Abstract
The most common cause of fungal meningitis in humans, Cryptococcus neoformans serotype A, is a basidiomycetous yeast with a bipolar mating system. However, the vast majority (>99.9%) of C. neoformans serotype A isolates possess only one of the two mating type alleles (MATα). Isolates with the other allele (MATa) were recently discovered and proven to mate in the laboratory. It has been a mystery whether and where C. neoformans strains undergo sexual reproduction. Here, we applied population genetic approaches to demonstrate that a population of C. neoformans serotype A clinical isolates from Botswana contains an unprecedented proportion of fertile MATa isolates and exhibits evidence of both clonal expansion and recombination within two partially genetically isolated subgroups. Our findings provide evidence for sexual recombination among some populations of C. neoformans serotype A from sub-Saharan Africa, which may have a direct impact on their evolution.
The population structure and biology of a microbial species are determined by the extent, if any, to which recombination contributes to its genetic variation. In clonal microorganisms, populations consist of one to several distinct clones or lineages, sexual reproduction is absent or limited, and evolution occurs only through the accumulation of favorable spontaneous mutations within independent lineages (28, 43, 44, 50). Sexual reproduction offers the opportunity to generate robust individuals more rapidly. In sexually reproducing populations, favorable combinations of mutations arising in different lineages may be lost through subsequent recombination (30). Many fungi exhibit a mixture of both clonal reproduction, which can be favored under uniform growth conditions, and sexual recombination, which can be advantageous in changing environments (9, 42, 50).
The frequency of recombination in natural populations of human pathogens can affect the evolution of infectious diseases and thereby have an impact on public health (21, 36). In predominantly clonal populations, clinically relevant phenotypes, such as virulence or resistance to antibiotics, are associated with particular clones or lineages, whereas in recombining populations, in the absence of inbreeding or assortative mating, clinically important traits are distributed among unrelated individuals.
Cryptococcus neoformans is a pathogenic yeast that often causes life-threatening infections, especially in immunocompromised individuals, such as patients with AIDS (10, 31). The five serotypes of C. neoformans differ in their global distributions: serotype A is ubiquitous, serotype D is global but more frequent in Europe, serotype AD is rare but global, and serotypes B and C are prevalent in tropical and subtropical regions. In addition to their geographical distributions, the serotypes also differ in their ecological characteristics, clinical manifestations, and several in vitro phenotypes (10, 31). The most common and serious clinical manifestation of infection is meningoencephalitis, which occurs with any of the serotypes. However, serotype A (C. neoformans var. grubii) is responsible globally for more than 90% of cryptococcal infections and more than 99% of cases in patients with AIDS (10, 31). The highest incidence of coinfection with human immunodeficiency virus and C. neoformans occurs in sub-Saharan Africa, where cryptococcosis is the leading cause of meningitis in patients with AIDS, among whom the incidence is approximately 45% and mortality approaches 40% (6, 17).
Cryptococcosis is acquired by inhalation of airborne cells of C. neoformans from the environment, but the source and nature of the infectious propagules have not been resolved. Serotypes A, D, and AD are isolated worldwide from avian excreta, soil, and vegetative debris, whereas serotypes B and C are thought to be associated with various tropical trees (10). Both desiccated yeast cells and basidiospores, which result from sexual reproduction or haploid “fruiting,” are small enough (<5 μm) to reach the alveoli to establish infection. The extent to which basidiospores are produced in nature is unknown, but experimentally produced basidiospores are highly infectious (37).
C. neoformans is a haploid basidiomycetous yeast with a bipolar mating system. Sexual reproduction is governed by a single mating type locus with two alternative alleles, MATa and MATα (26). However, the occurrence of sexual reproduction in natural populations is unknown. In an Australian population of serotype B, both mating types were found in equal proportions in the same environmental niche, yet this population was extensively clonal and exhibited little diversity and no evidence of genetic recombination (18).
Until 3 years ago, all clinical and environmental isolates of serotype A that had been examined possessed only the MATα allele, a fact which would seem to exclude the possibility of sexual reproduction. However, the population structure of serotype A could not be explained solely by mutation and clonal expansion (9, 52). Analyzing representatives of different multilocus genotypes for associations among loci provides a common method for distinguishing between the null hypothesis of recombination and the alternate hypothesis of clonality (9, 28, 41, 44). In a recombining population, loci that are physically unlinked are distributed randomly among the individuals. Conversely, clonal populations usually are characterized by linkage disequilibrium or nonrandom associations among physically unlinked loci. Population genetic analysis of multilocus genotypes of 222 serotype A isolates showed no statistically significant linkage disequilibrium among the loci, indicating that recombination might be occurring in this population (7, 9). Another commonly used method for distinguishing clonality from recombination is to compare gene genealogies, or phylogenetic trees, constructed from polymorphisms within gene sequences (22, 28, 52). In a clonal population, the genealogies of unrelated genes will be congruent, but in recombining organisms, the genealogies of unrelated genes may differ. Multiple gene genealogies of C. neoformans revealed significant incongruences, which might be explained by recombination and hybridization (51, 52). Recent reports revealed the existence of rare MATa isolates of serotype A and demonstrated their ability to undergo a sexual cycle (20, 27, 33, 45). However, only 3 of >2,000 isolates examined had mating type MATa; therefore, the occurrence of sexual reproduction in nature has been a mystery (33, 45, 46).
