Abstract
Glycosyl phosphatidylinositol (GPI)-anchored proteins are peripheral membrane proteins tethered to the cell through a lipid anchor. GPI-anchored proteins serve many functions in cellular physiology and cell signaling. The PIG-A gene codes for one of the enzymes of a complex that catalyzes the first step in anchor synthesis, and we have cloned the Paramecium tetraurelia pPIG-A gene using homology PCR. To understand the function of pPIG-A and the significance of GPI-anchored proteins in Paramecium, we reduced the mRNA for pPIG-A in transformed cells using an expression vector that transcribed antisense mRNA. The amount of transcript is reduced to ∼0.3% of the mRNA in control-transformed cells. Compared to control cells, cells transformed with the antisense pPIG-A vector show reduced synthesis of GPI anchor intermediates catalyzed in their endoplasmic reticula and a very few GPI-anchored proteins among the peripheral proteins that can be recovered from their surfaces. They also show specific defects in chemoresponse to glutamate and folate. Other cellular functions, such as growth and mating, seem to be normal.
There has been a recent explosion of interest in the role of lipids in signal transduction pathways and in a set of peripheral proteins, glycosyl phosphatidylinositol (GPI)-anchored proteins, that concentrate in special surface membrane lipid domains. Lipids modify signaling proteins, such as ras and src, which are targeted to the surface membrane. However, now more than ever we appreciate that lipids serve many more functions than modification. For example, lipid rafts are surface membrane microdomains characterized by high cholesterol and sphingolipids and by the congregation of many signaling proteins (39, 45). The study of lipid rafts, sometimes found as specializations for potocytosis called caveolae, is accelerating, and our appreciation of the role of these microdomains in cell function is similarly growing, especially in the realm of signal transduction (16, 36). Signaling molecules, such as tyrosine kinase receptors, kinases of the src class, small and trimeric G proteins, G protein-coupled receptors, and GPI-anchored proteins, concentrate in lipid rafts (3, 16). The role of the rafts in modulating signal transduction by these pathway components remains a matter of debate and study, but it is becoming clear that rafts play a role in organizing and regulating signaling molecules.
We have become interested in a class of surface proteins, the GPI-anchored proteins, that localize to lipid rafts in many cell types. These are peripheral proteins that adhere to the cell outer surface by virtue of a phosphatidyl moiety that embeds in the outer leaflet of the cell membrane. The phosphatidylinositol (PI), in turn, is linked to the protein at its C terminus through a glycan-carbohydrate complex that can differ with the cell type (11). A common aspect of GPI anchors is that they can be cleaved by PI-specific phospholipase C (PI-PLC) or by GPI-specific phospholipase D (PLD) (see reference 20 for a review). GPI-anchored proteins function to transport folate, activate T cells, inhibit plasminogen-activated cell death, guide axons, communicate between cells, and provide variety in surface markers to evade host defenses, among other functions (26). In particular, the GPI-anchored folate receptor has served as a convenient marker for lipid rafts and detergent-insoluble microdomains (46). The study of GPI-anchored proteins is intimately connected to the study of lipid rafts, because signal transduction through cholesterol depletion and the consequent disruption of lipid rafts also disrupts GPI-anchored protein function (15).
In Paramecium cells, many surface proteins are GPI-anchored proteins (4). These proteins include large surface glycoproteins that change as the cells' environment does, with temperature or pH, for example (13, 30, 32). However, there are many Paramecium tetraurelia and Paramecium primaurelia proteins of much smaller molecular mass that are also GPI anchored (4, 31). The core of the P. primaurelia GPI anchor, and presumably of the P. tetraurelia anchor, consists of ethanolamine phosphate, three mannoses, and glycosamine (GlcN)-PI, specifically, GlcN-inositol-phosphoceramide. Much is known about the P. primaurelia pathway for GPI synthesis and attachment to proteins (1, 2), and everything that we have learned about the P. tetraurelia pathway suggests that it is the same and also very much like the pathways in mammals and parasitic protozoa (31; J. Van Houten and J. Yano, unpublished observations).
Genes for all of the enzymes for GPI synthesis have been cloned from mammals, yeast, or parasitic protozoa, and most of them are highly conserved (38). A genetic approach to the study of these enzymes has led to the creation of knockout mice that are missing an enzyme for GPI anchor synthesis, but these animals generally die as embryos (23). Tissue culture cells that have mutated versions of the genes for one or more of the GPI anchor synthesis steps are viable but are not adequate for all purposes in the study of these proteins in signal transduction (38). Therefore, we attempted to develop strains of transformed P. tetraurelia that had little or no activity of crucial enzymes for GPI anchor synthesis in order to determine whether the GPI-anchored proteins of the Paramecium surface functioned in chemoresponse, among other important cell functions. We describe here the cloning of a gene, Paramecium tetraurelia pPIG-A, for an enzyme of GPI anchor synthesis and the results of its down-regulation through antisense techniques. The PIG-A protein, named for the PI glycan complementation group A, participates with two other enzymes in the catalysis of the first step of anchor synthesis (25, 38).
MATERIALS AND METHODS
Cells and cultures.
Strain 51-s of P. tetraurelia was cultured as described previously (34).
Genomic DNA.
Genomic DNA was prepared by the protocol of Forney et al. (13).
The PCR primers, with their corresponding cDNA nucleotide numbers, were as follows (where W = A + T, Y = C + T, V = A/C/G, B = C/G/T, and D = A/G/T): F, AGAACWGTBTTYACWGAYCA (349 to 368); R, AACTTCGGDATWCCWCCVAC (929 to 910); F1, TCCTTGTTTGCATTCAATGAT (370 to 390); F2, CTGTAATTCCAAATGCAGTT (518 to 537); F3, TTGCATAGCAATTGTTGAGGC (855 to 875); F4, AACAGGATATAATAAAGTTGATA (untranslated 5′ end; −40 to −20); R1, ATTAGTGGAAAGAACACACAA (909 to 889); R2, CAATTCAGTTTAATTTTATAGGT (771 to 749); R3, TGAATGATCAATTTCACACAA (450 to 530); and R4, ACTCTGAATTTATTATATCTA (extreme 3′ end; 1446 to 1426).
Genomic PCR.
The two conservative amino acid sequences, Q/RTVFTDH for the forward 5′ primer and VGGIPEV for the reverse 3′ primer, were selected from the human PIG-A gene (hPIG-A) (27) and the yeast GPI3 (12). The degenerative primers (F and R) were designed according to the P. tetraurelia codon usage frequency for genes sequenced in surface antigen A (37), calmodulin (22), plasma membrane Ca2+-ATPase (10), β-tubulin (9), protein kinase A regulatory subunit (7), and dihydrofolate reductase and thymidylate synthase (35). The genomic sequence was amplified under the following conditions: 94°C for 1 min, 35°C for 2 min, and 72°C for 2 min for 30 cycles and final extension at 72°C. The PCR products were ligated to the TOPO cloning vector (Invitrogen) for DNA sequencing. For obtaining the full length of the PIG-A coding sequence in the macronuclear DNA, the primers F4 and R4 were used.
