Cell 1:954-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lev, S., A. Sharon, R. Hadar, H. Ma, and B. A. Horwitz. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96:13542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loubradou, G., A. Brachmann, M. Feldbrugge, and R. Kahmann. 2001. A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40:719-730. [DOI] [PubMed] [Google Scholar]
- 33.Magee, B. B., M. Legrand, A. M. Alarco, M. Raymond, and P. T. Magee. 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 46:1345-1351. [DOI] [PubMed] [Google Scholar]
- 34.Mayorga, M. E., and S. E. Gold. 1998. Characterization and molecular genetic complementation of mutants affecting dimorphism in the fungus Ustilago maydis. Fungal Genet. Biol. 24:364-376. [DOI] [PubMed] [Google Scholar]
- 35.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 36.Mayorga, M. E., and S. E. Gold. 2001. The ubc2 gene of Ustilago maydis encodes a putative novel adaptor protein required for filamentous growth, pheromone response and virulence. Mol. Microbiol. 41:1365-1379. [DOI] [PubMed] [Google Scholar]
- 37.Muller, P., C. Aichinger, M. Feldbrugge, and R. Kahmann. 1999. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 38.Peter, M., A. Gartner, J. Horecka, G. Ammerer, and I. Herskowitz. 1993. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73:747-760. [DOI] [PubMed] [Google Scholar]
- 39.Peterson, A. J., M. Kyba, D. Bornemann, K. Morgan, H. W. Brock, and J. Simon. 1997. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 17:6683-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponting, C. P., and D. R. Benjamin. 1996. A novel family of Ras-binding domains. Trends Biochem. Sci. 21:422-425. [DOI] [PubMed] [Google Scholar]
- 41.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bolker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schultz, J., C. P. Ponting, K. Hofmann, and P. Bork. 1997. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6:249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schafer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 46.Song, D., J. W. Dolan, Y. L. Yuan, and S. Fields. 1991. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 5:741-750. [DOI] [PubMed] [Google Scholar]
- 47.Spellig, T., A. Bottin, and R. Kahmann. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503-509. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson, B. J., N. Rhodes, B. Errede, and G. F. Sprague, Jr. 1992. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 6:1293-1304. [DOI] [PubMed] [Google Scholar]
- 49.Szabo, Z., Tonnis, M., Kessler, H., and M. Feldbrugge. 2002. Structure-function analysis of lipopeptide pheromones from the plant pathogen Ustilago maydis. Mol. Genet. Genomics 268:362-370. [DOI] [PubMed] [Google Scholar]
- 50.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13:374-383. [DOI] [PubMed] [Google Scholar]
- 51.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 52.Tu, H., M. Barr, D. L. Dong, and M. Wigler. 1997. Multiple regulatory domains on the Byr2 protein kinase. Mol. Cell. Biol. 17:5876-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyers, M., and B. Futcher. 1993. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol. Cell. Biol. 13:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urban, M., R. Kahmann, and M. Bolker. 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251:31-37. [DOI] [PubMed] [Google Scholar]
- 55.Wedlich-Soldner, R., M. Bolker, R. Kahmann, and G. Steinberg. 2000. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19:1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, J. R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696-2706. [DOI] [PubMed] [Google Scholar]