Here, we analyzed the genetic structure of a population of 139 serotype A isolates from patients with AIDS in sub-Saharan Africa. Using multilocus genotypes and DNA sequence data, we provide evidence for population subdivisions and for both clonal expansion and recombination. This population included 14 fertile MATa serotype A isolates that undergo sexual reproduction and recombination in the laboratory. These findings provide supportive evidence that serotype A strains may be sexually recombining in sub-Saharan Africa, a finding which has considerable implications for the evolution of the pathogen C. neoformans.
MATERIALS AND METHODS
Yeast strains, maintenance, and serotyping.
The 139 strains used for this study were isolated from patients with AIDS in Botswana and obtained by L. Barth Reller, Duke University Medical Center. Isolates were colony purified, confirmed to be C. neoformans by standard morphological and physiological criteria (23), and maintained on yeast extract-peptone-dextrose (YPD) agar medium (Difco, Baltimore, Md.) at 30°C. Isolates were serotyped with commercial monoclonal antibodies (Iatron, Tokyo, Japan) and by PCR amplification with serotype-specific primers and amplified fragment length polymorphisms (AFLP) (see below). For mating assays, in addition to the clinical isolates from Botswana, the following laboratory tester strains of C. neoformans were used: H99, serotype A, MATα; KNA14, serotype A, MATa; JEC21, serotype D, MATα; and JEC20, serotype D, MATa (33). Additional laboratory strains of serotypes A, D, AD, B, and C were used as comparative controls for AFLP genotyping and phylogenetic analyses (52).
DNA manipulations.
A pure culture of each yeast isolate was grown on a YPD agar plate for 48 h. Genomic DNA was extracted from each isolate with a Camgen yeast genomic DNA purification kit (Whatman BioScience, Cambridge, United Kingdom) as prescribed by the manufacturer's instructions. The mating type of each isolate was identified by PCR with mating type- and serotype-specific primers that amplified portions of the STE20a or STE20α allele of serotype A or D (designated STE20aA, STE20αA, STE20aD, or STE20αD) (25, 27, 48, 53).
Mating assays.
To test for sexual reproduction and determine mating types, strains were crossed in the laboratory with reference strains (H99, KNA14, JEC21, and JEC20) as described previously (33). To isolate basidiospores, each serotype A MATa strain from Botswana was mated with the serotype A MATα reference strain (H99). Portions of cocultures producing basidiospores were excised and transferred to a fresh YPD agar plate, on which individual basidiospores were dissected by micromanipulation and cultured for subsequent evaluation.
Flow cytometry.
To establish the ploidy of isolates, the relative DNA content was measured by flow cytometry and compared to that of H99, the haploid reference strain of serotype A (11, 40).
AFLP.
AFLP markers were generated as described previously (4, 47) with the following modifications. Restriction and ligation reactions were carried out simultaneously overnight at 37°C with 25-μl volumes consisting of 1× ligation buffer (Invitrogen, Carlsbad, Calif.), 40 mM NaCl, 0.2 mg of bovine serum albumin/ml, 4 μM MseI adaptor, 0.4 μM EcoRI adaptor, 5 U of EcoRI (Promega, Madison, Wis.), 2 U of MseI (NEB, Beverly, Mass.), 0.2 U of ligase (Invitrogen), and 100 ng of genomic DNA. Samples from representative reactions were electrophoresed in 1% agarose to ensure that DNA was completely digested. Each reaction mixture was diluted 1:2, and 2 μl was used for the pre-PCRs, which were performed as described previously (4).
Two primer combinations, EcoRI-AC-FAM/MseI-G and EcoRI-TG-FAM/MseI-G, were used for the selective PCR. The selective EcoRI primers were labeled at the 5′ end with 6-carboxyfluorescein (6FAM). The primer sequences were as follows: EcoRI-AC-FAM, 5′-6FAM-GACTGCGTACCAATTCAC; EcoRI-TG-FAM, 5′-6FAM-GACTGCGTACCAATTCTG; and MseI-G, 5′-GATGAGTCCTGAGTAAG. The selective PCR conditions were 5 min at 95°C; 9 cycles at 94°C for 30 s, 65°C for 30 s, with a decrease of 1°C every cycle, and 72°C for 1 min; and 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min. Each PCR mixture contained 12 μl of 1× PCR buffer (Invitrogen), 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1 μM each primer, 0.065 μl of Taq DNA polymerase (Invitrogen,), and 2 μl of a 1:11 dilution of the DNA restriction-ligation mixture.