Total RNA.
Transformed cells were harvested by centrifugation from log-phase 100-ml cell cultures grown in wheat grass medium inoculated with bacteria and 100 μg of paromomycin/ml. The cell pellet was washed twice with Dryl's solution (1 mM Na2HPO4, 1 mM NaH2PO4, 1.5 mM CaCl2, 2 mM Na-citrate, pH 7.2). The total RNA was prepared by following the manufacturer's directions for the total RNA isolation system from Promega (Madison, Wis.).
RT-PCR for RACE.
The total RNA was treated with DNase I (Bio-Rad Laboratories [BRL]) to remove the genomic DNA. Using the oligo(dT)12-18 primer (BRL) for 3′ rapid amplification of cDNA ends (RACE) or the R1 primer for 5′ RACE, the first-strand cDNA was synthesized by reverse transcriptase (SuperScript RT; BRL), following the protocol from the manufacturer. In the reactions for cDNA synthesis, an RNase inhibitor was included (Rnasin; Promega).
In the first round of PCR of the 3′ RACE, the oligo(dT)13 primer with an added XhoI sequence and the F1 primer were used. In the second round of PCR, the primers F2 or F3 and oligo(dT)13 with an added XhoI sequence were used.
For 5′ RACE, the first-strand cDNA was purified using glass milk (Geneclean II kit; BIO 101, Inc.) after being boiled for 5 min and was then treated with terminal deoxynucleotidyl transferase (Gibco Invitrogen Technologies) to add a poly(A) tail. In the first round of PCR, the primers oligo(dT)13 with an XhoI sequence and R2 were used, and in the second round, oligo(dT)13 with an XhoI sequence and R3 were used. A single band of the expected size was obtained from the second PCR and ligated to the TOPO vector.
Sequencing and analysis.
The plasmid DNA was purified with the Wizard Plasmid Miniprep DNA purification system (Promega). DNA sequencing was carried out by the Nucleic Acid Analysis facility of the Vermont Cancer Center. The DNA sequences were analyzed using the Wisconsin Package program (Genomics Computer Group), MacVector, and National Center for Biotechnology Information BLAST.
Expression plasmids.
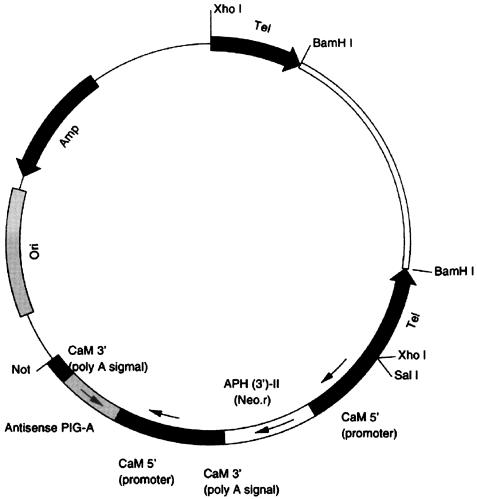
The expression vectors, pAntiPIG-A1 and -A2 (Fig. 1) and pNeo, were engineered from parts of the pPXV-Neo plasmid, provided by J. Haynes. The antisense-oriented sequence (anti-PIGA-1 or -2) of a part of PIG-A (−34 to +82 or +294 to +456) and the aminoglycoside 3′-phosphotransferase-II (neo) gene were each downstream from P. tetraurelia calmodulin promoters (kindly provided by J. Haynes) (21). The plasmid was cut with BamHI to produce a linear plasmid with Tetrahymena telomeres at both ends for microinjection.
FIG. 1.
Expression vector pAnti-PIG-A pNeotel (9.2 kb).
Microinjection and transformation.
BamHI-digested plasmid was suspended in water, and ∼10 μl of 1- to 3-μg/μl DNA was injected into the macronucleus of each cell. The injected cells were isolated in a bacteria culture medium containing 100 μg of paromomycin (Sigma)/ml. The paromomycin-resistant cells were grown as a clone for ∼1 week. The cells were maintained by continual transfer to fresh culture fluid in stock cultures, while some were grown in larger volume, e.g., 200 ml of culture medium containing 100 μg of paromomycin/ml. The cell cultures were grown at 22°C. The results shown here are from two separate transformations and were consistent with results from other transformations.
Immobilization tests.
For a quick check on the reduction of GPI-anchored proteins, cells were incubated in Dryl's solution containing both anti-A and -B sera (1/500 dilution; antisera kindly provided by J. Forney), because we had previously ascertained that the cells expressed primarily serotype B, and sometimes A, at 22°C. These serotypes refer to the large GPI-anchored glycoproteins mentioned in the introduction (13). The control cells were immobilized within 15 min in the antisera. In the putative antisense-transformed cell lines, the cells remained mobile for 20 to 30 min, the duration of the tests. For Western blot analysis and GPI biosynthesis assay, cells from the motile cell lines were cultured in 200 ml of culture medium containing 100 μg of paromomycin/ml for further testing.
Genomic PCR and RT-PCR.
For checking the relative amount of transfected plasmid or the reduction of PIG-A mRNA in the putative antisense-transformed cells, genomic DNA and total RNA were prepared from 100 cells, following the protocols described above. For genomic PCR, the primers F2 and R1 were used to detect the macronuclear pPIG-A gene, and the primers R2 and T7 (in the plasmid) were used to detect the antisense pPIG-A sequence of the plasmid. In semiquantitative RT-PCR, first-strand cDNA was diluted 101-, 102-, and 103-fold in distilled H2O. One microliter of the diluted cDNA was used as a template in 50 μl of reaction mixture, and the primers F2 and R1 were used for detecting pPIG-A cDNA. Calmodulin mRNA was assayed as a control.
Real-time RT-PCR.
RNA and first-strand cDNA were prepared as described above and analyzed using primers from Biosearch Technologies Inc. and TaqMan technology at the Vermont Cancer Center DNA Analysis Facility. Calmodulin transcripts were amplified as a control.
Real-time RT-PCR primers.
The real-time RT-PCR primers were as follows: for pPIG-A (GenBank no. AF267484; numbering starts from the open reading frame), pPIG-A TaqMan Probe (6-FAM TAAATACATCCTTGACGGAGGCCTTTTGC BHQ-1 [829 to 858 bp]), pPIG-A forward (CAGGTCATTAACTTAAAGATGTACTCAATAGA [785 to 806 bp]), and pPIG-A reverse (TGCAGCCTCAACAATTGCTATG [879 to 858 bp]); for Paramecium calmodulin (GenBank no. S680259; numbering starts from the open reading frame), Paramecium calmodulin (pCAM) TaqMan probe (6-FAM AATCGATTTCCCTGAATTTTATCTTGATGGCTA BHQ-1 [187 to 223 bp]), pCAM forward (TCGATGCTGACGGTAATGGA [168 to 187 bp]), and pCAM reverse (TTCTTCCTCAGAATCTTATTCCTTCAT [253 to 229 bp]).
Western blotting.
Cells (2 × 105) were collected by centrifugation and resuspended in 400 μl of bacterium-free culture medium; 240 μl of salt-ethanol solution (10 mM Na2HPO4, 150 mM NaCl, 30% ethanol) (31) was added to the samples, which were incubated for 1 h on ice. The cells were removed by centrifugation in a microcentrifuge at 6 × 103 rpm (2,940 × g). The supernatants were removed, and the same volume (640 μl) of cold acetone was added to the supernatants for protein precipitation. The samples were kept on ice for 10 min and then centrifuged by microcentrifuge at 14 × 103 rpm (16,000 × g) and 4°C for 10 min. The pellets were washed twice with cold acetone and dried. The pellets were resuspended in 50 μl of H buffer [20 mM Tris-(hydroxylmethyl)amino methane hydrochloride (Tris-HCl) (pH 7.4), 1 mM EDTA, 25 mM KCl, 50 mM sucrose] and then mixed with 2× sodium dodecyl sulfate (SDS) gel loading buffer (2% SDS, 1% β-mertcaptoethanol, 20% glycerol, 0.001% bromphenol blue, 125 mM Tris base, pH 6.8). The samples were boiled for 5 min and then loaded onto 10% SDS polyacrylamide gels, and the SDS gels were electroblotted onto nitrocellulose membranes (31). The membranes were developed with the polyclonal antiserum and anti-rabbit secondary antibody with alkaline phosphatase. Anti-A and -B sera and anti-cross-reactive determinant of the GPI anchor (anti-CRD; Oxford GlycoSystems) were used as primary antibodies. It should be noted that the Anti-A and -B antisera are not affinity purified for the large surface glycoproteins A and B, and the antisera recognize many proteins among the salt-ethanol wash proteins, which serve as the immunogen for antiserum production. We use these antisera to probe not only for the A and B antigens but also for other peripheral surface proteins. The dilutions of antisera are given in the figure legends.
GPI biosynthesis assay.
The cells (1 × 105 to 5 × 105) were collected by centrifugation from the mid-stationary growth phase and treated with 3 to 5 μg of tunicamycin/ml at 22°C for 3 h. The cells were washed with Dryl's solution and then suspended with 250 μl of HI buffer (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 25 mM KCl, 50 mM sucrose, 2 μg of leupeptin/ml, and 0.2 mM TLCK [tosyl-lysylchloromethane; Sigma]). The cells were sonicated three times for 20 s each time. The cellular debris was removed by centrifuging the suspension at 2,000 × g for 5 min at 4°C. The crude endoplasmic reticulum (ER) membranes were pelleted by centrifugation at 19,800 × g for 20 min at 4°C. The pellets were resuspended with HI buffer, rapidly frozen in liquid nitrogen, and kept at −70°C. The crude ER membranes were washed with 50 mM HEPES buffer, pH 7.4, containing 25 mM KCl, 5 mM MnCl2, 5 mM MgCl2, 1 μg of leupeptin/ml, 100 μM TLCK, and 0.1 μg of tunicamycin/ml. The membranes were suspended with 100 μl of the same buffer supplied with 1 mM ATP and 1 μCi of UDP N-acetyl-d-glucosamine-6-3H(N) (NEN Life Science Products) and incubated at room temperature for 1 h. The reaction was stopped by adding 650 μl of chloroform-methanol (1:1 [vol/vol]). The lipid residues were extracted, and the samples were dried under nitrogen gas. The lipid residues were extracted twice with n-butanol- water (1:1 [vol/vol]). The n-butanol phase was evaporated under nitrogen, and the lipid residues were dissolved in chloroform-methanol-water (10:10:3 [vol/vol]) and loaded on silica gel 60 plates (Merck). Chloroform-methanol-water (10:10:3) was used as the solvent system. After the plates were sprayed with En-3Hance, the radiolabeled compounds were detected by fluorography using Kodak XAR-5 film.
The lipids from the n-butanol phase were further incubated with 0.1 U of PI-PLC (Sigma) in 100 μl of 100 mM Tris-HCl (pH 7.4)- 0.2% Triton X-100, or 1 μl of PLD (from mouse serum; a gift from Paula Tracy) in 100 μl of 100 mM Tris-HCl (pH 7.4)- 0.2% Triton X-100- 0.2 mM CaCl2- 10 mM MgCl2 for 2 h at room temperature. After the reactions, the derivatives were extracted with n-butanol-water (1:1 [vol/vol]), and then the organic phase was analyzed by thin-layer chromatography (TLC).
Chemoresponse behavior in T mazes.
T-maze assays were carried out as described previously (44). The data were analyzed by the Mann-Whitney U test.
Nucleotide sequence accession number.
The sequence of the pPIG-A gene was deposited in GenBank under accession no. AF267484.
RESULTS
Cloning the pPIG-A gene.
Using primers designed from conserved regions of the yeast and human PIG-A genes, we cloned a partial sequence from P. tetraurelia genomic DNA. The full-length clone was obtained by RT-PCR using the strategy described in Materials and Methods above, and the sequence is shown in Fig. 2. The full-length cDNA is 1,503 bp, exclusive of the poly(A) tail. The open reading frame encoding the P. tetraurelia PIG-A protein (pPIG-A) extends from bases 56 to 1,384, with a TGA stop codon located immediately upstream from the initiation codon, ATG. pPIG-A codes for a protein of 442 amino acids, and the deduced protein sequence shows 46% identity and 61% similarity to the hPIG-A protein sequence (Fig. 2) and 43% identity and 54% similarity to yeast GPI3 (from the yeast gene Spt14). Moreover, the amino acid sequence between residues 279 and 311 has particularly high identity with the hPIG-A (88%) and yeast GPI3 (78%) protein sequences. This amino acid sequence is also similar to regions of bacterial glycosyltransferases: 65% similarity to both putative glycosyltransferase and mannosyltransferase in Escherichia coli, 67% similarity to galactosyltransferase in Archaeoglobus fulgidus, 72% similarity to glycosyltransferase in Bordetella pertussis, and 69% similarity to putative GlcNAc transferase in Bordetella parapertussis. pPIG-A is shorter than the hPIG-A protein by 31 amino acids in the N terminus. As in the hPIG-A and yeast GPI3 proteins, the putative transmembrane domain of pPIG-A is located near the C terminus, as surmised from Kyte-Doolittle hydrophobicity plots. The genomic DNA sequence corresponding to the full length of PIG-A cDNA has two small introns, which are consistent with the size of P. tetraurelia introns (33).
FIG. 2.
Theoretical protein sequence translated from cDNA sequence of P. tetraurelia gene pPIG-A compared with sequence from hPIG-A. Shading indicates identical residues, and dots indicate gaps.
Antisense pPIG-A transformants.