The selective PCR products were diluted 1:21 in sterile water and electrophoresed by using an automated sequencer (ABI 3700; Applied Biosystems, Foster City, Calif.) under the default run module for GeneScan version 3.7 according to the manufacturer's instructions. Data were collected and fragment sizes were determined by using GeneScan version 3.1 analysis software and a 500 TAMRA internal size standard (Applied Biosystems). Data were analyzed by using Genographer version 1.6 software (3) and scored manually. Polymorphic bands were defined as bands of the same size that were present in some but not all isolates. To assess the reproducibility of the AFLP method, DNA was extracted, and the AFLP reaction and analysis were performed on at least two separate occasions for each isolate. In replicate analyses, 92% of the AFLP bands were identical (data not shown). Only intense and reproducible bands were scored for the analysis of population structure.
Gene sequencing.
From a strain of each genotype, portions of the laccase (LAC1) gene and the intergenic spacer (IGS) region between the large subunit of ribosomal DNA and the 5S genes were obtained by PCR as described previously (12, 52). PCR products were purified and sequenced by using an ABI 3700 sequencer with Big Dye terminators (Applied Biosystems). Sequences were generated from both DNA strands, aligned, and optimized visually.
Data analysis.
The genetic relatedness among the isolates was evaluated by nonmetric multidimensional scaling (MDS) analysis with Euclidian distance measures through the use of Community Analysis Package 2.4 (PISCES Conservation Ltd., Hampshire, United Kingdom). We applied the method of Nei and Li to analyze genetic distances of restriction fragment data (32) and generated a dendrogram with the neighbor-joining algorithm by using PAUP (38). Phylogenetic analysis of the IGS and LAC1 genes was also performed by using PAUP. Maximum-parsimony trees were identified by using heuristic searches based on 100 random sequence additions for each region. Statistical support for phylogenetic groupings was assessed by bootstrap analysis with 500 replicate data sets. The parsimony tree length was tested by using 1,000 permutations (38). To evaluate the associations among loci in each sample, we used the index of association (IA), the new unbiased estimate of multilocus linkage disequilibrium (rd) (9, 28), and the parsimony tree length test (2, 9). For this text, maximum-parsimony trees were identified by using Dollo parsimony criteria (14). IA and rd values were calculated by using Multilocus 1.2 software, and 1,000 artificially recombined data sets were used to determine the statistical values of the test (1).
RESULTS
Unexpected proportion of MATa strains of C. neoformans serotype A in Africa.
To investigate the population genetic structure of C. neoformans in Africa, we analyzed 139 strains of serotype A (C. neoformans var. grubii) that were isolated in 2000 and 2001 from patients with AIDS in Botswana. AFLP analysis with two independent primer pairs confirmed that they were serotype A and that none was serotype D, AD, B, or C (4). PCR analysis of the mating type locus-specific STE20 gene revealed that 14 isolates were MATa. These isolates produced the appropriate amplicon only with the serotype A MATa (STE20aA) primer pair and failed to produce any PCR product when tested with the primer pairs specific for STE20aD, STE20αD, or STE20αA. Genomic DNA from each of the other 125 isolates was amplified only with the STE20αA primer pair (data not shown), indicating they were all serotype A MATα.
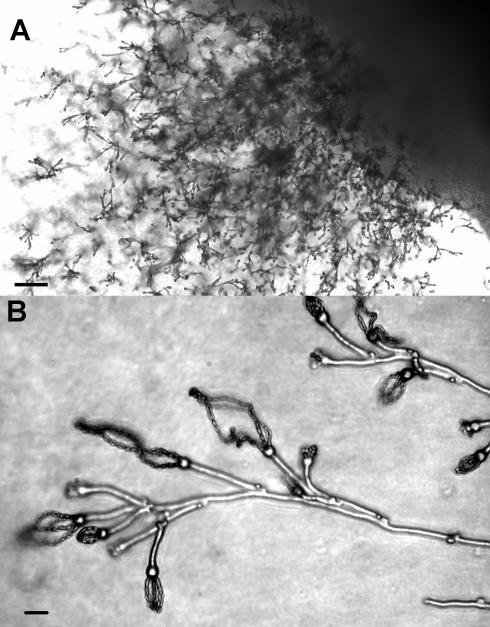
In mating assays, all 14 serotype A MATa isolates were fertile. When crossed on V8 or carrot juice agar with serotype A or D MATα tester strains, each produced dikaryotic hyphae and basidia with long chains of basidiospores (Fig. 1). Control crosses with MATa tester strains were all negative for hyphal formation and sexual reproductive structures (data not shown).
FIG. 1.
Microscopic view of basidia and basidiospores from the cross between Bt63(gen19) and H99. Scale bars 100 μm (A) and 5 μm (B).
Flow cytometry was used to determine the relative DNA contents and ploidy of the 14 MATa isolates as well as representative MATα strains from Botswana. Twelve MATa isolates and most MATα isolates were haploid. Two MATa isolates and three MATα isolates contained slightly larger amounts of DNA and thus might be aneuploid (data not shown).
Sexual reproduction of African isolates produces recombinant progeny.