To test for the function of pPIG-A, we created an expression vector in which part of the pPIG-A sequence (see Materials and Methods) was inserted in an orientation for expression of the antisense mRNA. We intended to use the expression of the antisense mRNA sequence to cause the loss of the endogenous pPIG-A mRNA and reduction of the pPIG-A protein.
About 30% of the cells injected with the pAntiPIG-A2 vector grew in the presence of 100 μg of paromomycin/ml and were tested further for immobilization by anti-A and -B antisera. This was our first screening method to determine whether to follow a cell line further. The A and B immobilization antigens of P. tetraurelia are GPI anchored (31). A test for the presence of these surface proteins has traditionally been the immobilization of swimming cells by antisera raised against proteins in salt-ethanol washes which included these surface proteins. Some cell lines were not immobilized within 20 to 30 min (the duration of the test) in the presence of antisera A and B, while the wild type and the control cells that were transformed with the empty vector were always immobilized within 15 min. From cells that remained motile for 20 to 30 min, we made clonal pAnti-PIG-A-transformed lines and went on to test them further by genomic PCR and RT-PCR. We monitored the cell numbers of the cultures daily and noted that the cell growth of the putative pAnti-PIG-A-transformed lines generally was the same as that of the control-transformed lines.
DNA was prepared from the control- and putative pAnti-PIG-A-transformed cells. If the cells retained the antisense vector gene for PIG-A, PCR using the R2 and R7 primers should result in a band of 500 bp. We noted a correlation between very intense PCR bands and transformed lines showing free swimming for >25 min in the immobilization tests. We also tested some lines for the presence of both the Neor gene and antisense PIG-A in a continuous sequence by using primers based on the Neor sequence and the anti-PIG-A sequence (see Materials and Methods), observing an expected band of 1.7 kb. Thus, the many copies of injected plasmids seem to be intact and maintained in the putative pAnti-PIG-A-transformed cell lines. Control-transformed cells did not show either of these bands when tested by PCR using the same primers.
We used a semiquantitative assay to determine whether there was a reduction in pPIG-A message, primarily as a screening procedure before we investigated the levels of GPI-anchored proteins and the cell behavior of transformed P. tetraurelia clones (see Materials and Methods). Figure 3 shows the results of these amplifications from a representative experiment. (These are typical, not exceptional, results.)
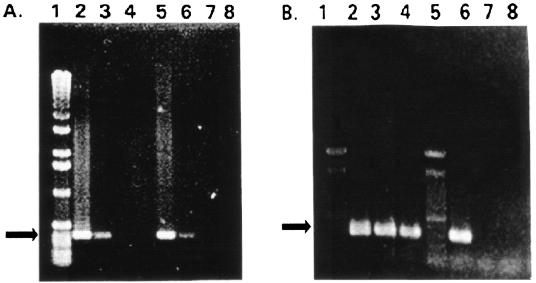
FIG. 3.
RT-PCR screening of PIG-A transcripts from control-transformed and pAnti-PIG-A-transformed cells. (A) Results of calmodulin PCR of dilutions of the cDNA template using calmodulin primers for an expected 400-bp product. The cDNA was diluted before the amplification of calmodulin sequences. Lanes 2, 3, and 4, results from 101-, 102-, and 103-fold dilutions of the cDNA from control-transformed cells; lanes 5, 6, 7, and 8, results from 101-, 102-, 103-, and 104-fold dilutions of pAnti-PIG-A-transformed cells. (B) A 550-bp sequence of the pPIG-A gene was amplified using a series of dilutions of the cDNA templates from the same cDNA as for panel A and an additional pAnti-PIG-A-transformed clone and primers F2 and R1. Lanes 2 and 6, PCR products from control-transformed cell cDNA diluted 10 and 100 times, respectively; lanes 3 and 4 and lanes 7 and 8, PCR products from two different pAnti-PIG-A-transformed lines, using cDNA template diluted 10 (lanes 3 and 4) or 100 (lanes 7 and 8) times. Lanes 1 contain molecular mass markers, and arrows point to expected band size.
Figure 3A shows the results of amplifications of calmodulin sequences by PCR with dilutions of the cDNA as templates and calmodulin primers for an expected 400-bp product. Note that the bands from cDNA templates from control-transformed cells (lanes 2 to 4) appear about the same as the bands from cDNAs from the pAnti-PIG-A cells (lanes 6 to 8). Results where both control and antisense-PIG-A cDNAs diluted 10 and 100 times supported the amplification of calmodulin sequences were very consistent. Therefore, the first strand of calmodulin cDNA seemed to be about equally synthesized from the total RNAs from only 100 cells of the control- and pAnti-PIG-A-transformed cell lines.
To detect whether PIG-A mRNA was reduced in the pAnti-PIG-A-transformed cells, we amplified a 550-bp sequence of the pPIG-A gene using a series of dilutions of the cDNA templates and primers F2 and R1. Figure 3B shows the results of a typical experiment, using the same cells as in Fig. 3A and a second pAnti-PIG-A-transformed clone. Figure 3B shows PIG-A PCR products from cDNA of control-transformed cells diluted 10- and 100-fold (lanes 2 and 6, respectively). Figure 3B also shows PIG-A PCR products from cDNAs from the two different pAnti-PIG-A-transformed lines. Again, the cDNA was diluted 10-fold (lanes 3 and 4) or 100-fold (lanes 7 and 8) before PCR. Note that there was no apparent product of the expected size when the cDNA from pAnti-PIG-A-transformed cells was diluted 100-fold (lanes 7 and 8) but that there was such a band when cDNA from control cells was similarly diluted. (Not shown are the results demonstrating that the expected band was obtained from cDNA diluted 103-fold from the control-transformed cells but not from pAnti-PIG-A cells.)
Following screening by RT-PCR, we also carried out real-time RT-PCR on one of the pAnti-PIG-A-transformed cell lines and found that the pPIG-A transcripts were present at only 0.3% of the level in control-transformed cells. The calmodulin control transcripts were represented in equal amounts in the pAnti-PIG-A- and control-transformed cells.
pPIG-A protein in transformed cells.
We examined the array of surface GPI-anchored proteins by using Western blots of proteins that we washed from the surface using a salt-ethanol mixture, which has been previously shown to remove not only the large GPI-anchored surface antigens but also many other GPI-anchored proteins (5, 31). For reasons that are not understood, the peripheral membrane proteins that are harvested with the salt-ethanol wash include GPI-anchored proteins with their anchors cleaved. Clearly, there is an activated lipase on the surfaces of the cells that makes this simple procedure a convenient way to analyze GPI-anchored proteins and to confirm whether they are indeed GPI anchored. Proteins harvested from 2 × 105 cells were separated by SDS-polyacrylamide gel electrophoresis and electroblotted for immunodetection using anti-A and -B sera and anti-CRD. The last antibody recognizes the form of the GPI anchor cleaved by PI-PLC and still attached to the protein. We also compared whole-cell homogenates with the proteins harvested from the surface to provide a loading control. The whole-cell homogenates are expected to show no cleaved anchors and hence no reactivity with anti-CRD antibodies.
Figure 4 shows the results of Western blotting of proteins from a control-transformed cell line and an pAnti-PIG-A-transformed line. Figure 4A shows proteins that react with the anti-A and -B antisera. Characteristically, in control cells, we find bands of many reactive proteins in whole-cell homogenates and salt-ethanol washes, including proteins of >200 (the highest doublet corresponding to the surface antigens) and 90 kDa and in the 35- to 60-kDa range. The 35-kDa band was always found on the blots of control cells, but the other bands were variable. The proteins from the whole-cell homogenates of the pAnti-PIG-A-transformed lines show bands like those from the control cells (Fig. 4A, compare lanes 2 and 3). However, the salt-ethanol washes of the pAnti-PIG-A cells always showed very few proteins that reacted with the anti-A and -B antisera (Fig. 4A, compare lanes 2 and 4).
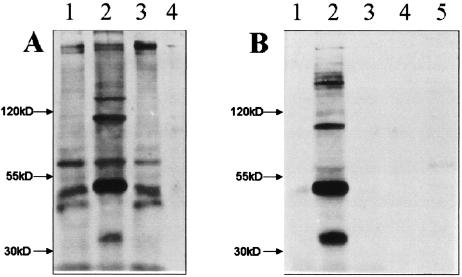
FIG. 4.
Western blot analysis of surface proteins from control- and pAnti-PIG-A-transformed cells. (A) Immunoblot developed with anti-A and -B antisera against whole-cell salt-ethanol washes. Lanes 1 and 3, proteins from whole-cell homogenates of the control-transformed cells and an pAnti-PIG-A-transformed line, respectively; lanes 2 and 4, results with salt-ethanol washes of control- and pAnti-PIG-A-transformed cells. (B) Immunoblots of the same kinds of protein preparations as in panel A developed with anti-CRD. Lanes 1 and 3 show no bands from whole-cell homogenates of control and pAnti-PIG-A lines, respectively, as expected. Lanes 2 and 4 show results with salt-ethanol washes from control cells and pAnti-PIG-A cells. Twice as much pAnti-PIG-A salt-ethanol wash product was loaded onto lane 5 as onto lanes 2 and 4.
The blots in Fig. 4B show the same kinds of protein preparations as those in Fig. 4A, but the blots are developed with anti-CRD that recognizes cleaved GPI anchors on proteins. In lanes B1 and B3, the anti-CRD antibody reacted with no whole-cell homogenate proteins for either transformed line, which was expected, since whole-cell homogenates should not have proteins with cleaved anchors. In lanes B2 and B4, blots of salt-ethanol wash proteins provide a more interesting comparison, since salt-ethanol washes are a way to harvest GPI-anchored proteins with cleaved anchors. The salt-ethanol wash from the control cells shows many anti-CRD-reactive (i.e., GPI-anchored) proteins, while the pAnti-PIG-A cells show none. In Fig. 5B, twice as much salt-ethanol wash was loaded onto lane 5 than onto lanes 2 and 4, with only a slight appearance of a band. These are typical results with the pAnti-PIG-A-transformed cells.
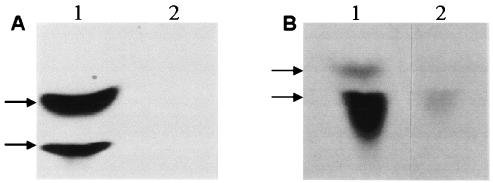
FIG. 5.
Products of PIG-A reaction in the ER. (A) 3H-labeled products of [3H]UDP-acetylglucosamine and an ER preparation from control-transformed cells separated by TLC. Lane 1, products N-acetylglucosamine-inositol phosphate-ceramide (bottom arrow) and glucosamine inositol phosphate-ceramide (top arrow); lane 2, products digested with PI-PLC prior to TLC to confirm identification. (B) 3H-labeled products N-acetylglucosamine-inositol phosphate-ceramide (bottom arrow) and glucosamine inositol phosphate-ceramide (top arrow) of [3H]UDP-acetylglucosamine from ER preparations from control-transformed cells (lane 1) and pAnti-PIG-A-transformed cells (lane 2).
GPI biosynthesis.
In order to determine whether the first step catalyzed by PIG-A in GPI anchor synthesis was defective or reduced in the pAnti-PIG-A-transformed cells, we assayed the first steps of GPI biosynthesis. Generally, GlcNAc is transferred to PI from UDP-GlcNAc by GPI-GlcNAc transferase, and GlcNAc-PI is de-N-acetylated to GlcN-PI. Unlike trypanosomes, humans, and yeast, P. primaurelia uses inositol-phosphoceramide instead of PI as a receptor of GlcNAC (1). Therefore, we asked whether the pAnti-PIG-A-transformed cells were defective in GPI-GlcNAc transferase activity by assaying for GlcNAc-inositol-phosphoceramide and the deacetylated GlcN-inositol-phosphoceramide.
We assayed the first step of GPI anchor synthesis using a crude ER preparation from the control or pAnti-PIG-A line and UDP-[3H]GlcNAC. Two resulting bands from the control cell ER were evident on TLC analysis, GlcNAc-inositol-phosphoceramide (Fig. 5A, lane 1, upper band) and GlcN-inositol-phosphoceramide (lower band). To confirm the identities of the bands, we digested them with PI-PLC and GPI-PLD, which should have led to their loss in the TLC assay (Fig. 5A, lane 2). In human and yeast systems, if the hPIG-A or GPI3 gene were mutated, the syntheses of GlcNAc-PI and GlcN-PI would be inhibited. In the Paramecium pAnti-PIG-A-transformed cells, the two ER products were missing or reduced (Fig. 5B, lanes 1 [control cells] and 2 [pAnti-PIG-A cells]). The results suggest that the reduction of pPIG-A mRNA causes the decrease in activity of GPI-GlcNAc transferase, suggesting a cause for the pAnti-PIG-A transformants' loss of GPI surface proteins.
Chemoresponse behavior.
After we had analyzed the proteins of the anti-PIG-A-transformed lines, we tested them for chemoresponse behavior in T mazes using a battery of attractant stimuli that would initiate three different signal transduction pathways in P. tetraurelia (42). Table 1 shows that the control-transformed cells are attracted, as expected, to glutamate, folate, acetate, and ammonium. Both pAnti-PIG-A-transformed lines tested show statistically significantly reduced attraction to the point of showing almost no attraction to glutamate and folate (an index of chemoresponse of 0.5 indicates a neutral response of the cells; >0.5 indicates attraction). There was no difference between the transformed lines in attraction to ammonium, which we expected, since ammonium does not appear to require a receptor for attraction (8). Ammonium serves as a convenient control for motility and non-receptor-mediated events. The response of the pAnti-PIG-A2 cells to acetate was significantly reduced, but not as severely affected as the responses to glutamate and folate.
TABLE 1.