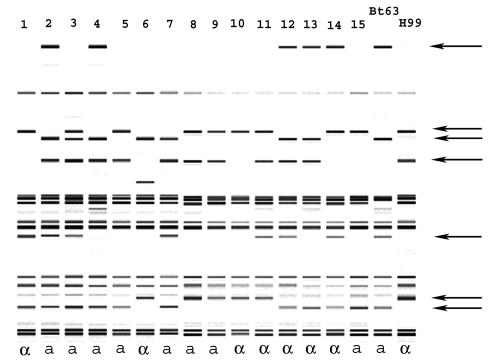
To confirm fertility, we crossed a Botswanan serotype A MATa isolate [Bt63(gen19)] with reference laboratory strain H99 (serotype A MATα), isolated single basidiospores by micromanipulation, and cultured them on fresh medium, where they germinated to produce individual progeny colonies. Subsequent analyses of 15 of these progeny colonies by PCR with mating type-specific primers (see above) and mating assays revealed a 1:1 segregation of the mating type alleles (χ2 = 0.067, P > 0.5) (Fig. 2). We also observed segregation of AFLP markers among the progeny; the parental and nonparental genotypes of the progeny are illustrated in Fig. 2. Five of seven additional crosses produced similar results (data not shown). The other two crosses, involving MATa isolates Bt131 and Bt142, produced progeny with AFLP genotypes that were identical to the parental H99 genotype (data not shown), which may have resulted from insufficient sampling or the induction of haploid fruiting by the proximity of H99 to these MATa isolates (49).
FIG. 2.
Computer-generated AFLP band patterns of recombinant progeny (lanes 1 to 15) from a cross between MATα tester strain H99 and MATa strain Bt63(gen19) from Botswana. Polymorphic bands are indicated by arrows. The MAT alleles of the progeny and the parents are shown below the lanes.
AFLP and MDS analyses reveal two distinct subpopulations.
AFLP genotypes were used to analyze the population structure and diversity of the clinical isolates of serotype A C. neoformans from Botswana. By use of 29 polymorphic markers, 34 AFLP genotypes that differed by two or more bands were identified (see Table 1 in the supplemental material). Twelve AFLP genotypes were unique, and 22 were represented more than once. The 14 MATa isolates consisted of eight different AFLP genotypes. Each of these 14 MATa isolates had identical MATa and/or MATα clones.
TABLE 1.
Statistical tests of associations among loci in the total population, the groups, and the subgroupsa
| Population | No. of:
|
Length of most-parsimonious tree for:
|
IA (P) | rd (P) | ||
|---|---|---|---|---|---|---|
| Isolates | Genotypes | Observed data (P) | Randomized data | |||
| Total | 139 | 34 | 87 (0.01) | 141 | 3.8 (<0.01) | 0.15 (<0.01) |
| Total, clone corrected | 2.4 (<0.01) | 0.09 (<0.01) | ||||
| Group I | 58 | 23 | 36 (0.01) | 50 | 0.84 (<0.01) | 0.05 (<0.01) |
| Group I, clone corrected | 0.3 (0.11) | 0.02 (0.11) | ||||
| Subgroup IA | 44 | 17 | 26 (0.32) | 27 | 0.4 (0.01) | 0.3 (0.01) |
| Subgroup IA, clone corrected | −0.23 (0.92) | −0.18 (0.92) | ||||
| Subgroup IB | 14 | 6 | 8 (1) | 8 | 2.4 (<0.01) | 0.2 (<0.01) |
| Subgroup IB, clone corrected | 0.15 (0.4) | 0.02 (0.4) | ||||
| Group II | 79 | 9 | 13 (0.44) | 14 | 0.37 (<0.01) | 0.05 (<0.01) |
| Group II, clone corrected | 0.12 (0.25) | 0.013 (0.25) | ||||
Results for samples for which a null hypothesis of recombination cannot be rejected are shown in bold type.
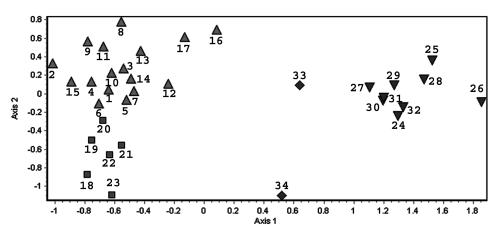
Nonmetric MDS provided a representation of the genetic relationships among the isolates (Fig. 3). MDS estimates the similarity relationships among isolates in Euclidian space, does not imply phylogenetic relationships among isolates, and has an advantage over clustering methods because genetically intermediate isolates are plotted as spatially intermediate (19). As shown in Fig. 3, the isolates grouped into two well-separated clusters, group I and group II. Group I could be subdivided into two smaller clusters, subgroup IA and subgroup IB. Two isolates with unique genotypes (gen33 and gen34) contained AFLP patterns characteristic of both major groups and may have arisen from recombination between groups I and II (Fig. 3); fluorescence-activated cell sorting analysis confirmed that these isolates were haploid. The same three clusters (IA, IB, and II) were also confirmed by two additional statistical treatments: the Nei-Li measure of genetic distance, which was designed to analyze data based on restriction fragment length polymorphism markers (32), and the neighbor-joining or unweighted pair-group method with arithmetic averages (UPGMA) clustering algorithm (39) (data not shown). All but one of the MATa isolates (gen24) was associated with group I, which was also characterized by a larger number of unique genotypes and greater diversity (Fig. 3 and 4).
FIG. 3.
Genetic diversity among isolates based on 29 polymorphic AFLP markers visualized by nonmetric MDS distances among genotypes. Symbols: ▴, subgroup IA; ▪, subgroup IB; ▾, group II; ♦, putative hybrid genotypes. Numbers above symbols identify each unique AFLP genotype (n = 34).
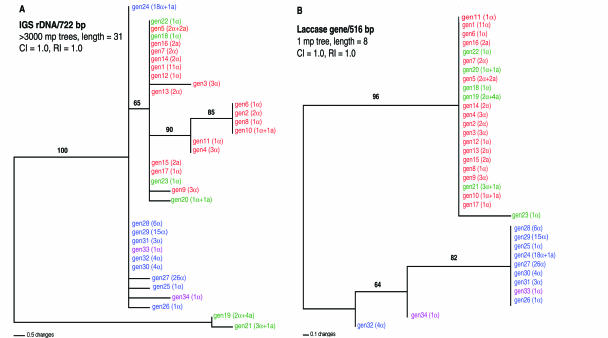
FIG. 4.
Most-parsimonious (mp) trees for 34 genotypes of C. neoformans based on sequence polymorphisms in the IGS region (A) and the LAC1 gene (B). CI, consistency index; RI, retention index. Numbers above each branch are bootstrap values (>50%) based on 500 replications. For the IGS tree, branches with >50% bootstrap support were also strict consensus branches. Notations in parentheses after each genotype indicate the number of isolates with the designated mating type. The three subgroups and putative hybrid genotypes identified by AFLP analyses (Fig. 3) are color coded: subgroup IA, red; subgroup IB, green; group II, blue; and putative hybrids, purple. rDNA, ribosomal DNA.
Multigene sequence analysis confirms two distinct subpopulations.
Our AFLP analysis delineated two major groups. As an independent assessment of the validity of these groups, we compared portions of two well-characterized nucleotide sequences of C. neoformans: the IGS region of ribosomal DNA (13) and the laccase (LAC1) gene (52). Both nucleotide sequences were obtained from a representative of each of the 34 AFLP genotypes. For the IGS region, 722 nucleotides could be aligned, and 20 were phylogenetically informative. For the LAC1 gene, 516 nucleotides could be aligned, and 7 were phylogenetically informative. The genealogy of each region was inferred by maximum-parsimony analysis (39), and the resulting trees are depicted in Fig. 4.
A single most-parsimonious tree was obtained for the LAC1 gene sequences. This tree comprises two well-supported clusters that accurately correspond to groups I and II, identified by AFLP analysis (Fig. 3 and 4B). The LAC1 sequence of one of the putative hybrid AFLP genotypes, gen33, was indistinguishable from those of the other group II genotypes, whereas the LAC1 sequence of the other putative hybrid AFLP genotype, gen34, was not associated with either major clade (Fig. 4B).
Among the 34 AFLP genotypes, the IGS region was significantly more divergent. More than 3,000 most-parsimonious trees were obtained. Nevertheless, the two major groups identified by both AFLP and LAC1 sequence analyses were clearly recognizable. With the exception of two isolates, isolates in AFLP genotypic groups I and II formed two distinct clades, and their separation was supported by a bootstrap value of 65% (Fig. 4A). The exceptions, isolates with genotypes gen19 and gen21, were associated with group I according to AFLP and LAC1 sequence analyses but were separate in the IGS region phylogeny (Fig. 3 and 4). Both of these genotypes include isolates with the MATa allele (Fig. 4A). Both putative hybrid AFLP genotypes (gen33 and gen34) clustered with group II (Fig. 4). Additional genealogical analyses of the IGS region and LAC1 gene by other methods (i.e., neighbor-joining, UPGMA, and maximum-likelihood methods) produced the same phylogenetic patterns as the maximum-parsimony method (data not shown).
Evidence of recombination within African populations.
Statistical analyses of linkage disequilibrium of the AFLP loci provided evidence for recombination within populations (8, 9, 28, 50). Separate analyses were performed on all 139 isolates (including redundant genotypes), as well as the clone-corrected sample, which included only one representative of each AFLP genotype. IA and rd were estimated for the entire sample and also for each group circumscribed by the MDS analysis (Fig. 3) (8, 9). The two hybrid genotypes, gen33 and gen34, were excluded from the subgroup analyses. Artificially recombined data sets were used to determine the statistical values of the test (9).
When all of the isolates were included in the analysis, the IA and rd values were significantly higher then zero (P < 0.01) for the entire data set as well as for the individual clusters, IA, IB, and II. IA and rd values that were significantly different from zero suggested that recombination is rare or absent (9, 28). For the clone-corrected data sets, the IA and rd values were still significantly higher than zero for the entire sample (P < 0.01), but both values were nearly zero when calculated independently for both group I and group II (P = 0.11 and P = 0.2, respectively) (Table 1). The results were even more statistically significant when calculated independently for both subgroup IA and subgroup IB (P = 0.95 and P = 0.4, respectively) (Table 1). Therefore, the null hypothesis of recombination was not rejected for the individual subgroups (28).