Chemoresponses of control and pAnti-PIG-A-transformed cells
| Strain | Chemoresponse toa:
|
|||
|---|---|---|---|---|
| Glutamate | Folate | Acetate | Ammonium | |
| Neo control | 0.81 ± 0.03 | 0.77 ± 0.02 | 0.81 ± 0.02 | 0.82 ± 0.03 |
| Anti-PIG-A1 | 0.55 ± 0.03b | 0.58 ± 0.03b | ND | ND |
| Anti-PIG-A2 | 0.58 ± 0.02b | 0.56 ± 0.02b | 0.69 ± 0.02 | 0.81 ± 0.03 |
Data are averages of 12 to 32 experiments ± 1 standard error of the mean. ND, not determined.
Significant difference from Neo control-transformed cells (P = 0.01).
DISCUSSION
In order to examine the functions of GPI-anchored proteins in P. tetraurelia, we cloned the gene for one of the enzymes of the first step in GPI anchor synthesis in other organisms. The gene that we cloned by homology PCR, pPIG-A, has a deduced amino acid sequence that shows ∼40% identity and 60% similarity to hPIG-A. Some domains of pPIG-A show even higher identity (78 to 88%) to hPIG-A and yeast GPI3, and also to bacterial glycosyltransferase or putative GlcNAc transferase. We believe that there are no close isoforms of the pPIG-A gene in the P. tetraurelia genome, based on our experience with RT-PCR, by which we cloned and sequenced many products in the process of cloning pPIG-A. However, we have no direct proof that pPIG-A is a unique sequence.
There are several methods in use to inhibit gene expression in Paramecium, including antisense oligodeoxynucleotide (ODN) transfection (49), gene silencing (see references 17 and 47 for examples), and RNA interference (6, 18). When we introduced antisense ODNs for calmodulin into cells by electroporation or microinjection, calmodulin decreased by 30%, chemoresponse to acetate was disrupted, and behavioral changes were consistent with reduced activity of the calcium-calmodulin-dependent Na+ channel (14, 49). To silence a gene in Paramecium, a large number of plasmids containing only the coding region of a gene are microinjected into the macronucleus, which causes a marked reduction in gene expression and subsequent phenotype change (17). For this study, we chose a fourth method, antisense RNA expression, to reduce expression of the pPIG-A gene. In general, plasmids expressing the antisense RNA suppress gene function by the formation of RNA-RNA hybrids in the nucleus, which then inhibit the transport of RNA to the cytoplasm, splicing, and protein translation and/or increase RNA turnover (6, 28, 48). For the purpose of expressing the antisense RNA from the pPIG-A gene, we constructed an expression plasmid from plasmids provided by J. Haynes and R. Hinrichsen. The pNeotel plasmid has part of the coding region of pPIG-A, under the control of the calmodulin promoter; Tetrahymena telomere sequences, which when linearized are located at the plasmid ends; and a Neor gene that is also regulated by a Paramecium calmodulin promoter. A similar plasmid with the coding region for green fluorescent protein has been used to transform cells that are both resistant to paromomycin and fluoresce green (J. Yano and J. L. Van Houten, unpublished data). The expressing plasmids are stable in the macronucleus, and cell lines can be maintained for several months.
We show here that the pAnti-PIG-A plasmid caused the reduction of pPIG-A mRNA and GPI-anchored proteins and did not affect the mRNA expression of the calmodulin gene that we used as a control. This is in contrast to other studies, in which a different plasmid, pAnti-Calmodulin, was purposely used to express antisense RNA for the P. tetraurelia calmodulin and to decrease the amount of calmodulin protein and was found to disturb the chemoresponse to acetate (J. Yano, W. E. Bell, and J. L. Van Houten, unpublished data).
We also present here the effects of antisense expression of pPIG-A on lipid intermediates in the ER, where the GPI anchor is synthesized. Generally, in the first steps of GPI biosynthesis, GlcNAc is transferred to PI from UDP-GlcNAc by GPI-GLcNAc transferase, and GlcNAc-PI is deacetylated to GlcN-PI in the mammalian, yeast, and protozoan cells (for a review, see reference 38). The genes coding for the enzymes for these steps in humans are hPIG-A, hPIG-C, hPIG-H, hGPI1, hPIG-J, and hPIG-L, and in yeast they are GPI3, GPI2, and GPI1. The complex consisting of the peptides from hPIG-A, -C, and -H and hPGI1 provides the enzyme activity GPI-GlcNAc transferase. For Paramecium, inositol-phosphoceramide instead of PI is the receptor of GlcNAC, and we reasoned that if the pPIG-A clone codes for a protein of the catalytic subunit of GPI-GlcNAc transferase, then the intermediates GlcNAc-inositol-phosphoceramide and deacylated GlcN-inositol-phosphoceramide should be reduced or missing in the antisense transformed cells. The antisense expression resulted in the reduction of these intermediates, whose identities were confirmed by PI-PLC digestion, compared to those from the ER preparations of the control-transformed cells. The losses of GPI-anchored proteins from the surface and the ER intermediates suggest strongly that the pPIG-A gene product is one of the components in the GPI-GlcNAc transferase of P. tetraurelia.
The loss of the GPI-anchored proteins from the cell surface can be profound in most of our pAnti-PIG-A-transformed cell lines, which implies that a 300-fold reduction in the apparent mRNA for pPIG-A leads to a reduction of the enzyme at the protein level. Therefore, pPIG-A does not appear to have a closely related gene with redundant gene function that is not also down-regulated by the antisense RNA. The protein precursors of the GPI-anchored proteins are synthesized in the pAnti-pPIG-A-transformed cells, as evidenced by the immunoblots of the whole-cell homogenates. The failure of the proteins to appear on the surfaces of these cells may be due to failure in trafficking to the surface or the secretion of the proteins into the medium for lack of the GPI anchor. Secretion of such proteins was shown in human and yeast mutants, in which proteins were cleaved at the ω site, where the anchor would normally be attached, but lacking their GPI anchors, the proteins were secreted (40).
The loss of many or most GPI-anchored proteins from the cell surface is not lethal for paramecia, since the transformed cell cultures grew as well as the transformed control cells. While the mating efficiency of the pAnti-PIG-A cells is low, they can mate (J. Yano, unpublished observation). Also, they can discharge trichocysts. As shown in this study, pAnti-PIG-A transformants have selectively altered chemoresponse, but they retain the ability to turn and swim normally in T mazes, as evidenced by their normal responses to ammonium, which, while not requiring a receptor for chemosensory signal transduction, does require the ability to modulate and execute turns while swimming (8). In all, we tested cells for chemoresponses to four stimuli, each of which utilizes a different signal transduction pathway and different second messengers for chemoresponse (43). While the cells behaved normally in ammonium, the pAnti-pPIG-A-transformed cells were not able to behave normally in glutamate or folate, showing almost no response at all. A GPI-anchored protein that is a candidate for the folate chemoreceptor has been found, which could explain the loss of the chemoresponse to folate with the loss of GPI-anchored proteins (31). The loss of response to glutamate implies that it, too, requires a GPI-anchored protein, perhaps as a receptor. The transformed cells responded with significant attraction to acetate, but this attraction was statistically significantly lower than the attraction of the control cells. Therefore, we surmise that there may be a GPI-anchored protein of the cell surface that functions in but is not essential for the chemosensory transduction pathway for acetate.