Nonrandom associations among alleles in the population was also evaluated by phylogenetic methods (41). The Dollo parsimony model does not allow parallel or convergent gains of the derived conditions and is appropriate for the estimation of phylogenetic relationships between closely related species or populations based on restriction site data (15, 39). When all 34 representatives of each multilocus AFLP genotype were included in this analysis, more then 3,000 most-parsimonious trees were found. A strict consensus tree generated from these trees was almost completely unresolved (data not shown); however, the observed length of this tree was significantly shorter than the length of the tree derived from the randomized data (P = 0.01) (Table 1). The parsimony tree length test with Dollo parsimony criteria was used to evaluate the associations among loci in a sample (2, 9). When genotypes from the three clusters (IA, IB, and II) were analyzed independently, the observed lengths of the trees were not significantly different from the lengths of the trees obtained from the randomized data (P = 0.32, P = 1, and P = 0.44 for subgroup IA, subgroup IB, and group II, respectively) (Table 1). This finding indicates that the null hypothesis of recombination cannot be rejected for any of the three clusters. When all of the isolates from group I were analyzed simultaneously, the length of the tree was significantly shorter than the length of the tree obtained from the randomized data (Table 1). This finding indicates stronger associations among alleles in subgroup IA and subgroup IB. Similar results were obtained when the parsimony tree length test with Wagner parsimony criteria was used (data not shown).
DISCUSSION
The population structure of a sample of clinical isolates of C. neoformans serotype A from AIDS patients in Botswana was determined, and the results support hypotheses for both clonal expansion and recombination in this population. Clonal reproduction was previously recognized in C. neoformans, as strains with identical genotypes were isolated from the environment and infected humans (5, 7, 16). The overrepresentation in the population of certain genotypes is a common feature of clonal structure (44). In the sample analyzed here, five genotypes comprised 45% of the total number of isolates (Fig. 4). Another indication of clonality is the calculation of considerable linkage disequilibrium (or nonrandom association) among the loci in the population. The IA and the rd are calculated estimates of linkage disequilibrium (or nonrandom association) among the loci; if there is no association between the loci, these values approach zero, whereas these values are much higher in clonal populations. When all of the isolates, including those with redundant genotypes, were included in the analyses, the IA and rd values of the entire sample as well as those of the individual subgroups were significantly higher than zero, consistent with clonality (28, 41, 42, 44). The existence of identical or nearly identical genotypes also might be explained by inbreeding within isolated populations (28). Indeed, the presence of MATa and MATα isolates with the same AFLP genotypes could reflect inbreeding. The possibility of inbreeding in subgroup IB is consistent with the large number of MATa and MATα isolates with identical genotypes and the presence of genetically distinct isolates (Fig. 3).
This investigation also provides evidence for recombination among the isolates in the population. The population contained isolates of both mating types that produced recombinant progeny in laboratory crosses. Nevertheless, MATa isolates represented only 10% of all of the isolates, a finding which would be unlikely for a population with substantial sexual reproduction (41). Several hypotheses could account for the unequal numbers of MATa and MATα isolates. Sexual reproduction might not occur in nature at the present time; that is, extant MATa isolates might reflect a history of sexual recombination (43). Alternatively, the biased ratio of MATa and MATα isolates could be attributed to selective pressure favoring the propagation of MATα isolates. Selection might not be strong enough to completely eliminate all MATa isolates from the population but might be sufficiently powerful to produce a biased distribution of MATα alleles (34). For example, the MATα allele has been linked to enhanced virulence of C. neoformans serotype D strains (24), and MATα strains therefore may have a selective advantage in the host. However, recently constructed congenic serotype A MATa and MATα strains were equally virulent in mice (33). Other factors, such as haploid fruiting of MATα strains (49), might contribute to the preponderance of this mating type. Environmental samples from sub-Saharan Africa should be analyzed to test this hypothesis. Last, the observed mating type bias might be artifactual: although MATa isolates represented only 10% of all of the isolates, they amounted to 36% of the isolates of subgroup IB; the latter value is not statistically different from a balanced segregation of mating types.
Analysis of the AFLP and recombination data divided this clinical sample of serotype A isolates into three partially distinct genetic clusters (IA, IB, and II). Sequence analysis of portions of the IGS region and the LAC1 gene supported only the two major groups (I and II); however, this apparent failure to discern subgroups IA and IB is not surprising, as AFLP genotypes are more discriminatory and thus more likely to reveal recent changes in the population structure (35, 42). The existence of genetically distinct subgroups within serotype A has been reported elsewhere (4, 29). The mechanisms that promote genetic isolation of these groups are not well understood. Since strains from our clusters could successfully mate in the laboratory, factors other than reproductive barriers, such as ecological or geographical separation, might have contributed to their isolation.
When one representative of each genotype (i.e., clone-corrected data sets) was used to calculate the IA and rd values for subgroups IA and IB and group II, both values were not significantly different from zero (Table 1). Therefore, the null hypothesis of recombination cannot be rejected for any subgroup, suggesting that recombination might have occurred between members of the same lineages but not between isolates of different, more distant groups (9, 28). The analysis of clone-corrected genotypes is appropriate because asexual reproduction is preponderant (10, 50). Therefore, the presence of multiple clones in the same environment is inevitable and, to detect occasional recombinational events between individuals in the population, clone-corrected samples are required (28).