If there were a GPI-anchored protein involved in swimming behavior, such as a component of the voltage-gated calcium channel which functions in the calcium-based swimming turn, we would expect that attraction to ammonium would likewise be affected, but it is not. Preliminary electrophysiological studies of the pAnti-pPIG-A cells shows that they can generate calcium action potentials but may have abnormal conductances in the only stimulus tested thus far, folate (R. R. Preston, unpublished observations). The somatic mutations of the hPIG-A gene (PI glycan complementation group A) in hematopoietic stem cells causes paroxysmal nocturnal hemoglobinuria, a disease in which the loss of expression of the hPIG-A gene results in the loss of GPI-anchored complement regulatory proteins, such as CD59 and CD55 (25). As a consequence, there is lysis of red blood cells through an alternative complement pathway. The knockout of the PIG-A gene in mice, specific to skin cells, showed that GPI anchoring is necessary for proper skin differentiation and maintenance (41). The GPI3 mutants of yeast demonstrated that loss of GPI anchoring affected morphogenesis and sporulation (24). Among parasitic protozoa, the GPI-anchored proteins are essential for Trypanosoma brucei but not for Leishmania mexicana to grow in mammalian cells (19, 29). PIG-A knockout mice do not survive until birth (23).
In contrast to the knockout mice, paramecia that have lost most or all GPI anchor synthesis do not die; rather, they appear to have lost the functions of large surface antigens and cell surface receptors that are involved in chemosensory transduction. In our continuing study of GPI-anchored proteins in Paramecium, we isolated the putative transamidase GPI8 gene (accession no. AY057228 [Yano et al., unpublished]), another enzyme in GPI anchor synthesis and transfer to proteins, and in preliminary studies of antisense expression, we found that reduction of GPI8 transcript has effects similar to those of anti-PIG-A expression on chemosensory responses, including a more severe change in the response to acetate.
Acknowledgments
This work was supported by Public Health Service grant GM-59988 from the Institute of General Medical Sciences and the Institute of Deafness and other Communication Disorders and by the Vermont Cancer Center NCI PHS 22435.
We thank Megan S. Valentine for assistance with preparation of the figures.
Footnotes
This paper is dedicated to Yvonne Capdeville, a pioneer in GPI anchoring in Paramecium.
REFERENCES
- 1.Azzouz, N., B. Striepen, P. Gerold, Y. Capdeville, and R. T. Schwarz. 1995. Glycosylinositol-phosphoceramide in the free-living protozoan Paramecium primaurelia: modification of core glycans by mannosyl phosphate. EMBO J. 14:4422-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzouz, N., P. Gerold, M. H. Kedees, H. Shams-Eldin, R. Werner, Y. Capdeville, and R. T. Schwarz. 2001. Regulation of Paramecium primaurelia glycosylphosphatidyl-inositol biosynthesis via dolichol phosphate mannose synthesis. Biochimie 83:801-809. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D., and K. Jacobson. 2001. Microdomains, lipid rafts and caveolae (San Feliu de Guixols, Spain, 19-24 May 2001). Traffic 2:668-672. [DOI] [PubMed] [Google Scholar]
- 4.Capdeville, Y., and A. Benwakrim. 1996. The major ciliary membrane proteins in Paramecium primaurelia are all glycosylphosphatidylinositol-anchored proteins. Eur. J. Cell Biol. 70:339-346. [PubMed] [Google Scholar]
- 5.Capdeville, Y., R. Charret, C. Antony, J. Delorme, P. Nahon, and A. Adoutte. 1993. Ciliary and plasma membrane proteins in Paramecium: description, localization, and intracellular transit, p. 181-226. In A. H. Tartakoff and H. Plattner (ed.), Advances in cell and molecular biology of membranes, vol. 2A. JAI Press, London, United Kingdom. [Google Scholar]
- 6.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9732-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson, G. L., and D. L. Nelson. 1996. The 44-kDa regulatory subunit of the Paramecium cAMP-dependent protein kinase lacks a dimerization domain and may have a unique autophosphorylation site sequence. J. Eukaryot. Microbiol. 43:347-356. [DOI] [PubMed] [Google Scholar]
- 8.Davis, D. P., J. F. Fiekers, and J. L. Van Houten. 1998. Intracellular pH and chemoresponse to NH4Cl in Paramecium. Cell Motil. Cytoskeleton 40:107-118. [DOI] [PubMed] [Google Scholar]
- 9.Dupuis, P. 1992. The beta-tubulin genes of Paramecium are interrupted by two 27 bp introns. EMBO J. 11:3717-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elwess, N. L., and J. L. Van Houten. 1997. Cloning and molecular analysis of the plasma membrane Ca2+-ATPase gene in Paramecium tetraurelia. J. Eukaryot. Microbiol. 44:250-257. [DOI] [PubMed] [Google Scholar]
- 11.Englund, P. T. 1993. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu. Rev. Biochem. 62:121-138. [DOI] [PubMed] [Google Scholar]
- 12.Fassler, J. S., W. Gray, J. P. Lee, G. Y. Yu, and G. Gingerich. 1991. The Saccharomyces cerevisiae STP14 gene is essential for normal expression of the yeast transposon, Ty, as well as for expression of the HIS4 gene and several genes in the mating pathway. Mol. Gen. Genet. 230:310-320. [DOI] [PubMed] [Google Scholar]
- 13.Forney, J., C. Epstein, L. Preer, B. Rudman, D. Widmayer, W. Klein, and J. Preer. 1983. Structure and expression of genes for surface proteins in Paramecium. Mol. Cell. Biol. 3:466-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraga, D., J. Yano, M. Reed, R. Chuang, W. E. Bell, J. L. Van Houten, and R. Hinrichsen. 1998. Introducing antisense oligonucleotides into Paramecium via electroporation. J. Eukaryot. Microbiol. 45:582-588. [DOI] [PubMed] [Google Scholar]
- 15.Futerman, A. H. 1995. Inhibition of sphingolipid synthesis: effects on glycosphingolipid-GPI-anchored protein microdomains. Trends Cell Biol. 5:377-379. [DOI] [PubMed] [Google Scholar]
- 16.Galbiati, F., B. Ranani, and M. P. Lisanti. 2001. Emerging themes in lipid rafts and caveolae. Cell 106:403-411. [DOI] [PubMed] [Google Scholar]
- 17.Galvani, A., and L. Sperling. 2001. Transgene-mediated post-transcriptional gene silencing is inhibited by 3′ non-coding sequences in Paramecium. Nucleic Acids Res. 29:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvani, A., and L. Sperling. 2002. RNA interference by feeding Paramecium. Trends Genet. 18:11-12. [DOI] [PubMed] [Google Scholar]
- 19.Hilley, J. D., J. L. Zawadzki, M. J. McConville, G. H. Coombs, and J. C. Mottram. 2000. Leishmania mexicana mutants lacking glycosylphosphatidylinositol (GPI):protein transamidase provide insights into the biosynthesis and functions of GPI-anchored proteins. Mol. Biol. Cell 11:1183-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooper, N. M. 2001. Determination of glycosyl-phosphatidylinositol membrane protein anchorage. Proteomics 1:748-755. [DOI] [PubMed] [Google Scholar]
- 21.Kanabrocki, J. A., Y. Saimi, R. R. Preston, J. W. Haynes, and C. Kung. 1991. Efficient transformation of Cam2, a behavioral mutant of Paramecium tetraurelia, with the calmodulin gene. Proc. Natl. Acad. Sci. USA 88:10845-10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kink, J. A., M. E. Maley, K. Y. Ling, J. A. Kanabrocki, and C. Kung. 1991. Efficient expression of the Paramecium calmodulin gene in Escherichia coli after four TAA to CAA changes through a series of polymerase chain reactions. J. Protozool. 38:661-667. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita, T., M. Bessler, and J. Takeda. 2002. Animal models of PNH, p. 139-158. In N. S. Young and J. Moss (ed.), PNH and the GPI-linked proteins. Academic Press, New York, N.Y.