Both population genetic and phylogenetic approaches detected no linkage disequilibrium among the alleles of strains within the genetic subdivisions of our sample (Table 1). The data support the hypothesis that recombination may occur in genetically isolated subpopulations of serotype A C. neoformans. Members of these separate populations also may come into contact to produce hybrid strains, such as gen33 or gen34 strains (Fig. 3). Since only clinical isolates were examined, we cannot determine their origins. Analysis of environmental isolates of C. neoformans from Botswana will clarify the impact of geographical and other ecological factors on the formation of subpopulations.
There are several, nonexclusive explanations for the inordinate prevalence of cryptococcosis in Africa. For cultural, financial, and public health reasons, it is likely that the diagnosis is not timely and treatment is less available. In addition, AIDS patients in Africa may be more susceptible or more exposed to C. neoformans than AIDS patients in other parts of the world. Another possibility is that strains of C. neoformans in Africa are more virulent. Our findings demonstrate that clinical isolates of serotype A from patients in Botswana are genetically more diverse, separable into genetic subdivisions, and capable of sexual reproduction and recombination. The population structure and discovery of a high proportion of MATa isolates imply sexual reproduction and recombination as a mechanism for generating diversity and possibly the emergence of strains with enhanced virulence. These results also suggest that similar mechanisms to generate diversity may function in other human pathogenic fungi capable of sexual reproduction, such as Candida albicans, Histoplasma capsulatum, and Blastomyces dermatitidis.
Supplementary Material
Acknowledgments
We thank L. Barth Reller for the Botswanan isolates, Lori Kestenbaum for technical assistance with processing the strains, and Dmitri Kazmin, Leonid Dzantiev, and Joan Henson for critically reading the manuscript.
This investigation was supported by the following grants from the National Institutes of Health: AI44975, AI25783, and AI50113. Joseph Heitman is a Burroughs Wellcome Scholar in molecular pathogenic mycology and an Investigator of the Howard Hughes Medical Institute.
Footnotes
The supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Agapow, P.-M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1:101-102. [Google Scholar]
- 2.Archie, J. W. 1989. A randomization test for phylogenetic information in systematic data. Syst. Zool. 38:239-252. [Google Scholar]
- 3.Benham, J., J.-U. Jeung, M. Jasieniuk, V. Kanazin, and T. Blake. 1 May 1999, posting date. Genographer: a graphical tool for automated fluorescent AFLP and microsatellite analysis. J. Agric. Genomics 4:15-19. [Online.] http://www.ncgr.org/jag. [Google Scholar]
- 4.Boekhout, T., B. Theelen, M. R. Diaz, J. W. Fell, W. C. Hop, E. C. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. [DOI] [PubMed] [Google Scholar]
- 5.Boekhout, T., A. van Belkum, A. C. A. P. Leenders, H. A. Verbrugh, P. Mukamurangwa, D. Swinne, and W. A. Scheffers. 1997. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int. J. Syst. Bacteriol. 47:432-442. [DOI] [PubMed] [Google Scholar]
- 6.Bogaerts, J., D. Rouvroy, H. Taelman, A. Kagame, M. A. Aziz, D. Swinne, and J. Verhaegen. 1999. AIDS-associated cryptococcal meningitis in Rwanda (1983-1992): epidemiologic and diagnostic features. J. Infect. 39:32-37. [DOI] [PubMed] [Google Scholar]
- 7.Brandt, M. E., L. C. Hutwagner, R. J. Kuykendall, R. W. Pinner, and Cryptococcal Disease Active Surveillance Group. 1995. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. J. Clin. Microbiol. 33:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, A. H. D., M. W. Feldman, and E. Nevo. 1980. Multilocus structure of natural populations of Hordeum spontaneum. Genetics 96:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burt, A., V. Koufopanou, and J. W. Taylor. 2000. Population genetics of human pathogenic fungi, p. 229-244. In R. C. A. Thompson (ed.), Molecular epidemiology of infectious diseases. Arnold Publishers, London, England.
- 10.Casadevall, A., and J. R. Perfect. 1999. Cryptococcus neoformans, p. 1-541. ASM Press, Washington, D.C.
- 11.Cogliati, M., M. C. Esposto, D. L. Clarke, B. L. Wickes, and M. A. Viviani. 2001. Origin of Cryptococcus neoformans var. neoformans diploid strains. J. Clin. Microbiol. 39:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz, M. R., T. Boekhout, B. Theelen, and J. W. Fell. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Syst. Appl. Microbiol. 23:535-545. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, M. R., and J. W. Fell. 2000. Molecular analyses of the IGS and ITS regions of rDNA of the psychrophilic yeasts in the genus Mrakia. Antonie Leeuwenhoek 77:7-12. [DOI] [PubMed] [Google Scholar]
- 14.Farris, J. S. 1977. Phylogenetic analysis under Dollo's law. Syst. Zool. 26:77-88. [Google Scholar]
- 15.Felsenstein, J. 2003. Inferring phylogenies, p. 230-231. Sinauer Associates, Inc., Sunderland, Mass.
- 16.Franzot, S. P., J. S. Hamdan, B. P. Currie, and A. Casadevall. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakim, J. G., I. T. Gangaidzo, R. S. Heyderman, J. Mielke, E. Mushangi, A. Taziwa, V. J. Robertson, P. Musvaire, and P. R. Mason. 2000. Impact of HIV infection on meningitis in Harare, Zimbabwe: a prospective study of 406 predominantly adult patients. AIDS 14:1401-1407. [DOI] [PubMed] [Google Scholar]
- 18.Halliday, C. L., and D. A. Carter. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hebert, P. D. N., A. Cywinska, S. L. Ball, and J. R. deWaard. 2002. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 270:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller, S. M., M. A. Viviani, M. C. Esposto, M. Cogliati, and B. L. Wickes. 2003. Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate. Microbiology 149:131-142. [DOI] [PubMed] [Google Scholar]
- 21.Klugman, K. P. 2003. The role of clonality in the global spread of fluoroquinolone-resistant bacteria. Clin. Infect. Dis. 36:783-785. [DOI] [PubMed] [Google Scholar]
- 22.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology, p. 397-446. Lea & Febiger, Philadelphia, Pa.
- 24.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lengeler, K. B., G. M. Cox, and J. Heitman. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating type locus. Infect. Immun. 69:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengeler, K. B., P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97:14455-14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maynard Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, W., A. Castañeda, S. Jackson, M. Huynh, E. Castañeda, and IberoAmerican Cryptococcal Study Group. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michod, R. E. 1995. Eros and evolution: a natural history philosophy of sex. Addison-Wesley Publishing Company, Reading, Mass.
- 31.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. The sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oudemans, P., H. M. Alexander, J. Antonovics, S. Altizer, P. H. Thrall, and L. Rose. 1998. The distribution of mating type bias in natural populations of the anther smut Ustilago violacea on Silene alba in Virginia. Mycologia 90:372-381. [Google Scholar]
- 35.Soll, D. R. 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13:332-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stefani, S., and A. Agodi. 2000. Molecular epidemiology of antibiotic resistance. Int. J. Antimicrob. Agents 13:143-153. [DOI] [PubMed] [Google Scholar]
- 37.Sukroongreung, S., K. Kitiniyom, C. Nilakul, and S. Tantimavanich. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36:419-424. [PubMed] [Google Scholar]
- 38.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0b. Sinauer Associates, Inc., Sunderland, Mass.
- 39.Swofford, D. L., G. J. Olsen, P. J. Waddell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics. Sinauer Associates, Inc., Sunderland, Mass.
- 40.Tanaka, R., H. Taguchi, K. Takeo, M. Miyaji, and K. Nishimura. 1996. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J. Med. Vet. Mycol. 34:299-301. [PubMed] [Google Scholar]
- 41.Taylor, J. W., D. M. Geiser, A. Burt, and V. Koufopanou. 1999. The evolutionary biology and population genetics underlying fungal strain typing. Clin. Microbiol. Rev. 12:126-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor, J. W., D. J. Jacobson, and M. C. Fisher. 1999. The evolution of asexual fungi: reproduction, speciation and classification. Annu. Rev. Phytopathol. 37:197-246. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, J. W., D. J. Jacobson, S. Kroken, T. Kasuga, D. M. Geiser, D. S. Hibbett, and M. C. Fisher. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21-32. [DOI] [PubMed] [Google Scholar]
- 44.Tibayrenc, M., F. Kjellberg, J. Arnaud, B. Oury, S. F. Brenière, M. L. Dardé, and F. J. Ayala. 1991. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc. Natl. Acad. Sci. USA 88:5129-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viviani, M. A., M. C. Esposto, M. Cogliati, M. T. Montagna, and B. L. Wickes. 2001. Isolation of a Cryptococcus neoformans serotype A MATa strain from the Italian environment. Med. Mycol. 39:383-386. [DOI] [PubMed] [Google Scholar]
- 46.Viviani, M. A., R. Nikolova, M. C. Esposto, G. Prinz, and M. Cogliati. 2003. First European case of serotype A MATa Cryptococcus neoformans infection. Emerg. Infect. Dis. 9:1179-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Freijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, P., C. B. Nichols, K. B. Lengeler, M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2002. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot. Cell 1:257-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, J., and T. G. Mitchell. 2002. Strain variation and clonality in Candida spp. and Cryptococcus neoformans, p. 739-749. In R. A. Calderone and R. L. Cihlar (ed.), Fungal pathogenesis: principles and clinical applications. Marcel Dekker, Inc., New York, N.Y.
- 51.Xu, J., and T. G. Mitchell. 2003. Comparative gene genealogical analyses of strains of serotype AD identify recombination in populations of serotypes A and D in the human pathogenic yeast Cryptococcus neoformans. Microbiology 149:2147-2154. [DOI] [PubMed] [Google Scholar]
- 52.Xu, J., R. J. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1482. [DOI] [PubMed] [Google Scholar]
- 53.Yan, Z., X. Li, and J. Xu. 2002. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 40:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.