- 24.Leidich, S. D., and P. Orlean. 1996. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycophosphatidylinositol anchor synthesis. J. Biol. Chem. 269:27829-27837. [DOI] [PubMed] [Google Scholar]
- 25.Luzzatto, L., and K. Nafa. 2000. Genetics of PNH, p. 21-48. In N. S. Young and J. Moss (ed.), PNH and the GPI-linked proteins. Academic Press, New York, N.Y.
- 26.McConville, M. J., and M. A. Ferguson. 1993. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 294:305-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyata, T., J. Takeda, Y. Iida, N. Yamada, N. Inoue, M. Takahashi, K. Maeda, T. Kitani, and T. Kinoshita. 1993. The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science 259:1318-1320. [DOI] [PubMed] [Google Scholar]
- 28.Murray, J. A. H., and N. Crockett. 1992. Antisense techniques: an overview, p. 1-49. In J. A. H. Murray (ed.), Antisense RNA and DNA. Wiley-Liss, New York, N.Y.
- 29.Nagamune, K., T. Nozaki, Y. Maeda, K. Ohishi, T. Fukuma, T. Hara, R. T. Schwarz, C. Süterlin, R. Brun, H. Riezman, and T. Kinoshita. 2000. Critical roles of glycosyl phosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 97:10335-10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, E., Y. You, and J. Forney. 1996. Cysteine residue periodicity is a conserved structural feature of variable surface proteins from Paramecium tetraurelia. J. Mol. Biol. 222:835-841. [DOI] [PubMed] [Google Scholar]
- 31.Paquette, C. A., V. Rakochy, A. Bush, and J. L. Van Houten. 2001. GPI anchored proteins in Paramecium tetraurelia: possible role in chemoresponse. J. Exp. Biol. 204:2899-2910. [DOI] [PubMed] [Google Scholar]
- 32.Preer, J. R., L. B. Preer, and B. M. Rudman. 1981. mRNAs for the immobilization antigens of Paramecium. Proc. Natl. Acad. Sci. USA 78:6776-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell, C. D., D. Fraga, and R. D. Hinrichsen. 1994. Extremely short 20-33 nucleotide introns are the standard length in Paramecium tetraurelia. Nucleic Acids Res. 22:1221-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasner, M., and J. L. Van Houten. 1989. Evidence for a Paramecium folate chemoreceptor. Chem. Senses 14:588-595. [Google Scholar]
- 35.Schlichtherle, I. M., D. S. Roos, and J. L. Van Houten. 1996. cloning and molecular analysis of bifunctional dihydrofolate reductase-thymidylate synthase gene in the ciliated protozoan Paramecium tetraurelia. Mol. Gen. Genet. 250:665-673. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder, R., A. M. Gallegos, B. P. Atshaves, S. M. Storey, A. L. McIntosh, A. D. Petrescu, H. Huang, O. Starodub, H. Chao, H. Yang, A. Frolov, and A. B. Kier. 2001. Recent advances in membrane microdomains: rafts, caveolae, and intracellular cholesterol trafficking. Exp. Biol. Med. 226:873-890. [DOI] [PubMed] [Google Scholar]
- 37.Scott, J., C. Leeck, and J. Forney. 1994. Analysis of the micronuclear B type surface protein gene in Paramecium tetraurelia. Nucleic Acids Res. 22:5079-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sevlever, D., R. Chen, and M. E. Medof. 2000. Synthesis of the GPI anchor, p. 199-220. In N. S. Young and J. Moss (ed.), PNH and the GPI-linked proteins. Academic Press, New York, N.Y.
- 39.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 40.Takami, N., K. Oda, and Y. Ikehara. 1992. Aberrant processing of alkaline phosphatase precursor caused by blocking the synthesis of glycophosphatidylinositol. J. Biol. Chem. 267:1042-1049. [PubMed] [Google Scholar]
- 41.Tarutani, M., S. Itami, M. Okabe, M. Ikawa, T. Tezuka, K. Yoshikawa, T. Kinoshita, and J. Takeda. 1997. Tissue-specific knockout of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc. Natl. Acad. Sci. USA 94:7400-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Houten, J. L. 1994. Chemoreception in microorganisms: trends for neuroscience? Trends Neurosci. 17:62-71. [DOI] [PubMed] [Google Scholar]
- 43.Van Houten, J. L. 1998. Chemosensory transduction in Paramecium. Eur. J. Protistol. 34:301-307. [Google Scholar]
- 44.Van Houten, J. L., E. Martel, and T. Kasch. 1982. Kinetic analysis of chemokinesis in Paramecium. J. Protozool. 29:226-230. [DOI] [PubMed] [Google Scholar]
- 45.VanMeer, G. 2002. The different hues of lipid rafts. Science 296:855-857. [DOI] [PubMed] [Google Scholar]
- 46.Varma, R., and S. Mayor. 1998. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394:798-801. [DOI] [PubMed] [Google Scholar]
- 47.Vayssié, L., N. G. de Loubresse, and L. Sperling. 2000. Growth and form of secretory granules involves stepwise assembly not differential sorting of a family of secretory proteins in Paramecium. J. Cell Sci. 114:875-886. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, E. G. H., and K. Flärdh. 2002. Antisense RNAs everywhere? Trends Genet. 18:223-226. [DOI] [PubMed] [Google Scholar]
- 49.Yano, J., D. Fraga, R. Hinrichsen, and J. L. Van Houten. 1996. Effects of calmodulin antisense oligonucleotides on chemoresponse in Paramecium. Chem. Senses 21:55-58. [DOI] [PubMed] [Google Scholar]